Genetic Diversity and Population Structure Assessed by SSR in a Peruvian Germplasm Collection of Loche Squash (Cucurbita moschata, Cucurbitaceae) †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples Examined and DNA Amplification
2.2. Data Analysis
3. Results and Discussion
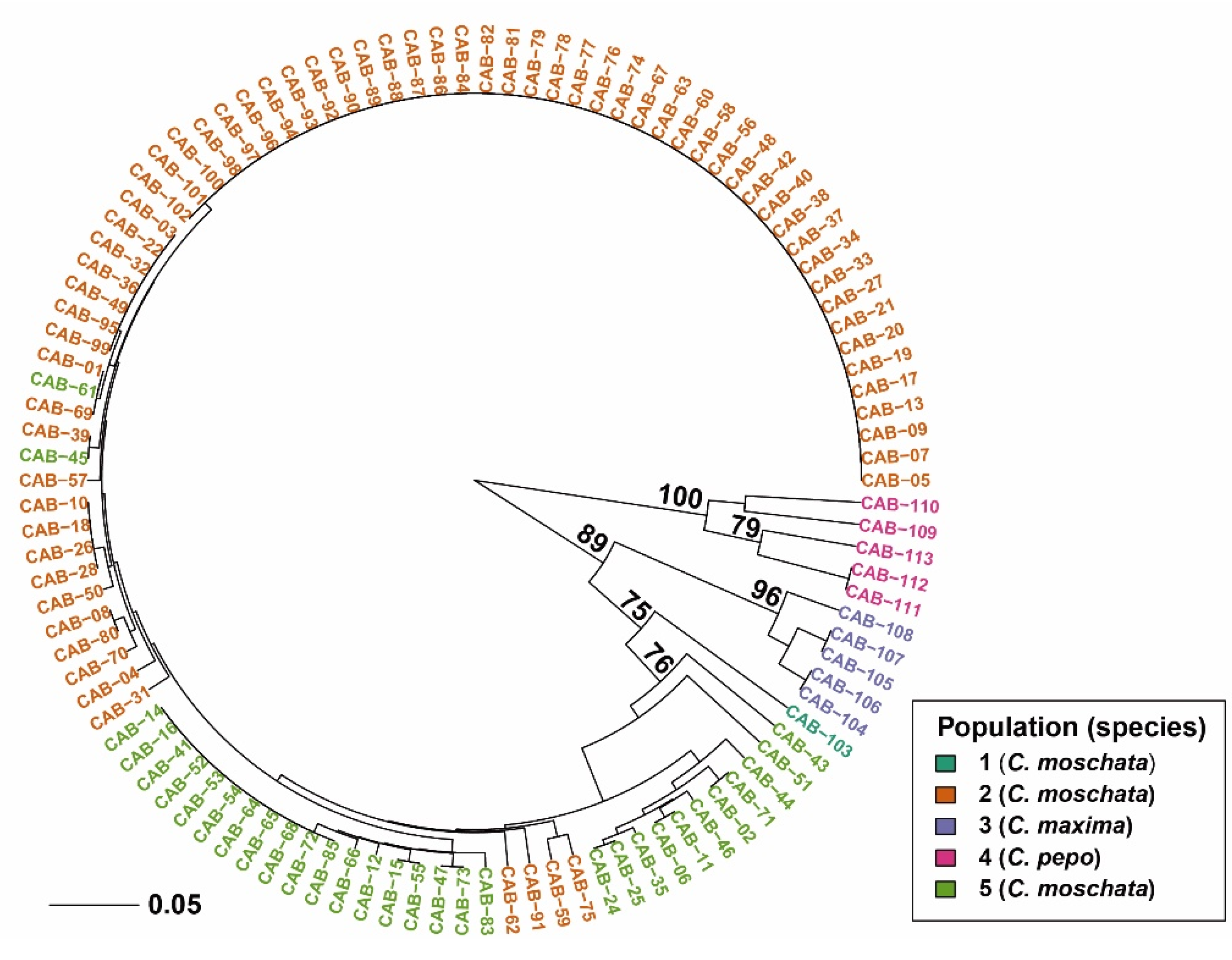
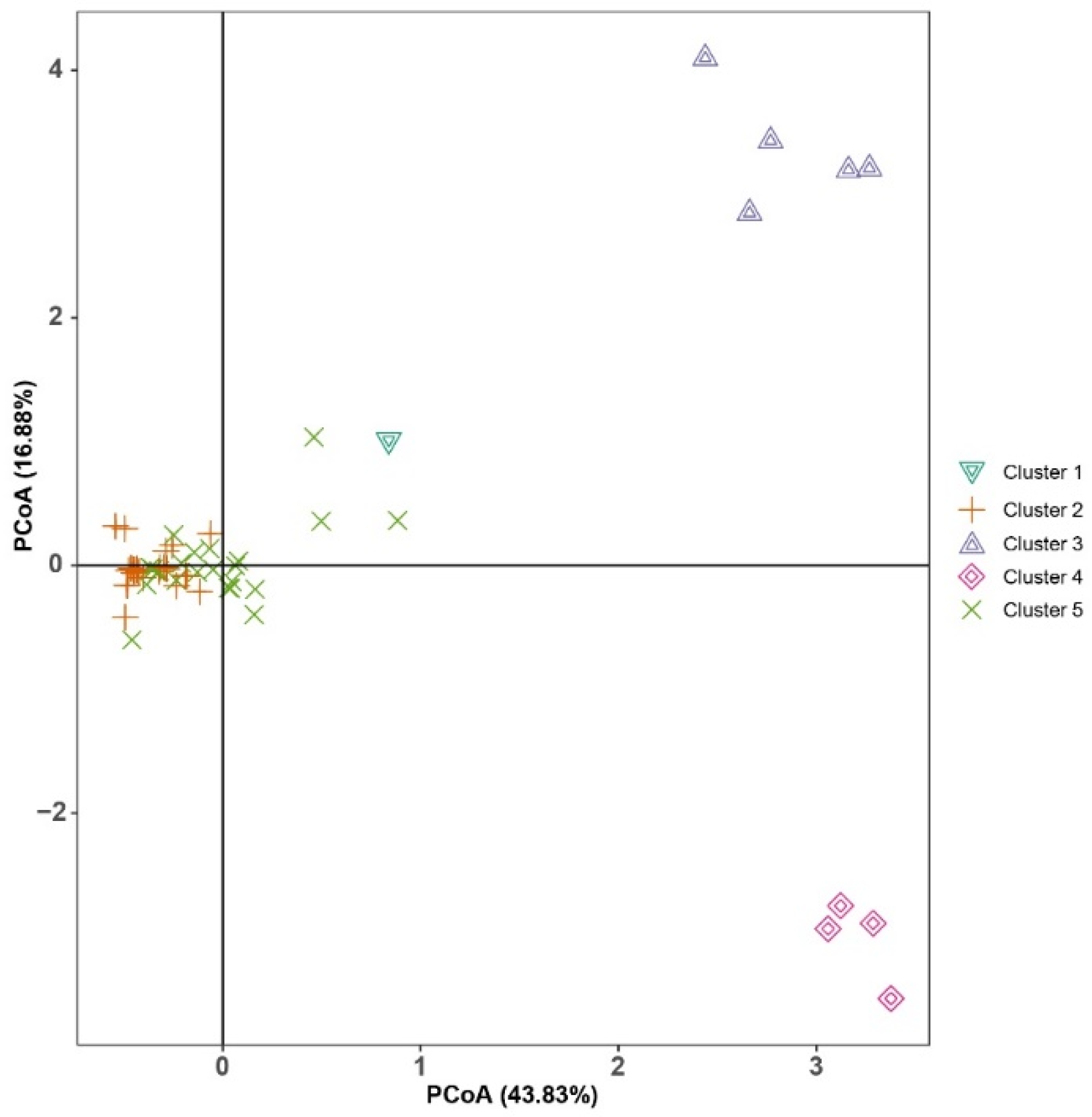
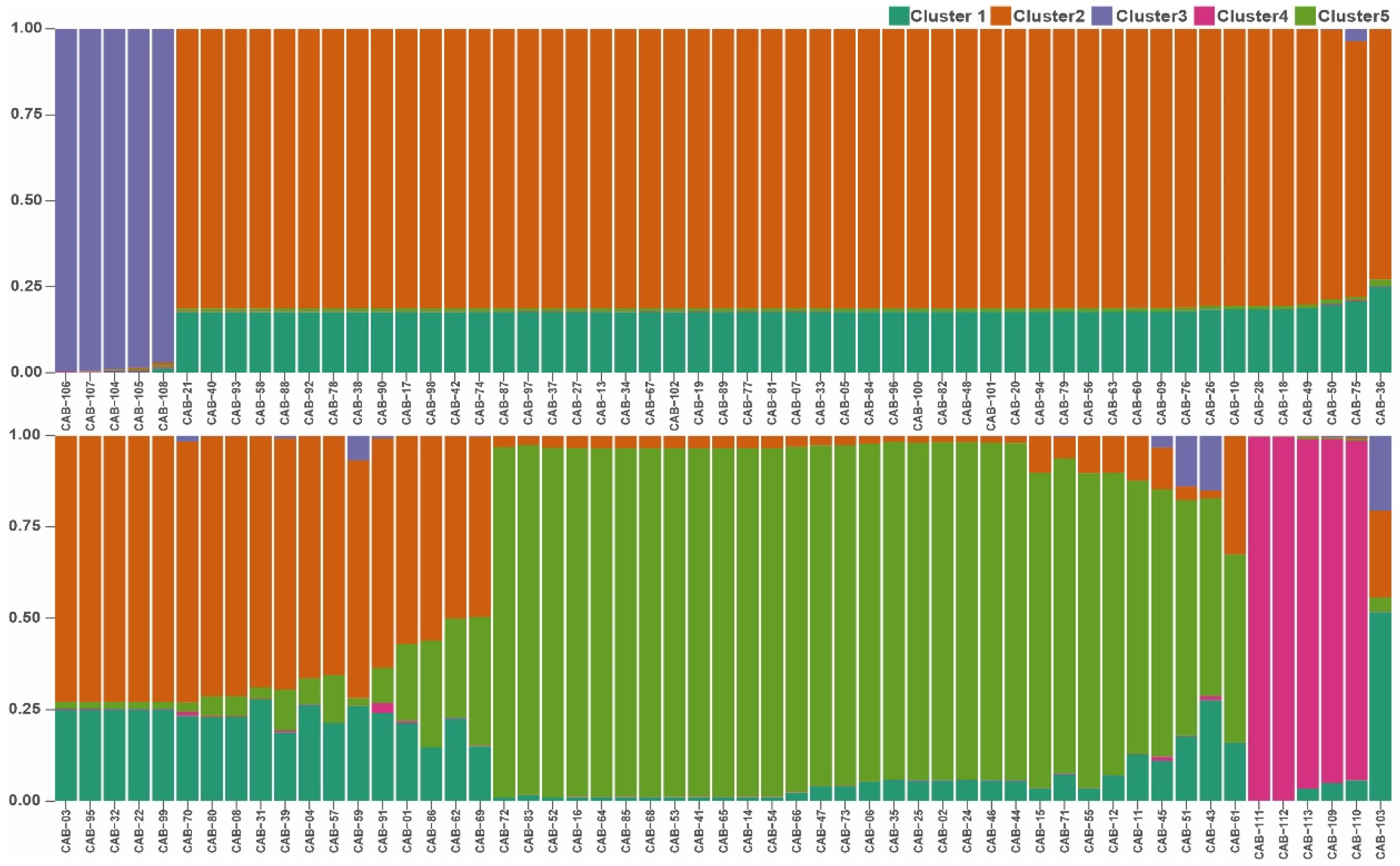
3.1. Data Analysis
3.2. Genetic Diversity Estimates and Population Structure Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andres, T.; Ugás, R.; Bustamente, F. Loche: A Unique Pre-Columbian Squash Locally Grown in North Coastal Peru; Holmes, G.J., Ed.; Proc. Cucurbitaceae; Universal Press: Raleigh, NC, USA, 2006; pp. 333–340. [Google Scholar]
- Saldaña, C.L.; Cancan, J.D.; Cruz, W.; Correa, M.Y.; Ramos, M.; Cuellar, E.; Arbizu, C.I. Genetic diversity and population structure of capirona (Calycophyllum spruceanum benth.) from the peruvian amazon revealed by rapd markers. Forests 2021, 12, 1125. [Google Scholar] [CrossRef]
- Calycophyllum, C.; Saldaña, C.L.; Rodriguez-grados, P.; Ch, J.C.; Feijoo, S.; Guerrero-abad, J.C.; Vásquez, H.V.; Maicelo, J.L.; Jhoncon, J.H.; Arbizu, C.I. Unlocking the Complete Chloroplast Genome of a Native Tree Spruceanum, Rubiaceae), and Its Comparative Analysis with Other Ixoroideae Species. Genes 2022, 13, 113. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A rapid isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2014, e281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bazo, S.I.; Espejo, J.R.; Palomino, A.C.; Flores, P.M.; Chang, L.M.; López, B.C.; Mansilla, S.R. Estudios de biología floral, reproductiva y visitantes florales en el “Loche” de Lambayeque (Cucurbita moschata DUCHESNE). Ecol. Apl. 2018, 17, 191–205. [Google Scholar] [CrossRef]
- Ferriol, M.; Picó, B.; Nuez, F. Morphological and Molecular Diversity of a Collection of Cucurbita maxima Landraces. J. Am. Soc. Hortic. Sci. 2004, 129, 60–69. [Google Scholar] [CrossRef]
- Wu, J.; Chang, Z.; Wu, Q.; Zhan, H.; Xie, S. Molecular diversity of Chinese Cucurbita moschata germplasm collections detected by AFLP markers. Sci. Hortic. 2011, 128, 7–13. [Google Scholar] [CrossRef]
- Barboza, N.; Albertazzi, F.J.; Sibaja-Cordero, J.A.; Mora-Umaña, F.; Astorga, C.; Ramírez, P. Analysis of genetic diversity of Cucurbita moschata (D.) germplasm accessions from Mesoamerica revealed by PCR SSCP and chloroplast sequence data. Sci. Hortic. 2012, 134, 60–71. [Google Scholar] [CrossRef]
- Yu, K.; Park, S.J.; Poysa, V. Abundance and variation of microsatellite DNA sequences in beans (Phaseolus and Vigna). Genome 1999, 42, 27–34. [Google Scholar] [CrossRef]
| Cluster | N | H | Lambda | He |
|---|---|---|---|---|
| 1 | 1 | 0 | 0 | 0 |
| 2 | 68 | 2.16 | 0.721 | 0.012 |
| 3 | 5 | 1.61 | 0.8 | 0.089 |
| 4 | 5 | 1.33 | 0.72 | 0.144 |
| 5 | 31 | 2.91 | 0.92 | 0.035 |
| Total | 110 | 3.26 | 0.886 | 0.0973 |
| Source | df | SS | MS | Est.Var. | % |
|---|---|---|---|---|---|
| Between clusters | 4 | 365.25 | 91.31 | 6.11 | 79.66% |
| Within clusters | 105 | 163.75 | 1.56 | 1.56 | 20.34% |
| Total | 109 | 528.99 | 4.85 | 7.67 | 100.00% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arbizu, C.I.; Blas, R.H.; Ugás, R. Genetic Diversity and Population Structure Assessed by SSR in a Peruvian Germplasm Collection of Loche Squash (Cucurbita moschata, Cucurbitaceae). Biol. Life Sci. Forum 2022, 15, 6. https://doi.org/10.3390/IECD2022-12420
Arbizu CI, Blas RH, Ugás R. Genetic Diversity and Population Structure Assessed by SSR in a Peruvian Germplasm Collection of Loche Squash (Cucurbita moschata, Cucurbitaceae). Biology and Life Sciences Forum. 2022; 15(1):6. https://doi.org/10.3390/IECD2022-12420
Chicago/Turabian StyleArbizu, Carlos I., Raúl H. Blas, and Roberto Ugás. 2022. "Genetic Diversity and Population Structure Assessed by SSR in a Peruvian Germplasm Collection of Loche Squash (Cucurbita moschata, Cucurbitaceae)" Biology and Life Sciences Forum 15, no. 1: 6. https://doi.org/10.3390/IECD2022-12420
APA StyleArbizu, C. I., Blas, R. H., & Ugás, R. (2022). Genetic Diversity and Population Structure Assessed by SSR in a Peruvian Germplasm Collection of Loche Squash (Cucurbita moschata, Cucurbitaceae). Biology and Life Sciences Forum, 15(1), 6. https://doi.org/10.3390/IECD2022-12420







