The Role of PME2 and PME3 in Arabidopsis Stomatal Development and Morphology † †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. DNA and RNA Extraction, Synthesis of cDNA and Gene Expression Analysis
2.3. Bioinformatic and Phylogenetic Analysis
2.4. Callose Localization
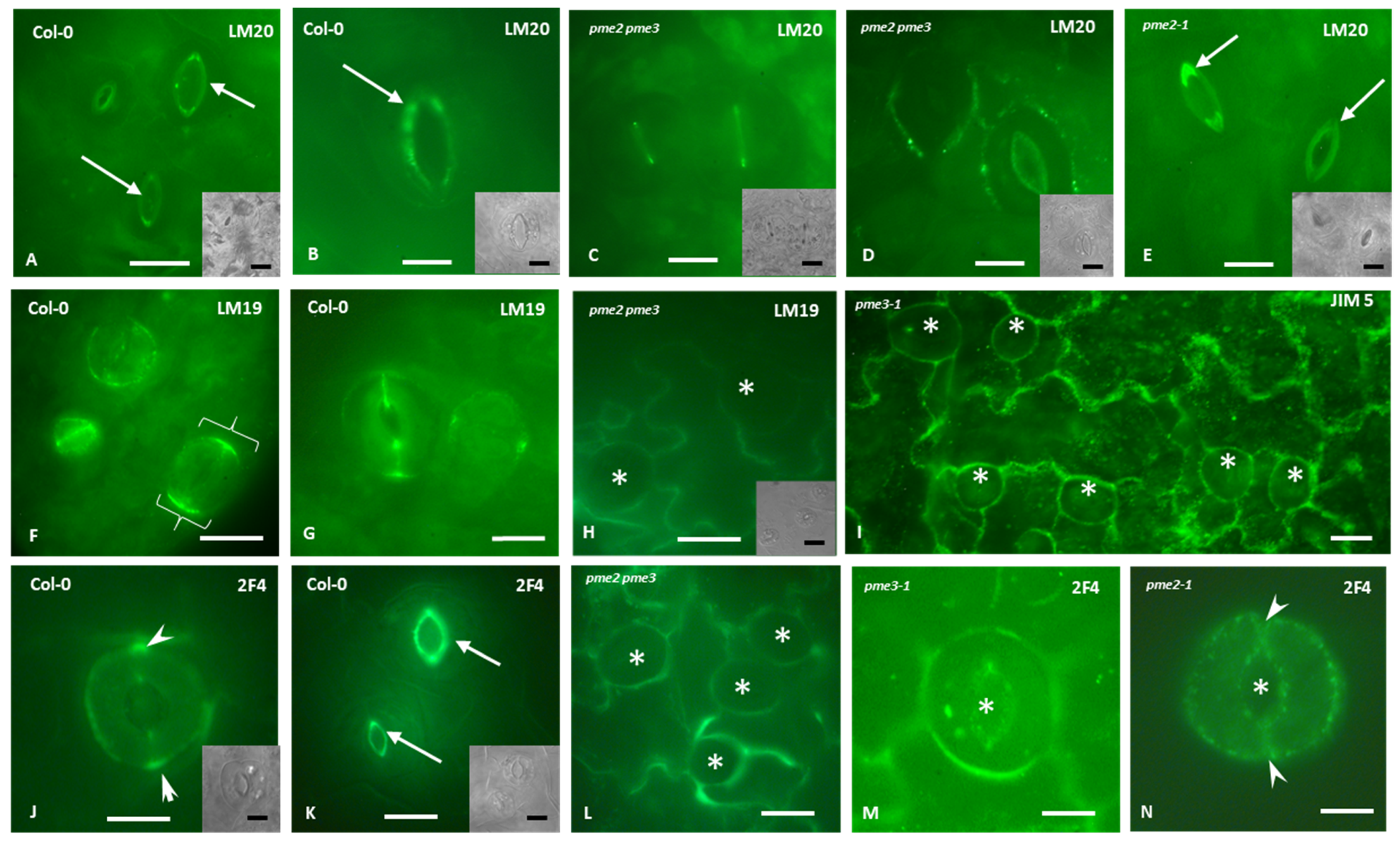
2.5. Immunolocalization of Homogalacturonans
2.6. Observation and Photography
3. Results
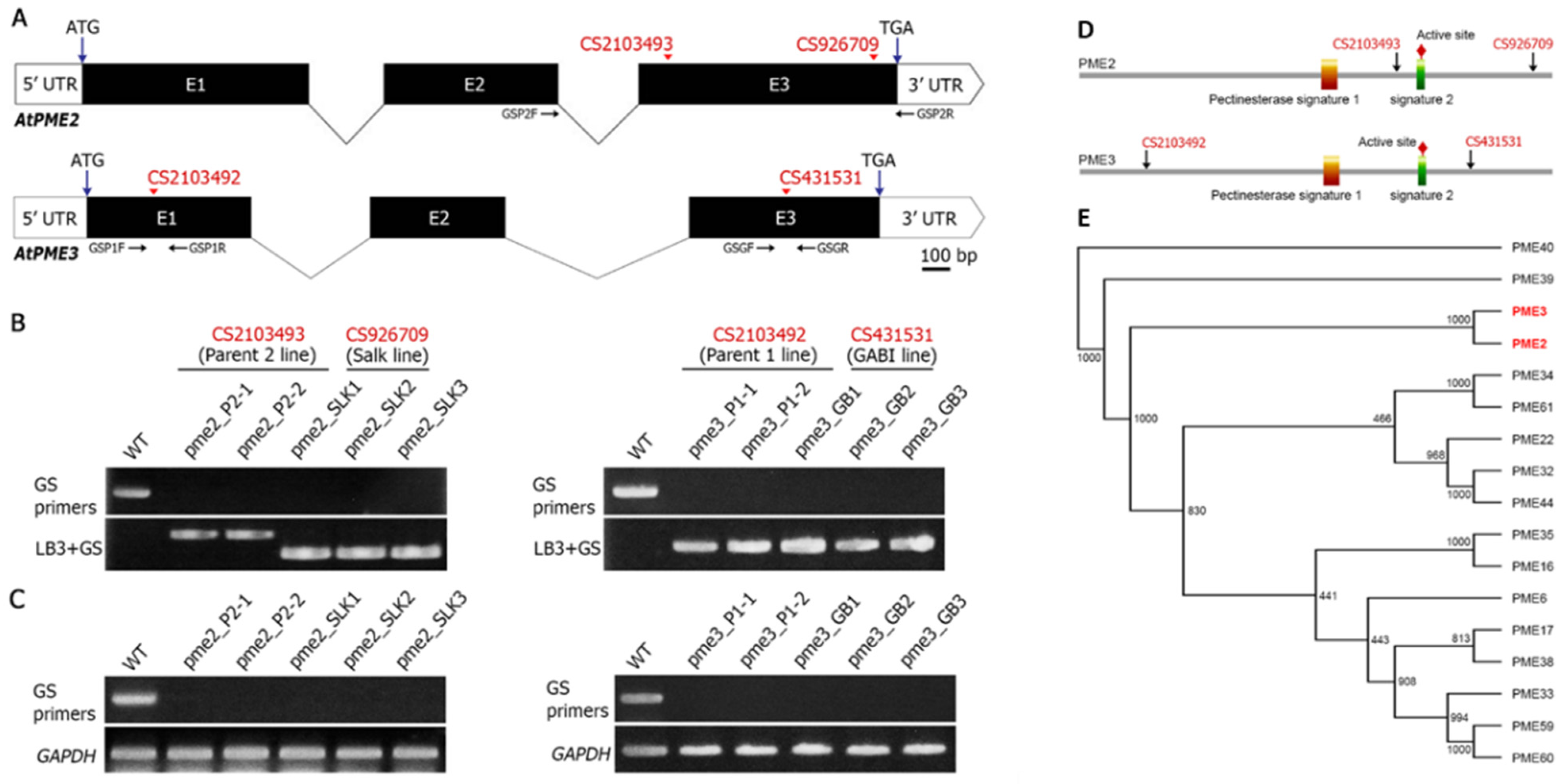
3.1. Analysis of PME Insertion Lines and Phylogeny
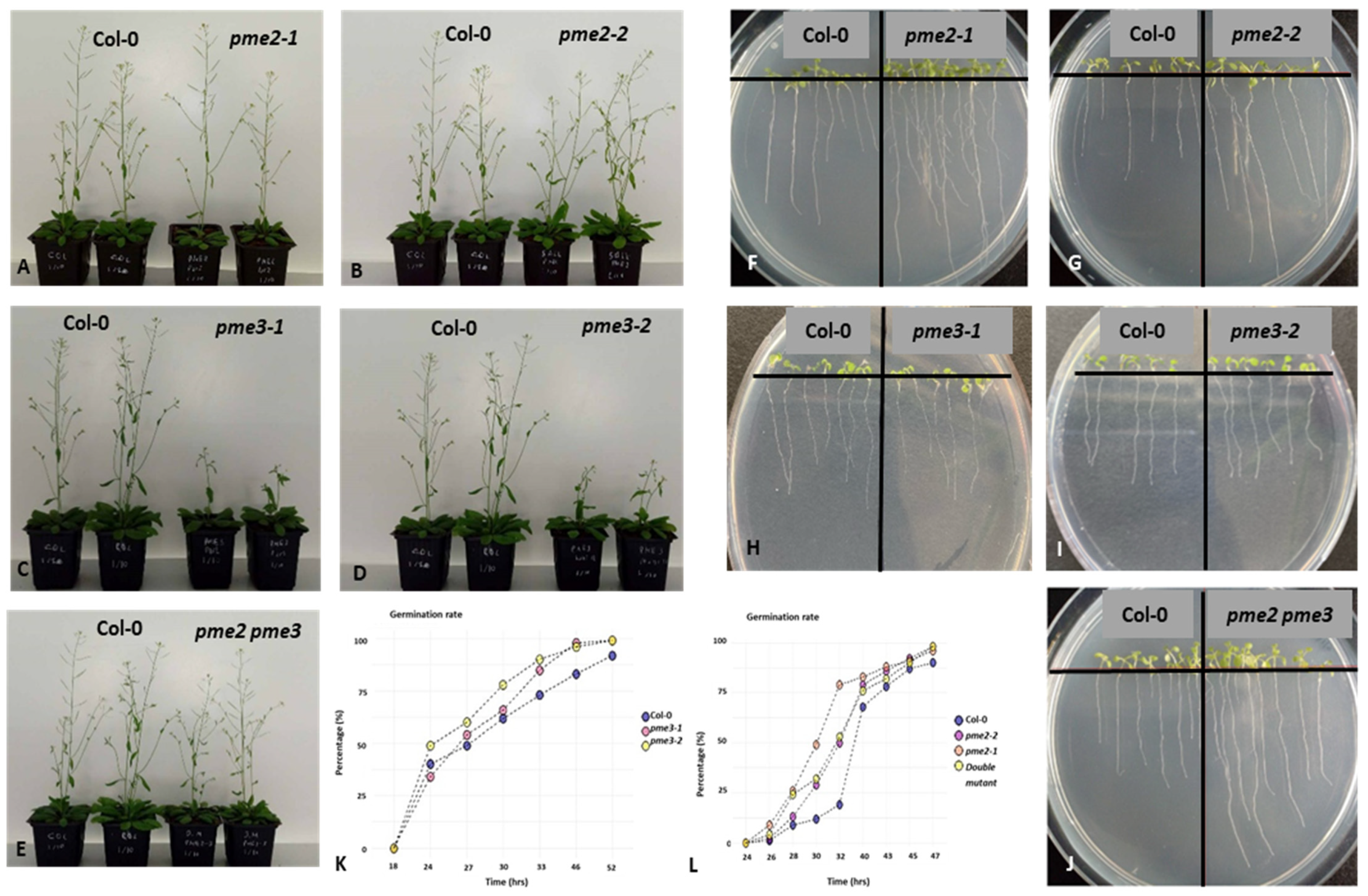
3.2. Macroscopic Phenotypes, Root Growth and Germination Rates of pme2, pme3 and pme2 pme3 Mutants
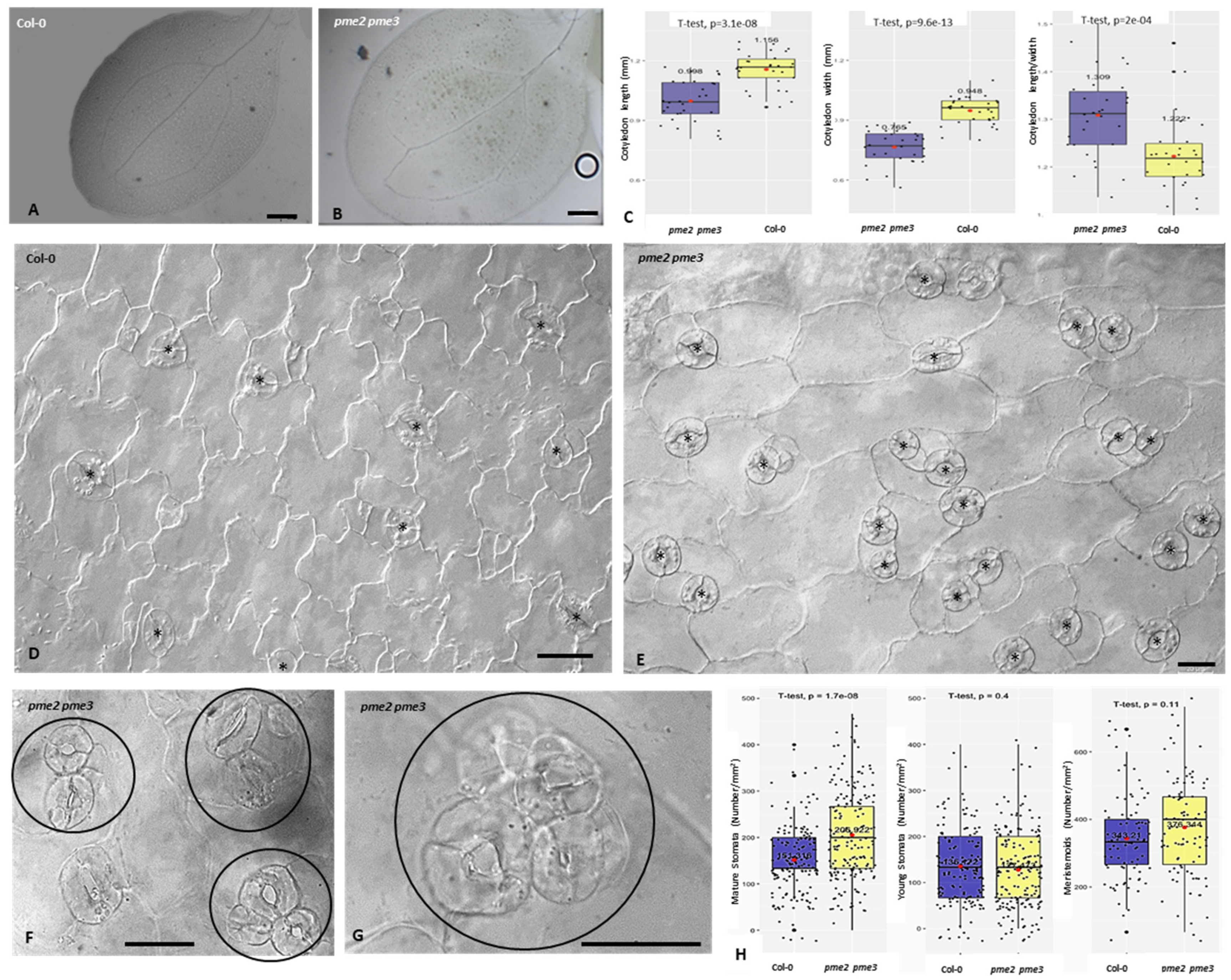
3.3. Cotyledons Shape, Pavement Cells Morphology and Stomatal Cells Patterning and Ontogenesis Are Affected in pme Mutants
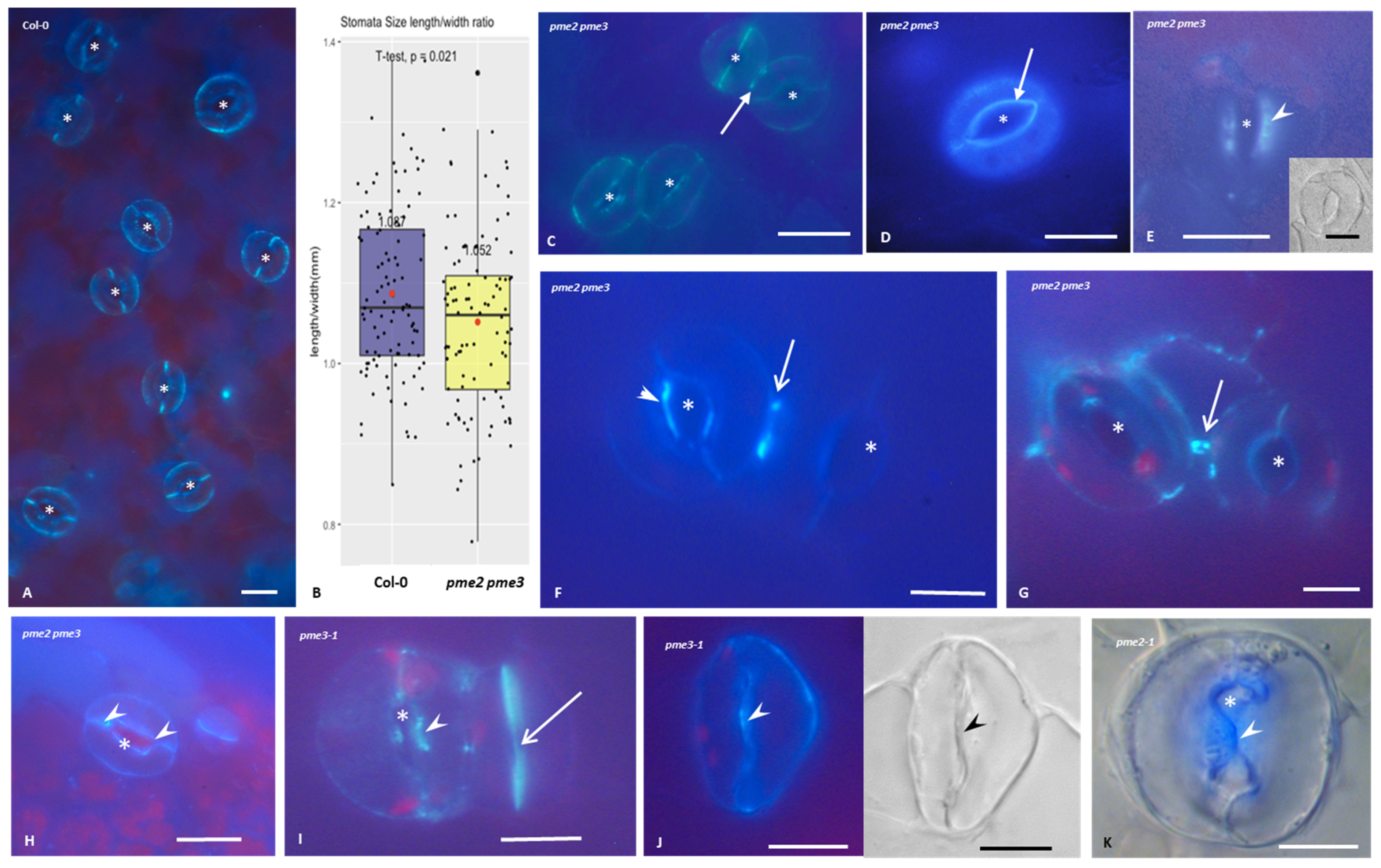
3.4. Callose Deposition in Stomatal Complexes of pme2, pme3 and pme2 pme3 Mutants
3.5. Homogalacturonans’ Deposition in Stomatal Complexes of pme2, pme3 and pme2 pme3 Mutants
4. Discussion
4.1. PMEs Role in Regulating Stomatal Patterning and Pavement Cells Shape
4.2. PMEs Role in Stomatal Shape, Stomatal Movement and Homogalacturonans Deposition
4.3. PMEs Affecting Callose and Stomatal Pore Formation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Appendix A
| Gene/Sequence | Primer Name | Sequence |
|---|---|---|
| AtPME2 | GSP2F | GACGGTAGCACCACTTTCCA |
| AtPME2 | GSP2R | CAGCCCGAGTTATGCATTGAC |
| AtPME3 | GSP1F | TCTGCCGGAGCTTCAAAAGC |
| AtPME3 | GSP1R | ACAGCCACCGTAGTTTCCTG |
| AtPME3 | GSGF | GGAGATGGTCGGACGAGAAC |
| AtPME3 | GSGR | CAAGGCGCAAAAACCCGCAT |
| T-DNA (Wisconsin) | p745 | AACGTCCGCAATGTGTTATTAAGTTGTC |
| T-DNA (Salk) | LBb1.3 | ATTTTGCCGATTTCGGAAC |
| T-DNA (Gabi) | o8474 | ATAATAACGCTGCGGACATCTACATTTT |
| T-DNA (Gabi) | o8409 | ATATTGACCATCATACTCATTGC |
| AtGAPDH | AtGAPDH-F | CTGTGAGTAACCCCATTCATTACATACCAAGC |
| AtGAPDH | AtGAPDH-R | GCAATGCATCTTGCACTACCACTGTCTTGC |
| Abbreviation | Tair ID | UniProtKB |
|---|---|---|
| PME2 | At1g53830 | Q42534 |
| PME3 | At3g14310 | O49006 |
| PME6 | At1g23200 | O49298 |
| PME16 | At2g43050 | Q9SKX2 |
| PME17 | At2g45220 | O22149 |
| PME22 | At3g05620 | Q9M9W7 |
| PME32 | At3g43270 | Q9LXK7 |
| PME33 | At3g47400 | Q9STY3 |
| PME34 | At3g49220 | Q9M3B0 |
| PME35 | At3g59010 | Q9LYT5 |
| PME38 | At4g00190 | O81320 |
| PME39 | At4g02300 | O81415 |
| PME40 | At4g02320 | O81301 |
| PME44 | At4g33220 | Q9SMY7 |
| PME59 | At5g51490 | Q9FHN5 |
| PME60 | At5g51500 | Q9FHN4 |
| PME61 | At5g53370 | Q9FK05 |
References
- Apostolakos, P.; Livanos, P.; Giannoutsou, E.; Panteris, E.; Galatis, B. The intracellular and intercellular cross-talk during subsidiary cell formation in Zea mays: Existing and novel components orchestrating cell polarization and asymmetric division. Ann. Bot. 2018, 122, 679–696. [Google Scholar] [CrossRef] [PubMed]
- Nadeau, J.A.; Sack, F.D. Control of stomatal distribution on the Arabidopsis leaf surface. Science 2002, 296, 1697–1700. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, M.R.; Ahamed, M.S.; Mas-Ud, M.A.; Islam, H.; Fatamatuzzohora, M.; Hossain, M.F.; Billah, M.; Hossain, M.S.; Matin, M.N. Stomatal development and genetic expression in Arabidopsis thaliana L. Heliyon 2021, 7, e07889. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, A.; Torii, K.U. Shouting out loud: Signaling modules in the regulation of stomatal development. Plant Physiol. 2021, 185, 765–780. [Google Scholar] [CrossRef]
- Han, S.K.; Torii, K.U. Lineage-specific stem cells, signals and asymmetries during stomatal development. Development 2016, 143, 1259–1270. [Google Scholar] [CrossRef] [Green Version]
- Pillitteri, L.J.; Dong, J. Stomatal development in arabidopsis. Arab. Book/Am. Soc. Plant Biol. 2013, 11, e0162. [Google Scholar] [CrossRef] [Green Version]
- Sotiriou, P.; Giannoutsou, E.; Panteris, E.; Galatis, B.; Apostolakos, P. Local differentiation of cell wall matrix polysaccharides in sinuous pavement cells: Its possible involvement in the flexibility of cell shape. Plant Biol. 2018, 20, 223–237. [Google Scholar] [CrossRef]
- Giannoutsou, E.; Apostolakos, P.; Galatis, B. Spatio-temporal diversification of the cell wall matrix materials in the developing stomatal complexes of Zea mays. Planta 2016, 244, 1125–1143. [Google Scholar] [CrossRef]
- Sotiriou, P.; Giannoutsou, E.; Panteris, E.; Apostolakos, P.; Galatis, B. Cell wall matrix polysaccharide distribution and cortical microtubule organization: Two factors controlling mesophyll cell morphogenesis in land plants. Ann. Bot. 2016, 117, 401–419. [Google Scholar] [CrossRef] [Green Version]
- Jones, L.; Milne, J.L.; Ashford, D.; McCann, M.C.; McQueen-Mason, S.J. A conserved functional role of pectic polymers in stomatal guard cells from a range of plant species. Planta 2005, 221, 255–264. [Google Scholar] [CrossRef]
- Giannoutsou, E.; Sotiriou, P.; Nikolakopoulou, T.L.; Galatis, B.; Apostolakos, P. Callose and homogalacturonan epitope distribution in stomatal complexes of Zea mays and Vigna sinensis. Protoplasma 2020, 257, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Marom, Z.; Shtein, I.; Bar-On, B. Stomatal opening: The role of cell-wall mechanical anisotropy and its analytical relations to the bio-composite characteristics. Front. Plant Sci. 2017, 8, 2061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woolfenden, H.C.; Bourdais, G.; Kopischke, M.; Miedes, E.; Molina, A.; Robatzek, S.; Morris, R.J. A computational approach for inferring the cell wall properties that govern guard cell dynamics. Plant J. 2017, 92, 5–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amsbury, S.; Hunt, L.; Elhaddad, N.; Baillie, A.; Lundgren, M.; Verhertbruggen, Y.; Scheller, H.V.; Knox, J.P.; Fleming, A.J.; Gray, J.E. Stomatal function requires pectin de-methyl-esterification of the guard cell wall. Curr. Biol. 2016, 26, 2899–2906. [Google Scholar] [CrossRef] [Green Version]
- Rui, Y.; Chen, Y.; Kandemir, B.; Yi, H.; Wang, J.Z.; Puri, V.M.; Anderson, C.T. Balancing strength and flexibility: How the synthesis, organization, and modification of guard cell walls govern stomatal development and dynamics. Front. Plant Sci. 2018, 9, 1202. [Google Scholar] [CrossRef] [Green Version]
- Rui, Y.; Chen, Y.; Yi, H.; Purzycki, T.; Puri, V.M.; Anderson, C.T. Synergistic pectin degradation and guard cell pressurization underlie stomatal pore formation. Plant Physiol. 2019, 180, 66–77. [Google Scholar] [CrossRef] [Green Version]
- Beris, D.; Podia, V.; Dervisi, I.; Kapolas, G.; Isaioglou, I.; Tsamadou, V.; Pikoula, L.; Rovoli, M.; Vallianou, A.; Roussis, A.; et al. RNAi silencing of the Arabidopsis thaliana ULCS1 gene results in pleiotropic phenotypes during plant growth and development. Int. J. Dev. Biol. 2021; in press. [Google Scholar] [CrossRef]
- Endo, H.; Torii, K.U. Stomatal development and perspectives toward agricultural improvement. Cold Spring Harb. Perspect. Biol. 2019, 11, a034660. [Google Scholar] [CrossRef] [Green Version]
- Komis, G.; Šamajová, O.; Ovečka, M.; Šamaj, J. Cell and Developmental Biology of Plant Mitogen-Activated Protein Kinases. Annu. Rev. Plant Biol. 2018, 69, 237–265. [Google Scholar] [CrossRef]
- Samakovli, D.; Komis, G.; Šamaj, J. Uncovering the Genetic Networks Driving Stomatal Lineage Development. Mol. Plant 2020, 10, 1355–1357. [Google Scholar] [CrossRef]
- Giannoutsou, E.; Sotiriou, P.; Apostolakos, P.; Galatis, B. Early local differentiation of the cell wall matrix defines the contact sites in lobed mesophyll cells of Zea mays. Ann. Bot. 2013, 112, 1067–1081. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; McEvoy, D.; Le, Y.; Ambrose, C. Live imaging of microtubule organization, cell expansion, and intercellular space formation in Arabidopsis leaf spongy mesophyll cells. Plant Cell 2021, 33, 623–641. [Google Scholar] [CrossRef] [PubMed]
- Sapala, A.; Runions, A.; Routier-Kierzkowska, A.L.; Gupta, M.D.; Hong, L.; Hofhuis, H.; Verger, S.; Mosca, G.; Li, C.B.; Hay, A.; et al. Why plants make puzzle cells, and how their shape emerges. eLife 2018, 7, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Woolfenden, H.C.; Baillie, A.L.; Gray, J.E.; Hobbs, J.K.; Morris, R.J.; Fleming, A.J. Models and mechanisms of stomatal mechanics. Trends Plant Sci. 2018, 23, 822–832. [Google Scholar] [CrossRef] [PubMed]
- Rui, Y.; Anderson, C.T. Functional analysis of cellulose and xyloglucan in the walls of stomatal guard cells of Arabidopsis. Plant Physiol. 2016, 170, 1398–1419. [Google Scholar] [CrossRef] [Green Version]
- Rui, Y.; Xiao, C.; Yi, H.; Kandemir, B.; Wang, J.Z.; Puri, V.M.; Anderson, C.T. Polygalacturonase involved in expansion3: Functions in seedling development, rosette growth, and stomatal dynamics in Arabidopsis thaliana. Plant Cell 2017, 29, 2413–2432. [Google Scholar] [CrossRef] [Green Version]
- Carter, R.; Woolfenden, H.; Baillie, A.; Amsbury, S.; Carroll, S.; Healicon, E.; Sovatzoglou, S.; Braybrook, S.; Gray, J.E.; Hobbs, J.; et al. Stomatal opening involves polar, not radial, stiffening of guard cells. Curr. Biol. 2017, 27, 2974–2983.e2. [Google Scholar] [CrossRef] [Green Version]
- Apostolakos, P.; Livanos, P.; Galatis, B. Microtubule involvement in the deposition of radial fibrillar callose arrays in stomata of the fern Asplenium nidus L. Cell Motil. Cytoskel. 2009, 66, 342–349. [Google Scholar] [CrossRef]
- Apostolakos, P.; Livanos, P.; Nikolakopoulou, T.L.; Galatis, B. The role of callose in guard cell wall differentiation and stomatal pore formation in the fern Asplenium nidus L. Ann. Bot. 2009, 104, 1373–1387. [Google Scholar] [CrossRef] [Green Version]
- Apostolakos, P.; Livanos, P.; Nikolakopoulou, T.L.; Galatis, B. Callose implication in stomatal opening and closure in the fern Asplenium nidus. New Phytol. 2010, 186, 623–635. [Google Scholar] [CrossRef]
- Galatis, B.; Apostolakos, P. A new callose function: Involvement in differentiation and function of fern stomatal complexes. Plant Sign Behav. 2010, 5, 1359–1364. [Google Scholar] [CrossRef] [Green Version]
- Paraskevopoulou, D.; Anezakis, N.; Giannoutsou, E.; Sotiriou, P.; Adamakis, I.-D.S. The Stomata of the Katanin Mutants, fra2, lue1 and bot1. Biol. Life Sci. Forum 2021, 4, 30. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsakali, A.; Asitzoglou, I.-C.; Basdeki, V.; Podia, V.; Adamakis, I.-D.S.; Giannoutsou, E.; Haralampidis, K.
The Role of PME2 and PME3 in Arabidopsis Stomatal Development and Morphology
Tsakali A, Asitzoglou I-C, Basdeki V, Podia V, Adamakis I-DS, Giannoutsou E, Haralampidis K.
The Role of PME2 and PME3 in Arabidopsis Stomatal Development and Morphology
Tsakali, Amalia, Ioannis-Christos Asitzoglou, Vassiliki Basdeki, Varvara Podia, Ioannis-Dimosthenis S. Adamakis, Eleni Giannoutsou, and Kosmas Haralampidis.
2022. "The Role of PME2 and PME3 in Arabidopsis Stomatal Development and Morphology
Tsakali, A., Asitzoglou, I.-C., Basdeki, V., Podia, V., Adamakis, I.-D. S., Giannoutsou, E., & Haralampidis, K.
(2022). The Role of PME2 and PME3 in Arabidopsis Stomatal Development and Morphology








