1. Introduction
The term ‘inflammatory bowel disease’ (IBD) is used to denote all recurrent inflammatory bowel disorders, of which Crohn’s disease (CD) and ulcerative colitis (UC) are the two most common. The etiology of IBD is not yet fully understood, and therefore, no effective prevention or cure is currently available [
1]. These disorders are considered to be heterogeneous conditions of multifactorial etiology, with a combination of genetic and environmental influences contributing to their development [
2].
The treatment of IBD is primarily focused on the administration of corticosteroids, such as prednisone, prednisolone, and budesonide, or aminosalicylic acid. In cases in which symptoms persist, the addition of immunomodulators, such as azathioprine and mercaptopurine, is advised. When these treatments prove ineffective or are contraindicated, methotrexate is recommended [
3]. Finally, for moderate to severe cases of IBD that do not respond to these therapies, biological treatment is employed, using tumor necrosis factor inhibitors (anti-TNFs) [
4,
5]. However, around one-third of patients do not respond to the initial treatment (primary non-responders), and an additional 30% experience a loss of effectiveness over time [
6].
Anti-TNF-α drugs are a class of therapeutic agents developed to neutralize TNF-α activity through a variety of mechanisms. These mechanisms are intended to prevent the involvement of TNF-α in the inflammatory cascade and the tissue damage that is associated with this response [
7]. The most extensively studied anti-TNF-α drugs are adalimumab (ADA) and infliximab (IFX). ADA is a recombinant human immunoglobulin (IgG1) monoclonal antibody that specifically binds to TNF-α and neutralizes its biological function by blocking its interaction with the p55 and p75 receptors for cell-surface TNF [
8]. IFX is a monoclonal antibody that binds with high affinity and selectivity to TNF-α in both its soluble and transmembrane forms [
9]. In order to understand the action of these drugs, it is first necessary to understand the role of TNF-α.
TNF-α is a cytokine that plays a central role in inflammatory and immune responses. It is synthesised by various cell types, with monocyte-macrophages being the predominant producers. However, it is also secreted by activated endothelial cells, fibroblasts, and chondrocytes of articular cartilage. To a lesser extent, it is also produced by keratinocytes and lymphocytes. TNF-α is initially synthesised as a transmembrane protein, and its conversion to the soluble form is catalyzed by the TNF alpha convertase enzyme (TACE), thus enabling it to circulate within the body. Once in its active form, TNF-α exerts its functions by binding to two specific receptors on the cell surface: TNF-RIp55 (TNFRSF1A), which is located on leukocytes and endothelial cells, and TNF-RIIp75 (TNFRSF1B), which is present on a wide variety of cells [
10,
11].
The interaction of TNF-α with its receptors activates three main intracellular signalling pathways: one that induces apoptosis by activating caspases; another that stimulates MAP kinases and the transcription of the AP-1 factor; and a third that activates NF-kB, which is a key protein in regulating the transcription of genes related to inflammation, immunity, and the expression of cell adhesion molecules. The functions of TNF-α are diverse and fundamental to the physiology of the immune system, as it promotes apoptosis or programmed cell death, favors the production of pro-inflammatory cytokines, such as IL6, IL8, and IL1β, induces the expression of adhesion molecules in the endothelium, facilitates the recruitment of leukocytes to inflamed tissues, and participates in the activation, proliferation, and differentiation of immune cells [
11].
In recent years, a range of other biologics have been authorized for the treatment of IBD. Vedolizumab (VLZ) and ustekinumab (UST) are two examples of this class of medications. VLZ is a humanized monoclonal antibody that targets the α4β7 heterodimer and inhibits the migration of lymphocytes specifically to the gut while sparing their migration to other areas, like the central nervous system (CNS) [
12,
13]. UST is a monoclonal antibody that targets the p40 subunit shared by the pro-inflammatory cytokines interleukin (IL) IL12 and IL23 and functions by inhibiting their binding to the IL12Rβ1 receptor, which is expressed on the surface of T-lymphocytes [
14,
15]. Research conducted with real-world patient groups has demonstrated that approximately 40% of individuals do not respond initially to VLZ, while around 35% exhibit primary non-response to UST [
6].
Several studies have explored the potential of genetic variability among patients as a contributing factor to these observed variations in response to treatment. The potential of various genomic biomarkers, especially those that affect the response to anti-TNF therapies, has been explored. Research suggests that in addition to functional variants in
TNF-α and
TNFR, genetic differences within immune and cytokine signaling pathways may also significantly influence treatment outcomes. Consequently, numerous pharmacogenetic studies have been conducted to investigate the association between Single-Nucleotide Variants (SNVs) and the response to biological therapy in IBD [
3].
The objective of this study was to design a genotyping array with 60 SNVs associated with biological treatment response in IBD, as identified in the existing literature.
2. Methodology
A literature search was conducted in PubMed using the following terms: [“pharmacogenetic” or “genetic biomarkers” or “polymorphisms”] and [“response to biological treatments” or “anti-TNF” or “adalimumab” or “infliximab” or “vedolizumab” or “ustekinumab”] and [“inflammatory bowel disease” or “Crohn’s disease” or “ulcerative colitis”] (latest search date: 12 February 2024). The primary focus of this study was the most recent reviews published on the subject, from which all the SNVs that showed significant differences in response to treatment were selected for the array design. Subsequently, other studies presenting original data on the association between variants and response to biological treatment were selected to complete the 60 SNVs. Articles focused on the genetics of the disease rather than the response to treatment were excluded.
The analysis will be conducted via the use of OpenArray™ technology (Applied Biosystems, ThermoFisher, Waltham, MA, USA), which is based on real-time PCR and provides flexible solutions for configuring the plates for genotyping analysis. The available formats include 12, 26, 60, 120, 180, or 240 assays (SNVs) (
https://www.thermofisher.com/es/es/home/life-science/pcr/real-time-pcr/real-time-openarray.html, accessed on 10 February 2025). In this research, the 60-assay format was selected based on the literature search, the number of samples to be analyzed, and the budget.
In order to proceed with the design of the array, a total of 60 SNVs, which, according to previous studies, significantly influence the response to biological treatment and for which a TaqMan assay was available for genotyping, were selected.
The LDlink tool (
https://ldlink.nih.gov/?tab=home accessed on 10 June 2025) was used to check for linkage disequilibrium between the selected SNVs within the European and Latin American populations.
3. Results
Numerous pharmacogenetic studies identified a linkage between certain SNVs and response to biological therapy in IBD. In the present study, a comprehensive collection of three reviews [
3,
5,
16] and 14 original articles on the subject were selected [
12,
13,
14,
15,
16,
17,
18,
19,
20,
21,
22,
23,
24,
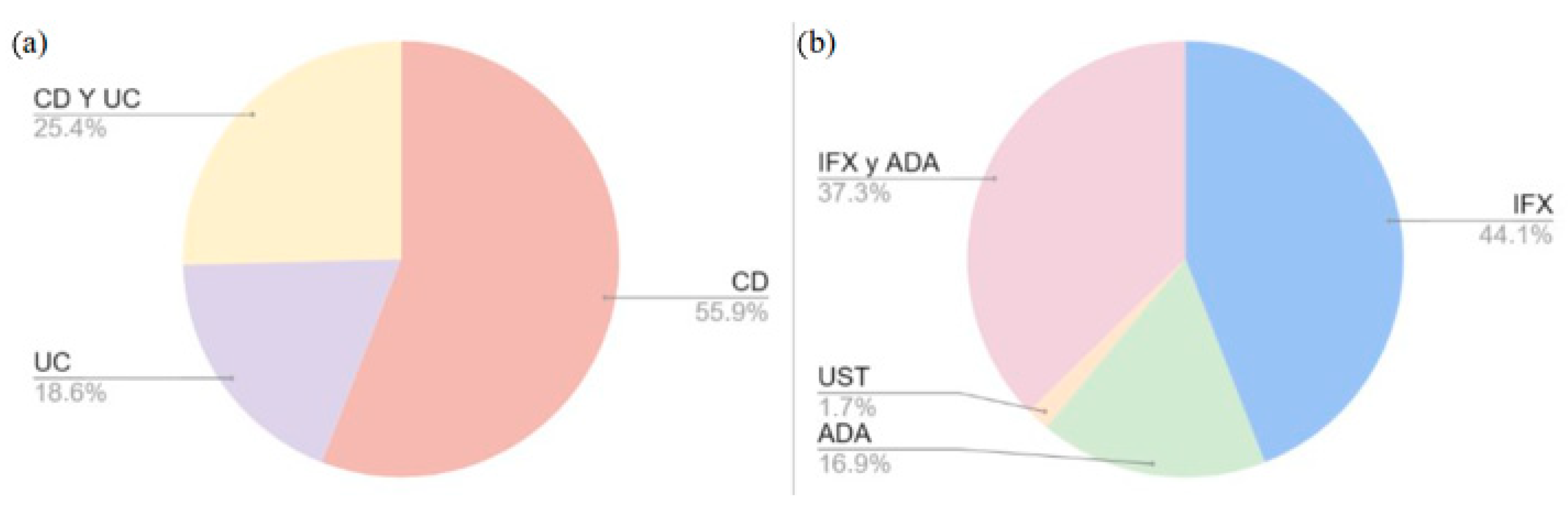
25]. As illustrated in
Figure 1, the percentages of selected SNVs are displayed according to two categories: the specific disease (a) and the biological drug used (b). With regard to the disease, 55.9% of the SNVs were studied in patients with CD, 18.6% were studied in patients with UC, and 25.4% were studied in patients with both pathologies. With respect to biologic therapy, 44.1% of SNVs were observed to influence the response to IFX, 16.9% influenced the response to ADA, and 37.3% influenced the response to both IFX and ADA; however, only one study (1.7%) reported an influence on the response to UST and none reported an influence on the response to VLZ.
A total of 38 genes were identified in the context of biological drug response; the genes and the variants that were selected are listed in
Table 1. The results of the linkage disequilibrium analysis indicated the non-existence of such disequilibrium among the selected SNVs within the European and Latin American populations. An array was designed to evaluate if these associations were maintained in the patient cohort under investigation. It should be noted that the array design contains 59 variants as a result of an error in its configuration. The TaqMan Assay used for the array design is displayed in the
Table S1.
The genes that were ultimately incorporated into our study have been linked to a variety of biological processes that may influence the effectiveness of anti-TNFs. The TNF signaling and regulation genes included TNF, TNFRSF1A, TNFRSF1B, TNFAIP3, and ADAM17. The interleukin signaling and regulation genes included IL1B, IL1R1, IL1RN, IL11, IL12B, IL17F, IL18, IL23R, IL27, and IL6 and the autophagy and inflammation regulation genes included ATG5 and ATG16L1. Furthermore, genes associated with apoptosis and cell survival, such as FAS, FASLG, CASP9, and TBX21, were covered, as well as genes involved in the interferon pathway and innate immune response, such as IFNGR1, IFNGR2, and JAK2, and genes associated with inflammasome signaling and response to molecular patterns, such as NLRP3, TLR2, TLR4, and TLR5. Moreover, genes associated with the regulation of nuclear factor kappa B (NF-κB), such as NFKBIA and CCNY, were considered. Finally, other genes of interest, such as HFE, TF, CRP, CD14, CD96, EMSY, PTPN2, and XBP1, were also included due to their possible involvement in modulating the anti-TNF response. The next section provides detailed information on each of these genes in alphabetical order.
3.1. ADAM17
This gene encodes a protein belonging to the Disintegrin and Metalloprotease Domain (ADAM) family, which is involved in numerous biological processes, including cell-to-cell and cell-to-matrix communication. Increased expression of this gene has been detected in certain cell types from individuals with psoriasis, rheumatoid arthritis, multiple sclerosis (MS), and CD, indicating that the protein it produces may contribute to the development or progression of autoimmune disorders (
https://www.ncbi.nlm.nih.gov/gene/6868, accessed on 14 February 2025).
A study was conducted on 222 Caucasian patients treated with IFX, including both luminal and fistulizing cases, to investigate several variants in the
ADAM17 gene. The haplotype defined by rs2001658TT, rs12469362CC, rs883399GG, rs1048610GG, rs2276338CC, rs1056204CC, rs10929587TT, rs10495565GG, rs4464248AA, and rs11684747AA was found to be significantly less prevalent in the clinical non-responders (
p = 0.0027) [
16,
17]. The genetic variant rs2001658 could not be included in the array design due to complications experienced during the probe fabrication process.
3.2. ATG16L1
Autophagy-related 16 like 1 (
ATG16L1) is responsible for encoding a protein necessary for autophagy. Alterations in this gene have been linked to an increased susceptibility to inflammatory bowel disease (
https://www.ncbi.nlm.nih.gov/gene/55054, accessed on 14 February 2025).
To investigate the influence of different SNVs associated with CD risk on ADA response, 33 SNVs in 31 genes, including
ATG16L1, were sequenced in 102 Slovakian CD patients. The T allele of the rs10210302 variant in the
ATG16L1 gene demonstrated the strongest correlation with response to ADA; patients carrying the TT genotype maintained a favorable therapeutic response, as measured using the Inflammatory Bowel Disease Questionnaire (IBDQ), at both 20 weeks (
p = 0.0461) and 30 weeks (
p = 0.0493) of treatment [
3,
16,
18]. Furthermore, the presence of the T allele in rs2241880 was linked to reduced treatment efficacy with IFX and ADA in a study involving 94 pediatric patients diagnosed with CD and UC [
3,
19].
3.3. ATG5
Autophagy-related 5 (
ATG5) encodes a protein that plays a key role in various cellular functions, such as the formation of autophagic vesicles, antigen presentation via MHC class II, and the regulation of apoptosis (
https://www.ncbi.nlm.nih.gov/gene/9474, accessed on 20 February 2025).
A prospective study was conducted to evaluate the role of different genes in predicting the response to ADA treatment in patients with CD. The findings indicated a significant association between the C allele (
p < 0.001), as well as the CC and CT genotypes (
p < 0.001) of the rs9373839 variant, and a favorable therapeutic response based on IBDQ scores. Individuals who carried at least one C allele were found to be 4.2 times more likely to exhibit a positive response to treatment. In addition, the CG haplotype, which was defined by the rs9373839 and rs510432 variants, showed a significant association (
p < 0.001) with a positive response to therapy. Patients with this particular haplotype were observed to have a 36% increased likelihood of a positive response to therapy [
20].
3.4. CASP9
The caspase 9 (
CASP9) gene codes for a member of the cysteine–aspartic acid protease (caspase) family, which is believed to play a key role in the apoptotic process and to function as a tumor suppressor (
https://www.ncbi.nlm.nih.gov/gene/842, accessed on 26 February 2025).
In a cohort of 287 patients diagnosed with CD who were treated with IFX, those carrying the TT genotype of the rs4645983 variant demonstrated a favorable response to therapy (
p = 0.04) in comparison to those with the CC or CT genotype [
3,
16,
21].
3.5. CCNY
Cyclins, an example of which is CCNY, are responsible for controlling cell division cycles and regulating cyclin-dependent kinases (
https://www.ncbi.nlm.nih.gov/gene/219771, accessed on 26 February 2025).
In the aforementioned study, in which
ATG16L1 was evaluated, it was observed that the CC genotype of the rs12777960 variant demonstrated a better response to therapy (OR: 3.26, 95% CI: 1.27–8.38,
p = 0.0156,
Pcorr = 0.515) [
16,
18].
3.6. CD14
The CD14 molecule (
CD14) gene encodes a surface antigen predominantly expressed on monocytes and macrophages. This protein works in conjunction with other molecules to mediate the innate immune response to bacterial lipopolysaccharides and viral infections (
https://www.ncbi.nlm.nih.gov/gene/929, accessed on 3 March 2025).
A study was conducted on a prior anti-TNF-naive Danish cohort of 738 patients with IBD. It revealed that the homozygous and the heterozygous genotypes of the rs2569190 variant were associated with non-response among patients with UC (ORunadj: 0.54, 95% CI: 0.30–0.98,
p = 0.04) [
5,
22].
3.7. CD96
The CD96 molecule (
CD96) gene codes for a protein that is part of the immunoglobulin superfamily and is believed to contribute to the adhesion of activated T cells and natural killer (NK) cells during the later stages of the immune response. Additionally, it has been proposed that this protein may play a role in antigen presentation (
https://www.ncbi.nlm.nih.gov/gene/10225, accessed on 4 March 2025).
A study of patients with CD who had received ADA therapy revealed a significant association between the clinical response to ADA and the A allele of the rs9828223 variant (OR = 1.77, 95% CI, 1.09–5.02,
p = 0.019). This allele was found to be associated with a lack of clinical response to ADA therapy [
23].
3.8. CRP
The C-reactive protein (
CRP) gene encodes a member of the pentraxin family, which plays a role in activating and amplifying the complement system through interactions with pattern recognition molecules. CRP contributes to host defense by recognizing foreign pathogens and damaged host cells, promoting their clearance through engagement with both humoral and cellular effector mechanisms in the bloodstream (
https://www.ncbi.nlm.nih.gov/gene/1401, accessed on 26 February 2025).
In the aforementioned study, in which
ATG5 was evaluated, it was observed that the C allele (
p < 0.001) of the rs1130864 variant exhibited a significant association with a positive response to therapy, as measured by the C-reactive protein (CRP) criterion. Patients who carried the C allele exhibited a fourfold elevated probability of responding positively to therapy [
20].
3.9. EMSY
The EMSY transcriptional repressor (
EMSY) is predicted to facilitate identical protein binding activity. In addition, it has been thought to be involved in DNA repair, chromatin organization, and the regulation of DNA-templated transcription (
https://www.ncbi.nlm.nih.gov/gene/56946, accessed on 3 March 2025).
In a previous study, in which
ATG16L1 was evaluated, it was observed that the CC genotype of the rs7927894 variant was associated with a better response to therapy at week 20 (
p = 0.006,
Pcorr = 0.192), compared with those carrying the T allele [
16,
18].
3.10. FAS
The Fas cell surface death receptor (
FAS) gene is responsible for encoding a protein that belongs to the TNF receptor superfamily. It has been demonstrated that this protein plays a fundamental role in the physiological regulation of programmed cell death, and it is involved in the pathogenesis of various malignancies and diseases of the immune system (
https://www.ncbi.nlm.nih.gov/gene/355, accessed on 3 March 2025).
In a cohort of 196 Polish patients with clinically confirmed CD who were receiving anti-TNF therapy, the CC genotype of the rs7896789 variant was significantly associated with a higher risk of non-response to treatment (OR: 15.22, 95% CI: 1.53–151.9,
p = 0.003). In contrast, the TT genotype seemed to confer a protective effect (OR: 0.07, 95% CI: 0.01–0.66,
p = 0.003) [
21,
24].
3.11. FASLG
The Fas ligand (
FASLG) gene codes for a transmembrane protein that belongs to the TNF superfamily, primarily functioning to induce apoptosis through its interaction with the FAS receptor. The FAS/FASLG signaling pathway plays a critical role in immune regulation, particularly in the activation of induced cell death (AICD) of T cells and in cytotoxic T cell-mediated apoptosis. The dysregulation of this pathway has been linked to the development of various cancers, and mutations in
FASLG have been associated with certain forms of systemic lupus erythematosus (
https://www.ncbi.nlm.nih.gov/gene/356, accessed on 3 March 2025).
In the abovementioned study, in which
CASP9 was evaluated, it was observed that patients with luminal CD who presented with the CC/CT genotype of the rs763110 variant demonstrated a response rate of 75%, in comparison to the 38% response rate of those carrying the TT genotype (OR = 0.11; 95% CI: 0.08–0.56;
p = 0.002). In the cohort of patients diagnosed with fistulising CD, the CC/CT genotype was identified as the only predictor of response (OR = 1.66, 95% CI: 1.21–2.29,
p = 0.002). In summary, the C allele of the rs763110 variant demonstrated a better response to IFX in both luminal and fistulizing CD patients [
3,
5,
16,
21].
3.12. HFE
The homeostatic iron regulator (
HFE) gene codes for a membrane protein resembling MHC class I molecules, which interacts with beta-2-microglobulin (β2M). This protein is believed to play a role in controlling iron absorption by modulating the binding between the transferrin receptor and transferrin (
https://www.ncbi.nlm.nih.gov/gene/3077, accessed on 3 March 2025).
In a study of 68 refractory CD patients from Slovenia who were treated with ADA, it was observed that those who carried the G allele exhibited a better response in comparison to those who possessed the A allele at the rs2071303 variant after 20 weeks (
p = 0.008) [
25].
3.13. IFNGR1 and IFNGR2
The interferon gamma receptor 1 (
IFNGR1) gene is responsible for encoding the ligand-binding chain (alpha) of the gamma interferon receptor. The human interferon-gamma receptor is a heterodimer composed of IFNGR1 and IFNGR2. Genetic variation in
IFNGR1 was observed to be associated with an increased susceptibility to
Helicobacter pylori infection (
https://www.ncbi.nlm.nih.gov/gene/3459, accessed on 3 March 2025). The interferon gamma receptor 2 (
IFNGR2) gene is responsible for encoding the non-ligand-binding beta chain of the gamma interferon receptor. Mutations in the IFNGR2 gene have been identified as the cause of Mendelian susceptibility to mycobacterial disease (MSMD), also referred to as familial disseminated atypical mycobacterial infection (
https://www.ncbi.nlm.nih.gov/gene/3460, accessed on 3 March 2025).
A study involving 482 and 256 previously untreated Danish patients with CD and UC, respectively, who received anti-TNF treatment, revealed that the heterozygous (TC) genotype of the rs2234711 variant in
IFNGR1 was associated with non-response among UC patients (OR: 0.29, 95% CI: 0.11–0.78,
p = 0.01). The same study also established an association between the heterozygous genotype of rs2234711 and non-response among patients with IBD (CD or UC) (OR: 0.57, 95% CI: 0.34–0.96,
p = 0.04). The homozygous (CC) variant genotype of the rs8126756 variant in
IFNGR2 was found to be associated with non-response among patients with CD (OR: 0.09, 95% CI: 0.01–0.65,
p = 0.02) [
26].
3.14. IL11
The interleukin 11 (
IL11) gene codes for a cytokine that belongs to the gp130 family. IL-11 has been shown to promote T cell-dependent differentiation of B cells into immunoglobulin-producing cells and to enhance the proliferation of hematopoietic stem cells and megakaryocyte progenitors (
https://www.ncbi.nlm.nih.gov/gene/3589, accessed on 3 March 2025).
In a study comprising 350 patients with active CD who received a minimum of three induction doses of IFX, the influence of rs1126760 and rs1042506 variants in the
IL11 gene on response to IFX was investigated. The results of the study demonstrated that the CT haplotype, defined by the rs1126760 and rs1042506 variants, was associated with a better response to IFX (OR: 1.72, 95% CI: 0.91–3.21,
p = 0.068) [
16,
27]. The genetic variant rs1126760 was not included in the array design because a TaqMan assay was not available for genotyping purposes.
3.15. IL12B
The interleukin 12 (
IL12B) gene encodes a subunit of interleukin-12, a cytokine that influences both T cells and NK cells and is involved in various immune functions. Produced by activated macrophages, IL-12 plays a key role in promoting the differentiation of Th1 cells. The elevated expression of this gene has been observed in the central nervous system of individuals with MS, indicating its possible involvement in disease pathogenesis. Additionally, a promoter polymorphism in
IL12B has been linked to the severity of both atopic and non-atopic asthma in children (
https://www.ncbi.nlm.nih.gov/gene/3593, accessed on 3 March 2025).
In the aforementioned study, in which
IFNGR1 was evaluated, the combined homozygous (CC) and heterozygous (GC) genotypes of the rs3212217 variant (OR: 0.33, 95% CI: 0.15–0.69,
p = 0.004) were associated with non-response among patients with UC [
26].
3.16. IL17F
The interleukin 17F (
IL17F) gene encodes a cytokine closely related in sequence to IL-17. Produced by activated T cells, IL-17F promotes the expression of various other cytokines. Additionally, it has been shown to inhibit angiogenesis in endothelial cells and to stimulate these cells to produce IL-2, TGF-β1, and monocyte chemoattractant protein-1 (MCP-1) (
https://www.ncbi.nlm.nih.gov/gene/112744, accessed on 3 March 2025).
A study was conducted on 113 Japanese patients with CD to investigate their response to IFX. The frequency of the heterozygous GA genotype or the homozygous AA genotype of rs766748 was found to be significantly lower in responders in comparison to non-responders (OR = 0.203,
p = 0.019). In fact, there was almost a 4.9-fold reduction in response to IFX in non-responders with these genotypes after one year of treatment compared to responders. Conversely, the presence of a homozygous GG genotype of rs766748 was associated with a 4.9-fold increase in response to IFX in the responders with this genotype, as compared to the non-responders [
3,
28].
3.17. IL18
The interleukin 18 (
IL18) gene encodes a proinflammatory cytokine member of the IL-1 family. It is synthesized as an inactive precursor within the cytoplasm of various cell types, such as macrophages and keratinocytes. IL-18 has been associated with tissue damage in multiple organs and is implicated in life-threatening conditions that involve cytokine storms (
https://www.ncbi.nlm.nih.gov/gene/3606, accessed on 4 March 2025).
In the aforementioned study, in which
IFNGR1 was evaluated, both the homozygous and heterozygous genotypes of the rs1946518 variant were significantly associated with a favorable therapeutic response in patients with UC (OR: 2.14, 95% CI: 1.06–4.32,
p = 0.03) [
26].
Moreover, in a study conducted on 587 CD and 458 UC patients treated with anti-TNF, the combined homozygous and heterozygous genotypes of the rs187238 variant were associated with a beneficial response among patients with CD (OR: 1.35, 95% CI: 1.00–1.82,
p = 0.047), and the combined homozygous and heterozygous genotypes of the rs1946518 variant were associated with a beneficial response among patients with IBD (OR: 1.24, 95% CI: 1.01–1.53,
p = 0.04) [
29].
3.18. IL1B
The IL-1 beta (
IL1B) gene is responsible for encoding a protein belonging to the IL-1 cytokine family. This cytokine plays a pivotal role as an inflammatory response mediator, and it is involved in a variety of cellular activities, including cell proliferation, differentiation, and apoptosis. Furthermore, IL1B has been implicated in the pathogenesis of human osteoarthritis (
https://www.ncbi.nlm.nih.gov/gene/3553, accessed on 4 March 2025).
A study of 47 patients (29 CD and 18 UC) who received IFX showed that the C allele in the rs1143634 variant was more common in non-responders (86.4%) than in responders (65.0%) [
3,
30].
On the other hand, in the aforementioned study, in which
CD14 was evaluated, the homozygous and heterozygous genotypes of the rs4848306 variant were associated with a beneficial response among patients with UC (ORadj: 2.69, 95% CI: 1.04–6.94,
p = 0.04) and patients with IBD (ORadj: 1.85, 95% CI: 1.05–3.27,
p = 0.03). The A allele in rs4848306 was associated with a better response to IFX [
3,
22].
3.19. IL1R1
The IL-1 receptor type 1 (
IL1R1) gene encodes a cytokine receptor that belongs to the interleukin-1 receptor family. IL-1α, IL-1β, and the IL-1 receptor antagonist bind to this receptor, thus playing a key role in mediating immune and inflammatory responses triggered by cytokines (
https://www.ncbi.nlm.nih.gov/gene/3554, accessed on 4 March 2025).
In a previous study, in which
FAS was evaluated, 33 patients (16.8%) were found to be non-responders to therapy. This lack of response was associated with the presence of the G allele in the rs2041747 variant of the
IL1R1 gene. (OR: 3.72,
p = 0.009) [
24].
3.20. IL1RN
The interleukin 1 receptor antagonist (
IL1RN) gene codes for a protein belonging to the IL-1 cytokine family. It functions as an antagonist that inhibits the activities of IL-1 alpha (IL1A) and IL-1 beta (IL1B). It plays a regulatory role in various IL-1-mediated immune and inflammatory responses, especially during the acute phases of infection and inflammation. Furthermore, certain polymorphisms in this gene have been linked to an increased risk of osteoporotic fractures and gastric cancer (
https://www.ncbi.nlm.nih.gov/gene/3557, accessed on 4 March 2025).
In the aforementioned study, in which
IL18 was evaluated, the combined homozygous and heterozygous genotypes of the rs4251961 variant were associated with non-response among patients with IBD (OR: 0.81, 95% CI: 0.66–1.00,
p = 0.049) [
29].
3.21. IL23R
The interleukin 23 receptor (
IL23R) gene encodes a component of the receptor complex for IL-23 (IL23A/IL23). This protein forms a heterodimer with IL12RB1 (also known as IL12Rβ1), and together, they mediate IL-23 signaling. IL23R is constitutively linked to Janus kinase 2 (JAK2), and it interacts with the transcription factor STAT3 in response to ligand binding (
https://www.ncbi.nlm.nih.gov/gene/149233, accessed on 4 March 2025).
Genetic variants in
IL23R have been demonstrated to be associated with a response to IFX in patients suffering from moderate to severe UC (
n = 90) at week 14 in previous research. The percentage of responders (74.5%) within the group of homozygous carriers of the A allele for rs1004819 and rs10889677 variants was higher than that of non-responders (25.5%). Furthermore, the AA genotype for both rs1004819 and rs10889677 was found to increase the probability of response to IFX. In contrast, for rs7517847 and rs11465804 variants, the proportion of non-responders (65.4%) exceeded that of responders (34.6%) among homozygous carriers of the G allele. For instance, the probability of response to the drug was decreased in individuals with a GG genotype for both rs7517847 and rs11465804 [
5,
31].
3.22. IL27
Interleukin 27 (
IL27) codes for the subunits of a heterodimeric cytokine complex. Moreover, it interacts with Epstein–Barr virus-induced gene 3 (EBI3), which produces a protein related to interleukin 12B (IL12B). This complex has been demonstrated to promote the rapid proliferation of naive CD4(+) T cells (
https://www.ncbi.nlm.nih.gov/gene/246778, accessed on 26 February 2025).
In a study of 102 patients suffering from CD, an association between the rs8049439 variant and the response to ADA after 4 weeks was observed. The T allele was observed to be more prevalent among subjects who demonstrated a positive response, in comparison with those who exhibited the CC genotype (
p = 0.01) [
16,
18].
3.23. IL6
The interleukin 6 (
IL6) gene encodes a cytokine that plays a key role in inflammation and the maturation of B cells. This protein also acts as an endogenous pyrogen that triggers fever in individuals with autoimmune conditions or infections. The gene’s function has been linked to various inflammatory disorders, including increased susceptibility to diabetes mellitus and systemic juvenile rheumatoid arthritis (
https://www.ncbi.nlm.nih.gov/gene/3569, accessed on 4 March 2025).
In the aforementioned study, in which
IL18 was evaluated, the combined homozygous and heterozygous genotypes of the rs10499563 variant were found to be borderline associated with a beneficial response among patients with IBD (OR: 1.31, 95% CI: 0.99–1.71,
p = 0.05) [
3,
29].
3.24. JAK2
The Janus kinase 2 (
JAK2) gene is responsible for encoding a non-receptor tyrosine kinase that plays a fundamental role in the process of cytokine and growth factor signaling. Mutations in the
JAK2 gene were identified as a contributing factor to the development of various inflammatory diseases and malignancies (
https://www.ncbi.nlm.nih.gov/gene/3717, accessed on 4 March 2025).
In the abovementioned study, in which
IFNGR1 was evaluated, the homozygous genotype of the rs12343867 variant was associated with non-response among patients with UC (OR: 0.17, 95% CI: 0.03–0.85,
p = 0.03) [
26].
3.25. NFKBIA
The NFKB inhibitor alpha (
NFKBIA) gene is responsible for encoding a member of the NF-κB inhibitor family. The encoded protein is involved in inflammatory responses. Variants in this gene have been identified in cases of anhidrotic ectodermal dysplasia with T cell immunodeficiency autosomal dominant disease (
https://www.ncbi.nlm.nih.gov/gene/4792, accessed on 4 March 2025).
In a previous study, in which
IL18 was evaluated, the combined homozygous and heterozygous genotypes of the rs696 variant were found to be associated with a favorable response among patients with UC (OR: 1.45, 95% CI: 1.06–2.00,
p = 0.02) and IBD patients (CD or UC) (OR: 1.25, 95% CI: 1.01–1.54,
p = 0.04) [
29].
3.26. NLRP3
The NLR family pyrin domain-containing 3 gene (
NLRP3) encodes a pyrin-like protein involved in regulating inflammation, immune responses, and apoptosis. Genetic variants in this gene have been associated with several disorders, such as familial cold autoinflammatory syndrome, Muckle–Wells syndrome, chronic infantile neurological cutaneous and articular syndrome, neonatal-onset multisystem inflammatory disease, keratoendotheliitis fugax hereditaria, and autosomal dominant deafness type 34, which may occur with or without inflammation (
https://www.ncbi.nlm.nih.gov/gene/114548, accessed on 4 March 2025).
In the aforementioned study, in which
IFNGR1 was evaluated, the combined homozygous and heterozygous genotypes of the rs10754558 variant were associated with a beneficial response among patients with IBD (OR: 1.60, 95% CI: 1.02–2.52,
p = 0.04) [
26]. Furthermore, in a previous study, in which
IL18 was evaluated, the combined homozygous and the heterozygous genotypes of the rs4612666 variant were associated with non-response among patients with UC (OR: 0.63, 95% CI: 0.44–0.91,
p = 0.01) and patients with IBD (CD or UC) (OR: 0.73, 95% CI: 0.57–0.95,
p = 0.02) [
29].
3.27. PTPN2
The protein encoded by the protein tyrosine phosphatase non-receptor type 2 (
PTPN2) gene is a member of the protein tyrosine phosphatase (PTP) family. These enzymes act as signaling molecules involved in regulating numerous cellular functions, such as cell growth, differentiation, cell cycle progression, and oncogenic transformation (
https://www.ncbi.nlm.nih.gov/gene/5771, accessed on 4 March 2025).
The impact of the rs7234029 variant was examined in an uncontrolled monocentric retrospective observational study of 379 patients with moderate to severe CD. The study identified an association between the presence of this variant and a lack of response to UST treatment [
3,
32].
3.28. TBX21
The T-box transcription factor 21 (
TBX21) gene belongs to a family of genes that are phylogenetically conserved and share a common DNA-binding domain, known as the T-box. T-box genes are known to encode transcription factors that play a role in the regulation of developmental processes (
https://www.ncbi.nlm.nih.gov/gene/30009, accessed on 4 March 2025).
In the abovementioned study, in which
IFNGR1 was evaluated, the homozygous genotype of the rs17250932 variant was associated with non-response among patients with UC (OR: 0.06, 95% CI: 0.01–0.80,
p = 0.03) [
26].
3.29. TF
The transferrin (TF) gene encodes a glycoprotein responsible for transporting iron from the intestine, reticuloendothelial system, and liver cells to proliferating cells throughout the body. Furthermore, this protein is thought to function as a granulocyte/pollen binding protein (GPBP), potentially aiding in the clearance of specific organic substances and allergens from the bloodstream (
https://www.ncbi.nlm.nih.gov/gene/7018, accessed on 4 March 2025).
In the aforementioned study, in which
HFE was evaluated, a positive response to ADA treatment was observed after 12 and 20 weeks in 87.5% of patients with the T allele of the rs1799852 variant, in comparison to 65% of patients with the C allele. However, statistical significance was only achieved at the borderline level (
p = 0.058) [
25].
3.30. TLR2, TLR4, and TLR5
In a previous study, in which
IL18 was evaluated, the homozygous genotype of the rs11938228 variant in
TLR2 was associated with non-response among patients with UC (OR: 0.55, 95% CI: 0.33–0.92,
p = 0.02) [
29]. It was also observed that the homozygous genotype of the rs1554973 variant in
TLR4 was associated with non-response among patients with UC (OR: 0.49, 95% CI: 0.27–0.90,
p = 0.02). Furthermore, the combined homozygous and heterozygous genotypes were associated with non-response among patients with IBD (OR: 0.80, 95% CI: 0.65–0.98,
p = 0.03). It was also observed that the homozygous genotype of the rs5030728 variant was associated with a favorable response among patients with UC (OR: 2.23, 95% CI: 1.24–4.01,
p = 0.01 and IBD patients (CD or UC) (OR: 1.46, 95% CI: 1.01–2.11,
p = 0.04) [
3,
29].
Furthermore, in a previous study, in which
IFNGR1 was evaluated, the heterozygous genotype of the rs5744174 variant in
TLR5 was associated with non-response among patients with CD (OR: 0.36, 95% CI: 0.16–0.81,
p = 0.01) [
26].
3.31. TNF
The TNF gene codes for a versatile proinflammatory cytokine member of the TNF superfamily. Mainly produced by macrophages, this cytokine plays a key role in regulating numerous biological processes, such as cell proliferation, differentiation, apoptosis, lipid metabolism, and blood coagulation. Its involvement in various conditions, including autoimmune disorders, insulin resistance, psoriasis, rheumatoid arthritis, ankylosing spondylitis, tuberculosis, autosomal dominant polycystic kidney disease, and cancer, has been extensively documented (
https://www.ncbi.nlm.nih.gov/gene/7124, accessed on 4 March 2025).
Several studies have observed that the presence of the A allele in the rs1800629 variant is associated with non-response to IFX treatment. For example, in 82 Spanish CD and UC patients, an increased frequency of the A allele was found in non-responders (
p < 0.05) [
3,
33], and in a prospective cohort study of 121 patients, the presence of the A allele was associated with a three-fold increased likelihood of being a non-responder (
p = 0.049) [
3,
34].
A comprehensive meta-analysis of 532 studies revealed that the G allele at the rs361525 variant, which is a common allele, was associated with a favorable response to anti-TNF therapy in the overall population (
p = 0.011), as well as in Caucasians (
p = 0.016) [
3,
35].
3.32. TNFAIP3
The TNF alpha-induced protein 3 (
TNFAIP3) gene is rapidly activated by tumor necrosis factor (TNF) stimulation. Its encoded protein inhibits NF-kappa B activation and TNF-induced apoptosis, playing a crucial role in regulating cytokine-driven immune and inflammatory responses (
https://www.ncbi.nlm.nih.gov/gene/7128, accessed on 4 March 2025).
In the abovementioned study, in which
CD14 was evaluated, the homozygous and heterozygous genotypes for the rs6927172 variant were associated with non-response among patients with UC (ORadj: 0.34, 95% CI: 0.13–0.90,
p = 0.03) and IBD patients (ORadj: 0.62, 95% CI: 0.42–0.92,
p = 0.02) [
5,
22].
3.33. TNFRSF1A
The TNF receptor superfamily member 1A (
TNFRSF1A) gene is responsible for encoding a member of the TNF receptor superfamily of proteins. The encoded receptor has been demonstrated to play a role in cell survival, apoptosis, and inflammation. Furthermore, it has been observed that mutations in this gene are associated with MS in human patients (
https://www.ncbi.nlm.nih.gov/gene/7132, accessed on 4 March 2025).
In the aforementioned study, in which
IL18 was evaluated, the homozygous genotype of the rs4149570 variant was associated with a beneficial response among patients with CD (OR: 1.92, 95% CI: 1.02–3.60,
p = 0.04) [
29].
Moreover, an investigation was conducted to analyze the efficacy of anti-TNF inhibitors in a cohort of 81 CD patients. The results of this study indicated that individuals who carried the G allele of the rs767455 variant exhibited a lower response to IFX compared with those who carried the AA genotype (
p < 0.01) [
3,
36].
3.34. TNFRSF1B
The TNF receptor superfamily member 1B (
TNFRSF1B) gene encodes a protein that is part of the TNF-receptor superfamily. Together with TNF-receptor 1, it forms a heterocomplex that aids in the recruitment of two anti-apoptotic proteins (
https://www.ncbi.nlm.nih.gov/gene/7133, accessed on 4 March 2025).
In a study involving 297 CD patients from seven centers across Spain with known responses to IFX, the A allele of the rs1061624 variant was found at a significantly higher frequency in non-responders (53.6%) compared to responders (41.5%) (OR: 1.63, 95% CI: 1.05–2.51,
p = 0.02). Additionally, an analysis of the rs3397 variant showed that the minor genotype (CC) was more common among IFX responders than in non-responders, reaching statistical significance (OR: 3.19, 95% CI: 0.95–16.78,
p = 0.05 [
3,
37].
Furthermore, an observational cohort study was conducted on 124 Caucasian patients diagnosed with CD who were undergoing IFX maintenance therapy. The study revealed that the presence of the C allele of the rs976881 variant was associated with a loss of response (OR: 3.3, 95% CI 1.2–9.1;
p = 0.014), particularly in cases of homozygosity (
p = 0.006) [
3,
38].
3.35. XBP1
The X-box binding protein 1 (
XBP1) gene is responsible for encoding a transcription factor that regulates MHC class II genes. This process occurs through the binding of the transcription factor to a promoter element, which is referred to as an X box (
https://www.ncbi.nlm.nih.gov/gene/7494, accessed on 4 March 2025).
In a study of 570 patients with IBD, carriers of the G allele of the rs35873774 variant exhibited a weaker response to IFX compared to non-carriers (OR; 3.7, 95% CI 1.2–10.8;
p = 0.016) [
39].
4. Discussion
IBD is the result of a complex interplay of genetic, environmental, and immunoregulatory factors. Despite the absence of a definitive etiological agent, IBD is characterized by an excessive immune response to the gut microbiota, driven by cytokines such as TNF, IL1, and IL6, which promote the activation of T helper (Th) 1 and Th17 cells [
5].
In individuals with a genetic predisposition for IBD, both innate and adaptive immune systems contribute to the inflammatory response. Th1 cells, which predominate in CD, and Th2 cells, which are more prevalent in UC, play distinct roles, while Th17 cells, which are regulated by IL23, further amplify the inflammatory cascade. The process of microbial recognition triggers Th1 differentiation via IL12 and interferon-gamma (IFN-γ), thereby stimulating macrophages and dendritic cells to release TNF, IL6, IL23, and IL1. TLRs, which mediate pathogen recognition, also influence immune activation. Variants in these receptors are known to affect anti-TNF therapy responses in IBD patients. The secretion of IFN-γ and TNF by activated Th1 cells exacerbates inflammation. Furthermore, IFN-γ may enhance pathogenic Th17 expansion, thereby promoting autoimmunity. In the context of UC, non-classical NK T cells produce IL13, which is a phenomenon that may disrupt the Th1 and Th17 pathways. Th17 cells, which are sustained by IL23, release IL17A and IL17F, thereby inducing TNF production and driving fibrosis and tissue damage in CD [
5].
In recent decades, significant efforts have been made to identify pharmacogenetic biomarkers with the capacity to predict the response to biologic therapy in patients with IBD. Numerous genes have been studied, and some variants, particularly those in the TNF family and interleukins, might be promising. However, the results remain inconclusive [
16].
When considering the mechanism of action of anti-TNF-α drugs and the functions of TNF-α, it can be proposed that variants in genes encoding enzymes or receptors involved in these mechanisms might impact the response to anti-TNFα drugs. For instance, genes involved in apoptosis, such as ATG5, CASP9, FAS, FASLG, IL1B, NLRP3, TNF, TNFAIP3, TNFRSF1A, and TNFRSF1B, are good options for studying treatment response. Furthermore, the study identified additional genes that code for cytokines or cytokine receptors or mediate their production that may also be related to the outcome of treatment with these drugs. These include IL11, IL12B, IL17F, IL18, IL1B, IL1R1, IL1RN, IL23R, IL27, IL6, and TLR4. Finally, other genes involved in various cellular functions have also been studied, such as those involved in cell damage (CRP); DNA repair (EMSY); cell-to-cell and cell-to-matrix interactions (ADAM17); autophagy (ATG16L1); cell division (CCNY); and the immune and/or inflammatory response (CD14, CD96, NFKBIA, NLRP3, TLR2, and TLR5).
A significant number of studies have investigated the effect of variants in these genes on the response to anti-TNFα drugs in patients diagnosed with CD and/or UC. To date, no reliable biomarkers have been identified that can predict the response to IFX or ADA. While some of these biomarkers were demonstrated to be relevant in the prediction of the response to biologic therapy in other inflammatory diseases, the impact on IBD patients remains uncertain [
16]. The absence of such associations may be attributable to a number of factors, including the underrepresentation of a particular genotype, which is influenced by population frequencies or potential study biases, or the limited sample size. The majority of the SNVs documented in this research have been demonstrated to be associated with treatment response in a limited number of studies.
Furthermore, the classification of patients as responders or non-responders is determined by diverse criteria across each study. For instance, the following indices and measurements may be taken into consideration when assessing the activity of CD or UC: the Crohn’s disease activity index (CDAI) calculation [
17,
21,
24,
28,
30], CRP measurement [
17,
18,
20,
21,
38,
39], the IBDQ index [
18,
20,
25], Harvey Bradshaw Index (HBI) activity scores [
19,
23,
27,
33,
36,
37], partial Mayo scores [
19], the simple three-step scale [
22,
26,
29], the colitis activity index (CAI) [
31], and medical criteria [
32,
34].
To the best of our knowledge, there is a lack of research publications concerning VLZ, and our research has revealed only one article related to UST. However, there is a possibility that the biomarkers implicated could be the same as those associated with ADA and IFX, which would be worth investigating. Given the heterogeneity observed in the studies, and in the event that an association is observed between an SNV and treatment efficacy, it is imperative that further research be conducted in order to validate this association. The subsequent phase of our research involves the validation of this array within our study population, a cohort of over 400 patients diagnosed with Crohn’s disease or ulcerative colitis who are undergoing treatment with ADA, IFX, UST, or VLZ. The patients will be categorized as either good responders or non-responders according to the same standard criteria. Therefore, it is essential to continue with large and detailed studies in the field of pharmacogenetics applied to IBD therapy.
5. Study Limitations
The selection of variants for the array design is subject to certain limitations. Firstly, although an extensive review of the literature was conducted, due to the limitations of the array design, it was necessary to reduce the number of variants to 60, selecting those considered most relevant and interesting for future research, according to the established criteria. Secondly, the literature review was largely based on the latest published reviews, so it is possible that information not included in them was missed. It is also possible that clinically significant variants have not yet been investigated. Therefore, larger studies are needed to confirm the relevance of the association found between variants and response to anti-TNF-α treatment in IBD, as well as to discover new SNVs. It should be noted that the present study is focused on anti-TNF drugs, as there are no studies that have investigated the association between SNVs and VLZ response in patients with IBD, and only one study has been conducted on the response to UST and its relationship with a PTPN2 variant.
6. Conclusions
An array of 38 genes and 59 SNVs was designed to validate the biomarkers described in the literature in a future study involving more than 400 patients suffering from IBD who are undergoing biologic drug treatment.