Environmental DNA Detection in Marine Macrophyte Ecosystems as a Potential Blue Carbon Source in Sediments
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Site and Sediment Collection
2.2. eDNA Extraction, PCR Amplification and Sequencing
2.3. Data Analysis
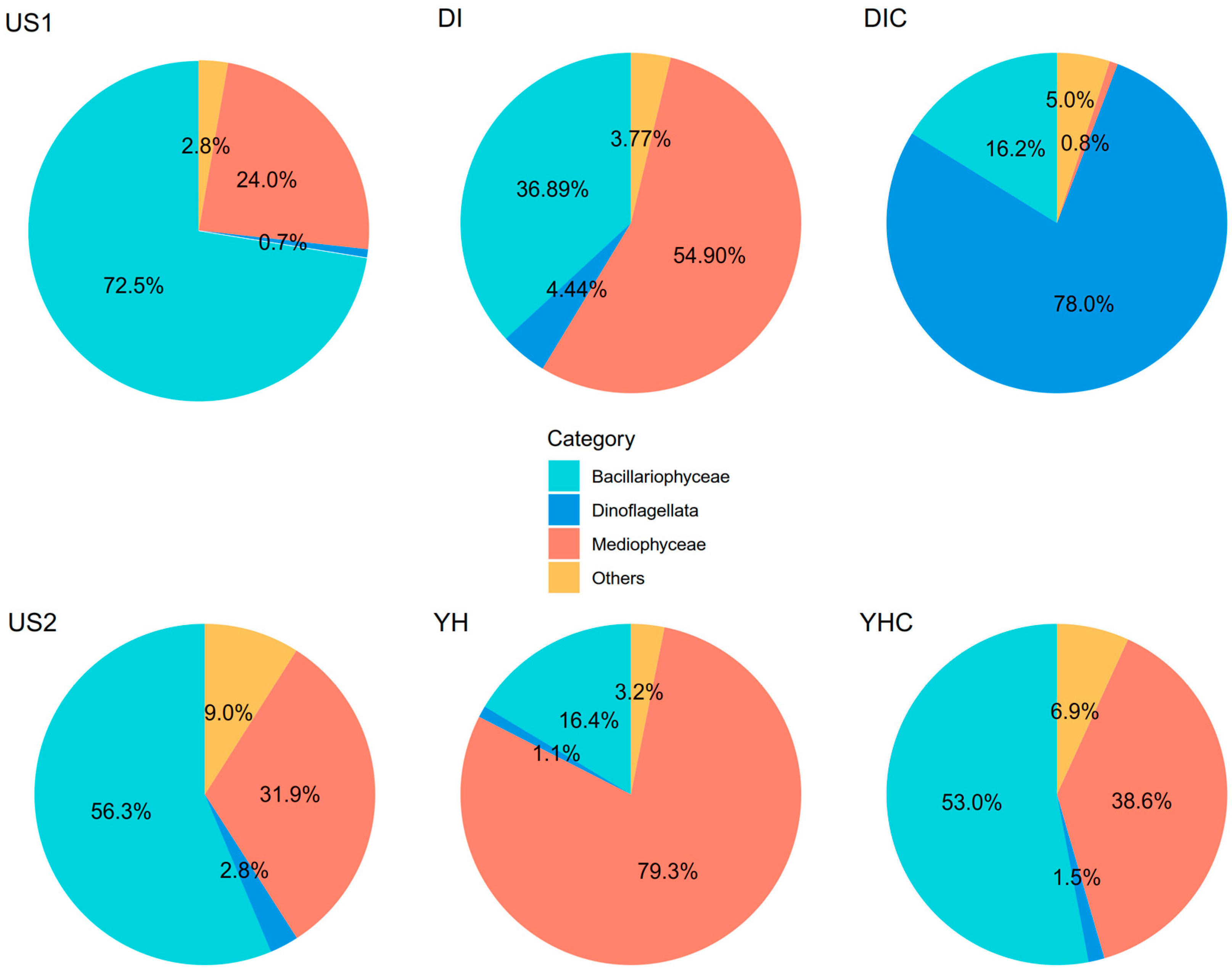
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Legg, S. IPCC, 2021: Climate Change 2021-the physical science basis. Interaction 2021, 49, 44–45. [Google Scholar]
- Grubb, M.; Okereke, C.; Arima, J.; Bosetti, V.; Chen, Y.; Edmonds, J.; Gupta, S.; Köberle, A.; Kverndokk, S.; Malik, A.; et al. Introduction and Framing. In Proceedings of the IPCC, 2022: Climate Change 2022: Mitigation of Climate Change. Contribution of Working Group III to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change, IPCC, Sharm el-Sheikh, Egypt, 6–20 November 2022. [Google Scholar]
- Nellemann, C.; Corcoran, E.; Duarte, S.M.; Valdés, L.; DeYoung, C.; Fonseca, L.; Grimsditch, G. Blue Carbon: The Role of Healthy Oceans in Binding Carbon. A Rapid Response Assessment; UNEP/FAO/UNESCO/IUCN/CSIC; Birkeland Trykkeri AS: Aust-Agde, Norway, 2009.
- Herr, D.; Landis, E. Coastal Blue Carbon Ecosystems. Opportunities for Nationally Determined Contributions; Policy brief. IUCN; UN Environment Program: Nairobi, Kenya, 2016. [Google Scholar]
- Macreadie, P.I.; Costa, M.D.; Atwood, T.B.; Friess, D.A.; Kelleway, J.J.; Kennedy, H.; Lovelock, C.E.; Serrano, O.; Duarte, C.M. Blue carbon as a natural climate solution. Nat. Rev. Earth Environ. 2021, 2, 826–839. [Google Scholar] [CrossRef]
- Sánchez-Arcilla, A.; Cáceres, I.; Le Roux, X.; Hinkel, J.; Schuerch, M.; Nicholls, R.J.; Otero, D.M.; Staneva, J.; de Vries, M.; Pernice, U.; et al. Barriers and enablers for upscaling coastal restoration. Nat. Based Solut. 2022, 2, 100032. [Google Scholar] [CrossRef]
- Friess, D.A.; Shribman, Z.I.; Stankovic, M.; Iram, N.; Baustian, M.M.; Ewers Lewis, C.J. Restoring Blue carbon ecosystems. Camb. Prism. Coast. Futures 2024, 2, e9. [Google Scholar] [CrossRef]
- Taberlet, P.; Bonin, A.; Zinger, L.; Coissac, E. Environmental DNA: For Biodiversity Research and Monitoring; Oxford University Press: Oxford, UK, 2018. [Google Scholar]
- Valentini, A.; Taberlet, P.; Miaud, C.; Civade, R.; Herder, J.; Thomsen, P.F.; Bellemain, E.; Besnard, A.; Coissac, E.; Boyer, F.; et al. Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Mol. Ecol. 2016, 25, 929–942. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Willerslev, E. Environmental DNA—An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Schadewell, Y.; Adams, C.I.M. Forensics meets ecology—Environmental DNA offers new capabilities for marine ecosystem and fisheries research. Front. Mar. Sci. 2021, 8, 668822. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Bonin, A.; Miaud, C. Population genetics reveals origin and number of founders in a biological invasion. Mol. Ecol. 2008, 17, 773–782. [Google Scholar] [CrossRef]
- Ortega, A.; Geraldi, N.R.; Duarte, C.M. Environmental DNA identifies marine macrophyte contributions to blue carbon sediments. Limnol. Oceanogr. 2020, 65, 3139–3149. [Google Scholar] [CrossRef]
- Hamaguchi, M.; Miyajima, T.; Shimabukuro, H.; Hori, M. Development of quantitative real-time PCR for detecting environmental DNA derived from marine macrophytes and its application to a field survey in Hiroshima Bay, Japan. Water 2022, 14, 827. [Google Scholar] [CrossRef]
- Zeng, Y.; Wang, X.; Liu, J.; Cao, J.; Sun, Y.; Zhao, S.; Chen, Z.; Kim, J.K.; Zhang, J.; He, P. Harnessing the power of eDNA technology for macroalgal ecological studies: Recent advances, challenges, and future perspectives. Algal Res. 2023, 77, 103340. [Google Scholar] [CrossRef]
- Amaral-Zettler, L.A.; McCliment, E.A.; Ducklow, H.W.; Huse, S.M. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS ONE 2009, 4, e6372. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2022, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Q.; Chen, B.; Yu, Y.; Wang, T.; Xu, N.; Fan, X.; Penuelas, J.; Fu, Z.; Deng, Y.; et al. Global biogeography of microbes driving ocean ecological status under climate change. Nat. Commun. 2024, 15, 4657. [Google Scholar] [CrossRef]
- Chávez-Sánchez, T.; Piñón-Gimate, A.; Serviere-Zaragoza, E.; Sánchez-González, A.; Hernández-Carmona, G.; Casas-Valdez, M. Recruitment in Ulva blooms in relation to temperature, salinity and nutrients in a subtropical bay of the Gulf of California. Bot. Mar. 2017, 60, 257–270. [Google Scholar] [CrossRef]
- Maggs, C.; Hommersand, M.H. Seaweeds of the British Isles; Volume 1 Rhodophyta, Part 3A Ceramiales; Pelagic Publishing: London, UK, 1993. [Google Scholar]
- Vaudrey, J.M.P.; Kremer, J.N.; Branco, B.F.; Short, F.T. Eelgrass recovery after nutrient enrichment reversal. Aquat. Bot. 2010, 93, 237–243. [Google Scholar] [CrossRef]
- Bae, J.I.; Shin, H.C.; Hwang, S.I.; Lee, J.H. Distribution of sedimentation environments and benthic macro-fauna communities in habitats and non-habitats of Zostera marina on the Yeongheung-do tidal flats, West Coast of Korea. Korean J. Environ. Biol. 2018, 36, 107–116. [Google Scholar] [CrossRef]
- Rozaimi, M.; Zainee, N.F.A.; Raynusha, C.; Arina, N.; Hidayah, N.; Hengjie, T.; Tangang, F. Carbon and Nitrogen Deposits of Macroalgal Origin on a Tropical Seagrass Meadow. Ecosyst. Health Sustain. 2024, 10, 0157. [Google Scholar] [CrossRef]
- van Patten, M.S.; Yarish, C. Seaweeds of Long Island Sound (2nd edition). In Connecticut Sea Grant College Program; NOAA: Silver Spring, MD, USA, 2009. Available online: https://repository.library.noaa.gov/view/noaa/45755 (accessed on 18 January 2024).
- Kanagawa, T. Bias and artifacts in multitemplate poly-merase chain reactions (PCR). J. Biosci. Bioeng. 2003, 96, 317–323. [Google Scholar] [CrossRef]
- Fuller, N.J.; Campbell, C.; Allen, D.J.; Pitt, F.D.; Le Gail, F.; Vaulot, D.; Scanlan, D.J. Analysis of photosynthetic picoeukaryote diversity at open ocean sites in the Arabian Sea using a PCR biased towards marine algal plastids. Aquat. Microb. Ecol. 2006, 43, 79–93. [Google Scholar] [CrossRef]
- Capriulo, G.M.; Smith, G.; Troy, R.; Wikfors, G.; Pellet, J.; Yarish, C. The Planktonic Food Web Structure of a Temperate Zone Estuary, and its Alteration Due to Eutrophication. Hydrobiologia 2002, 475/476, 263–333. [Google Scholar] [CrossRef]
- Lopez, G.; Carey, D.; Carlton, J.; Cerato, R.; Dam Guerrero, H.; Digiovanni, C.; Elphick, C.; Frisk, M.; Gobler, C.; Hice, L.; et al. Biology and Ecology of Long Island Sound, In Long Island Sound: Prospects for the Urban Sea; Latimer, J.S., Tedesco, M., Swanson, R.L., Yarish, C., Stacey, P., Garza, C., Eds.; Springer: New York, NY, USA, 2014; pp. 285–479. [Google Scholar]
- Saunders, G.W. Applying DNA barcoding to red macro-algae: A preliminary appraisal holds promise for future applications. Philos. Trans. R. Soc. 2005, 360, 1879–1888. [Google Scholar] [CrossRef] [PubMed]
- Saunders, G.W.; Kucera, H. An evaluation of rbcL, ufA, UPA, LSU and ITS as DNA barcode markers for the marine green macroalgae. Cryptogam. Algol. 2010, 31, 487. [Google Scholar]
- Krause-Jensen, D.; Lavery, P.; Serrano, O.; Marbà, N.; Masque, P.; Duarte, C.M. Sequestration of macroalgal carbon: The elephant in the Blue Carbon room. Biol. Lett. 2018, 14, 20180236. [Google Scholar] [CrossRef]



| Location | Substrate | Vegetation Type |
|---|---|---|
| Dae Isac Do, Incheon, Republic of Korea (DI) | Muddy | Seagrass bed |
| Dae Isac Do, Incheon, Republic of Korea (DIC) | Muddy | No vegetation |
| New Haven, CT, USA (US1) | Muddy | Ulva blooms |
| Avery Point, CT, USA (US2) | Sandy/Rocky | Highly diverse seaweed community (fucoid dominant)/Seagrass bed |
| Yeong Heung Do, Incheon, Republic of Korea (YH) | Muddy | Seagrass bed |
| Yeong Heung Do, Incheon, Republic of Korea (YHC) | Muddy | No vegetation |
| Lineage | Taxa | DI | DIC | US1 | US2 | YH | YHC |
|---|---|---|---|---|---|---|---|
| Chlorophyta | Blidingia dawsonii | - | - | - | 0.32 | - | - |
| Collinsiella tuberculata | - | - | - | - | - | 0.31 | |
| Ruthnielsenia tenuis | - | - | - | - | - | 0.6 | |
| Ulva sp. | - | - | 2.59 | 0.33 | - | - | |
| Rhodophyta | Ceramium diaphanum | - | - | 0.91 | 0.23 | - | - |
| Ceramium sp. | - | - | - | - | 2.01 | 0.22 | |
| Gracilaria sp. | - | - | 0.49 | - | - | - | |
| Phaeophyta | Cantina marsupialis | - | - | - | - | 0.21 | 0.57 |
| Sargassum sp. | 0.3 | 1.24 | 0.19 | 0.46 | 0.14 | 0.18 | |
| Ectocarpus siliculosus | - | - | 0.36 | 4.25 | - | - | |
| Scytosiphon lomentaria | - | - | - | 0.84 | 58.14 | 19.55 | |
| Antarctosaccion applanatum | 0.11 | - | - | - | - | 0.41 | |
| Seagrass | Zostera marina | - | 0.44 | - | 0.78 | 0.34 | 0.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, Q.; Kim, S.J.; Yarish, C. Environmental DNA Detection in Marine Macrophyte Ecosystems as a Potential Blue Carbon Source in Sediments. Coasts 2024, 4, 687-696. https://doi.org/10.3390/coasts4040036
Xing Q, Kim SJ, Yarish C. Environmental DNA Detection in Marine Macrophyte Ecosystems as a Potential Blue Carbon Source in Sediments. Coasts. 2024; 4(4):687-696. https://doi.org/10.3390/coasts4040036
Chicago/Turabian StyleXing, Qikun, Samuel J. Kim, and Charles Yarish. 2024. "Environmental DNA Detection in Marine Macrophyte Ecosystems as a Potential Blue Carbon Source in Sediments" Coasts 4, no. 4: 687-696. https://doi.org/10.3390/coasts4040036
APA StyleXing, Q., Kim, S. J., & Yarish, C. (2024). Environmental DNA Detection in Marine Macrophyte Ecosystems as a Potential Blue Carbon Source in Sediments. Coasts, 4(4), 687-696. https://doi.org/10.3390/coasts4040036







