Gingipain Genotyping as a Potential Predictor for the Assessment of Periodontal Health and Disease Condition
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Amplification of kgp and rgpA Gene
2.3. Restriction Digestion for kgp and rgpA Genotyping
2.4. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lindhe, J.; Ranney, R.; Lamster, I.; Charles, A.; Chung, C.P.; Flemmig, T.; Kinane, D.; Listgarten, M.; Loe, H.; Schoor, R.; et al. Consensus Report: Chronic Periodontitis. Ann. Periodontol. 1999, 4, 38. [Google Scholar] [CrossRef]
- Bullon, P.; Newman, H.N.; Battino, M. Obesity, diabetes mellitus, atherosclerosis and chronic periodontitis: A shared pathology via oxidative stress and mitochondrial dysfunction. Periodontol. 2000 2014, 64, 139–153. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Darveau, R.P.; Curtis, M.A. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 2012, 10, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G.; Lamont, R.J. Breaking bad: Manipulation of the host response by Porphyromonas gingivalis. Eur. J. Immunol. 2014, 44, 328–338. [Google Scholar] [CrossRef]
- Holt, S.C.; Kesavalu, L.; Walker, S.; Genco, C.A. Virulence factors of Porphyromonas gingivalis. Periodontol. 2000 1999, 20, 168–238. [Google Scholar] [CrossRef]
- Eichinger, A.; Beisel, H.G.; Jacob, U.; Huber, R.; Medrano, F.J.; Banbula, A.; Potempa, J.; Travis, J.; Bode, W. Crystal structure of gingipain R: An Arg-specific bacterial cysteine proteinase with a caspase-like fold. EMBO J. 1999, 18, 5453–5462. [Google Scholar] [CrossRef] [PubMed]
- Allaker, R.P.; Aduse-Opoku, J.; Batten, J.E.; Curtis, M.A. Natural variation within the principal arginine-specific protease gene, prpR1, of Porphyromonas gingivalis. Oral Microbiol. Immunol. 1997, 12, 298–302. [Google Scholar] [CrossRef]
- Beikler, T.; Peters, U.; Ehmke, B.; Flemmig, T.F. Sequence analysis of kgp in Porphyromonas gingivalis isolates from periodontitis patients. Oral Microbiol. Immunol. 2003, 18, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Veith, P.D.; Talbo, G.H.; Slakeski, N.; Dashper, S.G.; Moore, C.; Paolini, R.A.; Reynolds, E.C. Major outer membrane proteins and proteolytic processing of RgpA and Kgp of Porphyromonas gingivalis W50. Biochem. J. 2002, 363, 105–115. [Google Scholar] [CrossRef]
- Olczak, T.; Dixon, D.W.; Genco, C.A. Binding specificity of the Porphyromonas gingivalis heme and hemoglobin receptor HmuR, gingipain K, and gingipain R1 for heme, porphyrins, and metalloporphyrins. J. Bacteriol. 2001, 183, 5599–5608. [Google Scholar] [CrossRef]
- Potempa, J.; Sroka, A.; Imamura, T.; Travis, J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: Structure, function and assembly of multidomain protein complexes. Curr. Protein Pept. Sci. 2003, 4, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Nguyen, K.A.; Potempa, J. Dichotomy of gingipains action as virulence factors: From cleaving substrates with the precision of a surgeon’s knife to a meat chopper-like brutal degradation of proteins. Periodontol. 2000 2010, 54, 15–44. [Google Scholar] [CrossRef]
- DeCarlo, A.A., Jr.; Windsor, L.J.; Bodden, M.K.; Harber, G.J.; Birkedal-Hansen, B.; Birkedal-Hansen, H. Activation and novel processing of matrix metalloproteinases by a thiol-proteinase from the oral anaerobe Porphyromonas gingivalis. J. Dent. Res. 1997, 76, 1260–1270. [Google Scholar] [CrossRef]
- Amano, A.; Nakagawa, I.; Okahashi, N.; Hamada, N. Variations of Porphyromonas gingivalis fimbriae in relation to microbial pathogenesis. J. Periodontal Res. 2004, 39, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Kawai, S.; Nakano, K.; Inaba, H.; Kuboniwa, M.; Nakagawa, I.; Tsuda, K.; Omori, H.; Ooshima, T.; Yoshimori, T.; et al. Virulence of Porphyromonas gingivalis is altered by substitution of fimbria gene with different genotype. Cell. Microbiol. 2007, 9, 753–765. [Google Scholar] [CrossRef]
- Kugaji, M.S.; Muddapur, U.M.; Bhat, K.G.; Joshi, V.M.; Kumbar, V.M.; Peram, M.R. Quantitative evaluation of Porphyromonas gingivalis in Indian subjects with chronic periodontitis by Real-Time Polymerase Chain Reaction. J. Adv. Oral Res. 2019, 10, 137–144. [Google Scholar] [CrossRef]
- Castro, S.A.; Collighan, R.; Lambert, P.A.; Dias, I.H.; Chauhan, P.; Bland, C.E. Porphyromonas gingivalis gingipains cause defective macrophage migration towards apoptotic cells and inhibit phagocytosis of primary apoptotic neutrophils. Cell Death. Dis. 2017, 8, e2644. [Google Scholar] [CrossRef] [PubMed]
- Kugaji, M.; Muddapur, U.; Bhat, K.; Joshi, V.; Manubolu, M.; Pathakoti, K.; Peram, M.R.; Kumbar, V. Variation in the Occurrence of fimA Genotypes of Porphyromonas gingivalis in Periodontal Health and Disease. Int. J. Environ. Res. Public Health 2020, 17, 1826. [Google Scholar] [CrossRef]
- Zhao, L.; Wu, Y.F.; Meng, S.; Yang, H.; OuYang, Y.L.; Zhou, X.D. Prevalence of fimA genotypes of Porphyromonas gingivalis and periodontal health status in Chinese adults. J. Periodontal Res. 2007, 42, 511–517. [Google Scholar] [CrossRef]
- Puig-Silla, M.; Dasí-Fernández, F.; Montiel-Company, J.M.; Almerich-Silla, J.M. Prevalence of fimA genotypes of Porphyromonas gingivalis and other periodontal bacteria in a Spanish population with chronic periodontitis. Med. Oral Patol. Oral Cir. Bucal 2012, 17, e1047–e1053. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, T.; Laine, M.L.; Van Winkelhoff, A.J.; Dahlen, G. Genotype variation and capsular serotypes of Porphyromonas gingivalis from chronic periodontitis and periodontal abscesses. FEMS Microbiol. Lett. 2007, 270, 75–81. [Google Scholar] [CrossRef]
- Abusleme, L.; Blanc, V.; Léon, R.; Gamonal, J.; Silva, N. Genotyping of rgpA and kgp genes coding for Porphyromonas gingivalis gingipains. Rev. Clin. Periodoncia Implantol. Rehabil. Oral 2012, 5, 135–138. [Google Scholar] [CrossRef]
- Cook, G.S.; Costerton, J.W.; Lamont, R.J. Biofilm formation by Porphyromonas gingivalis and Streptococcus gordonii. J. Periodontal Res. 1998, 33, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Ammann, T.W.; Belibasakis, G.N.; Thurnheer, T. Impact of early colonizers on in vitro subgingival biofilm formation. PLoS ONE 2013, 5, e83090. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, T.; Nakayama, K.; Yoshimura, F.; Okamoto, K.; Abe, N.; Yamamoto, K. Arg-gingipain acts as a major processing enzyme for various cell surface proteins in Porphyromonas gingivalis. J. Biol. Chem. 1998, 273, 29072–29076. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Miya, C.; Cueno, M.E.; Suzuki, R.; Maruoka, S.; Gon, Y.; Kaneko, T.; Yonehara, Y.; Imai, K. Porphyromonas gingivalis gingipains potentially affect MUC5AC gene expression and protein levels in respiratory epithelial cells. FEBS Open Bio 2021, 11, 446–455. [Google Scholar] [CrossRef]
- Putri, C.F.; Bachtiar, E.W. Infection of Porphyromonas gingivalis in Alzheimer’s Disease and the Suppression of Immunity. Dent. Hypotheses 2021, 12, 174–178. [Google Scholar]
- Kugaji, M.S.; Kumbar, V.M.; Peram, M.R.; Patil, S.; Bhat, K.G.; Diwan, P.V. Effect of Resveratrol on biofilm formation and virulence factor gene expression of Porphyromonas gingivalis in periodontal disease. APMIS 2019, 127, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Imamura, T. The role of gingipains in the pathogenesis of periodontal disease. J. Periodontol. 2003, 74, 111–118. [Google Scholar] [CrossRef]
- Ciancio, S.G. Clinical experiences with tetracyclines in the treatment of periodontal diseases. Ann. N. Y. Acad. Sci. 1994, 732, 132–139. [Google Scholar] [CrossRef]

| Gingipains | Primer Sequences (5′-3′) | Product Length in Base Pair | Reference |
|---|---|---|---|
| kgp forward | GAACTGACGAACATCATTG | 890 | [8] |
| kgp reverse | GCTGGCATTAGCAACAC | ||
| rgpA forward | AGTGAGCGAAACTTCGGA | 1700 | [7] |
| rgpA reverse | GGTATCACTGGGTATAACCTGT |
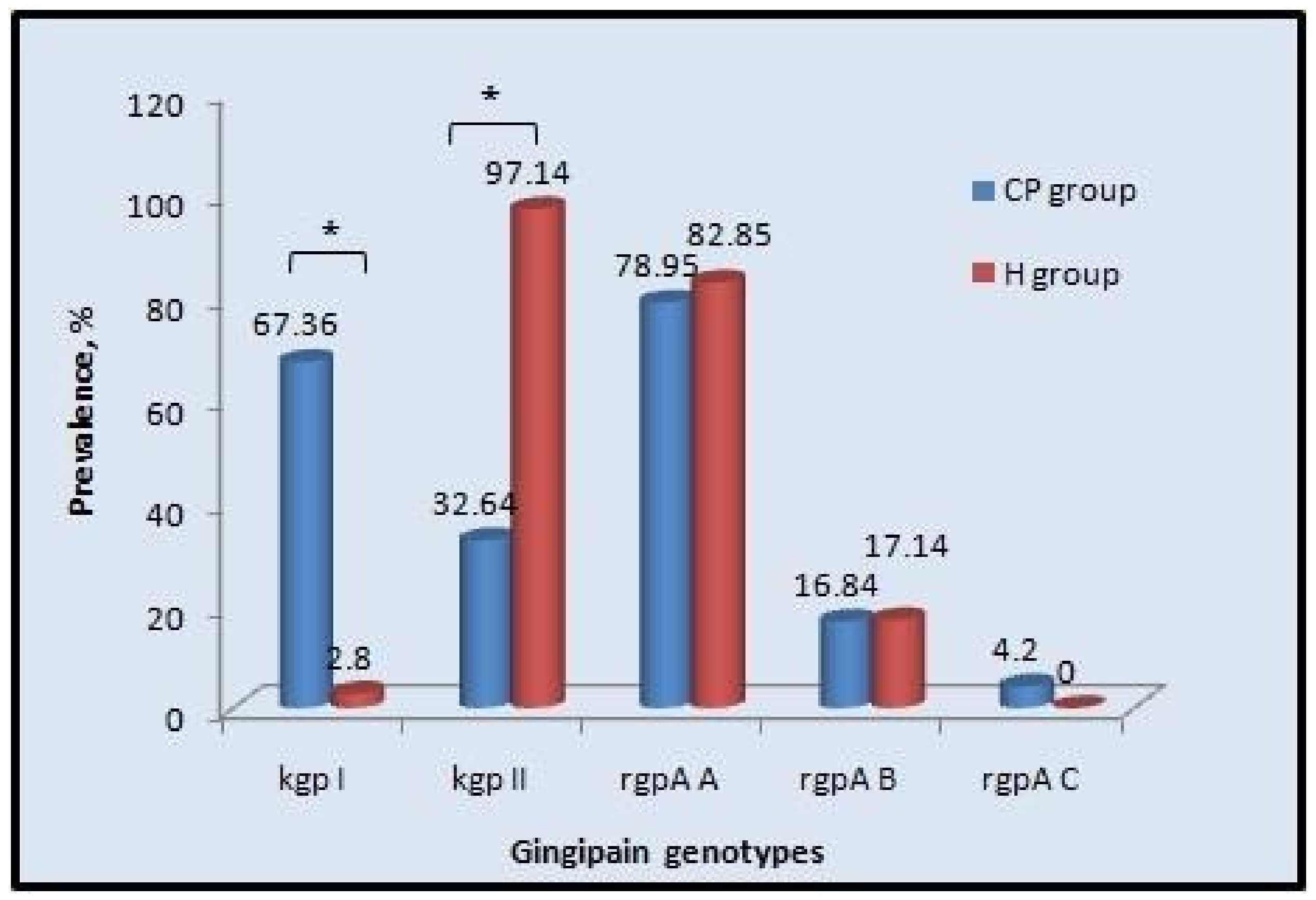
| Gingipain Genotypes | Chronic Periodontitis | Healthy | Odd Ratio | 95% Confidence Interval | p Value | ||
|---|---|---|---|---|---|---|---|
| N | % | N | % | ||||
| kgp-I | 64 | 67.36 | 1 | 2.8 | 65.65 | 19.33 to 222.9 | <0.0001 * |
| kgp-II | 31 | 32.64 | 34 | 97.14 | 0.015 | 0.004 to 0.05 | <0.0001 * |
| rgpA, type A | 75 | 78.95 | 29 | 82.85 | 0.77 | 0.37 to 1.56 | 0.8055 |
| rgpA, type B | 16 | 16.84 | 6 | 17.14 | 1 | 0.47 to 2.09 | 1 |
| Gingipain Genotypes | Present/ Absent | PD | CAL | PI | GI | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mean ± SD | p Value | Mean ± SD | p Value | Mean ± SD | p Value | Mean ± SD | p Value | ||
| kgp-I | Present | 5.88 ± 0.74 | <0.0001 * | 5.64 ± 1.27 | <0.0001 * | 2.51 ± 0.23 | 0.865 | 2.55 ± 0.29 | 0.437 |
| Absent | 5.45 ± 0.38 | 4.71 ± 1.20 | 2.51 ± 0.28 | 2.51 ± 0.35 | |||||
| kgp-II | Present | 5.35 ± 0.52 | <0.0001 * | 4.74 ± 1.49 | 0.004 * | 2.53 ± 0.38 | 0.5267 | 2.50 ± 0.52 | 0.3906 |
| Absent | 5.8 ± 0.56 | 5.37 ± 1.16 | 2.50 ± 0.20 | 2.54 ± 0.24 | |||||
| rgpA, A | Present | 5.70 ± 0.64 | 0.6495 | 5.35 ± 1.21 | 0.04 * | 2.54 ± 0.21 | 0.02 * | 2.56 ± 0.26 | 0.1133 |
| Absent | 5.65 ± 0.67 | 4.96 ± 1.58 | 2.46 ± 0.30 | 2.48 ± 0.40 | |||||
| rgpA, B | Present | 5.85 ± 1.63 | 0.1621 | 5.48 ± 2.60 | 0.2767 | 2.47 ± 0.55 | 0.3816 | 2.45 ± 0.80 | 0.1526 |
| Absent | 5.66 ± 0.48 | 5.16 ± 1.05 | 2.52 ± 0.19 | 2.54 ± 0.23 | |||||
| rgpA, C | Present | 5.40 ± 1.41 | 0.2673 | 4.67 ± 4.80 | 0.3114 | 2.30 ± 0.40 | 0.02 * | 2.35 ± 1.44 | 0.1361 |
| Absent | 5.69 ± 0.48 | 5.22 ± 0.99 | 2.52 ± 0.18 | 2.54 ± 0.22 | |||||
| Gingipain Genotypes | Presence/ Absence | Mean | ±SEM | Median | Interquartile Range (IQR) | p Value |
|---|---|---|---|---|---|---|
| kgp-I | Presence | 2.11 × 108 | 1.02 × 108 | 3.85 × 106 | 2.26 × 107 | <0.0001 * |
| Absence | 7.51 × 107 | 7.19 × 107 | 1.89 × 104 | 5.24 × 106 | ||
| Kgp-II | Presence | 1.36 × 108 | 1.30 × 108 | 4.53 × 106 | 1.01 × 107 | 0.1378 |
| Absence | 1.51 × 108 | 7.41 × 107 | 1.17 × 106 | 1.09 × 107 | ||
| rgpA, type A | Presence | 1.57 × 108 | 7.21 × 107 | 4.07 × 106 | 6.81 × 107 | <0.0001 * |
| Absence | 1.32 × 108 | 1.23 × 108 | 0.0000 | 1.23 × 108 | ||
| rgpA, type B | Presence | 3.68 × 108 | 3.44 × 108 | 4.13 × 106 | 3.40 × 108 | 0.0715 |
| Absence | 1.13 × 108 | 5.24 × 107 | 1.35 × 106 | 5.10 × 107 | ||
| rgpA, type C | Presence | 9.39 × 106 | 2.79 × 106 | 1.19 × 107 | 9.06 × 106 | 0.1378 |
| Absence | 1.52 × 108 | 6.63 × 107 | 1.59 × 106 | 6.47 × 107 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kugaji, M.; Bhat, K.; Muddapur, U.; Joshi, V.; Peram, M.R.; Kumbar, V. Gingipain Genotyping as a Potential Predictor for the Assessment of Periodontal Health and Disease Condition. Hygiene 2022, 2, 178-186. https://doi.org/10.3390/hygiene2040016
Kugaji M, Bhat K, Muddapur U, Joshi V, Peram MR, Kumbar V. Gingipain Genotyping as a Potential Predictor for the Assessment of Periodontal Health and Disease Condition. Hygiene. 2022; 2(4):178-186. https://doi.org/10.3390/hygiene2040016
Chicago/Turabian StyleKugaji, Manohar, Kishore Bhat, Uday Muddapur, Vinayak Joshi, Malleswara Rao Peram, and Vijay Kumbar. 2022. "Gingipain Genotyping as a Potential Predictor for the Assessment of Periodontal Health and Disease Condition" Hygiene 2, no. 4: 178-186. https://doi.org/10.3390/hygiene2040016
APA StyleKugaji, M., Bhat, K., Muddapur, U., Joshi, V., Peram, M. R., & Kumbar, V. (2022). Gingipain Genotyping as a Potential Predictor for the Assessment of Periodontal Health and Disease Condition. Hygiene, 2(4), 178-186. https://doi.org/10.3390/hygiene2040016







