Small-Scale Comparative Genomic Analysis of Listeria monocytogenes Isolated from Environments of Salmon Processing Plants and Human Cases in Norway
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates
2.2. Library Preparation and Sequencing
2.3. Core Genome Analysis
2.4. Identification of Virulence Genes
2.5. Selection of Reference Genomes
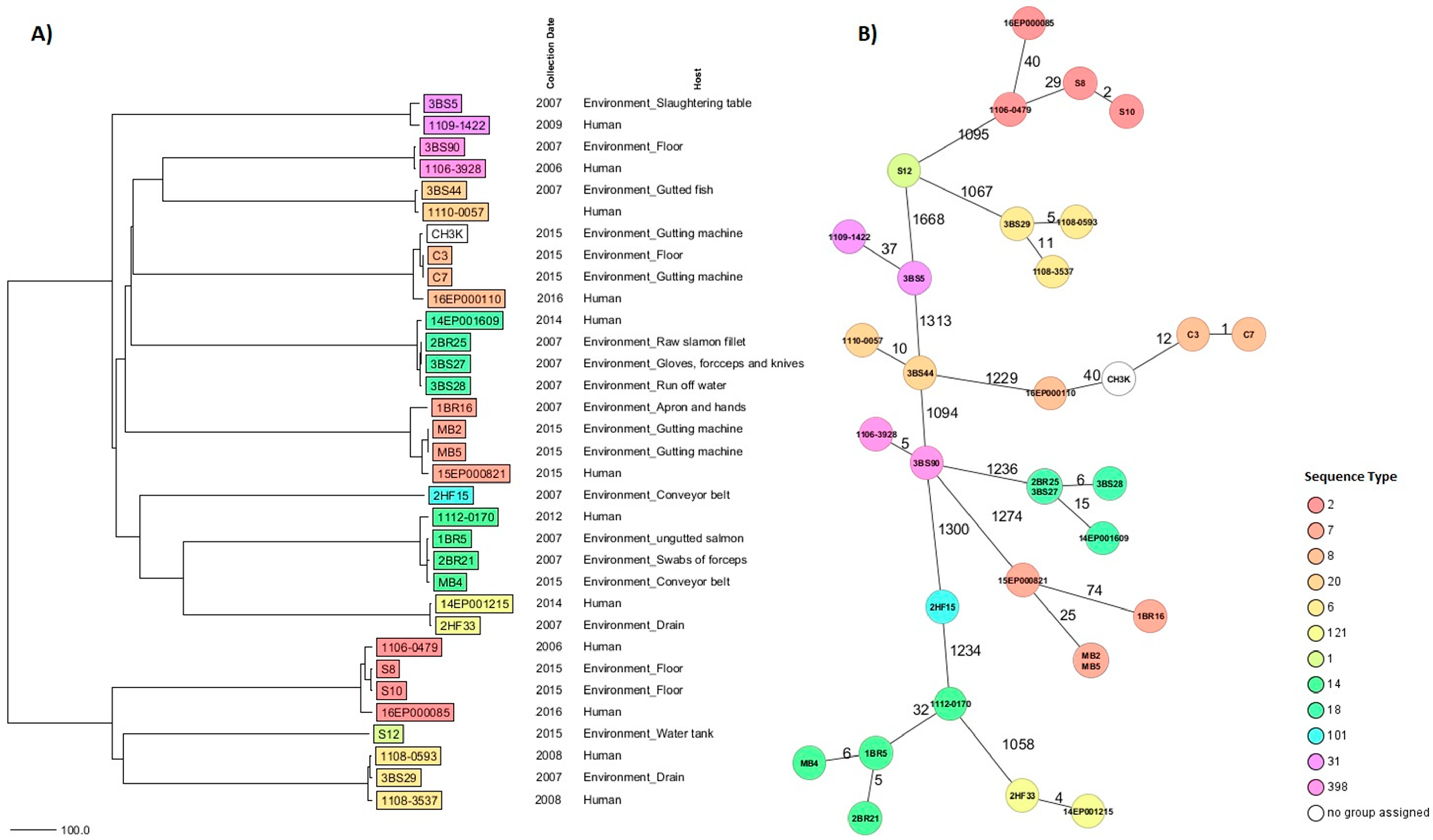
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- ANSES. Data Sheet on Foodborne Biological Hazards—Listeria Monocytogenes; ANSES—French Agency for Food, Environmental and Occupational Health & Safety: Maisons-Alfort, France, 2011; Available online: https://www.anses.fr/en/system/files/MIC2011sa0171FiEN.pdf (accessed on 4 March 2021).
- De Noordhout, C.M.; Devleesschauwer, B.; Angulo, F.J.; Verbeke, G.; Haagsma, J.; Kirk, M.; Havelaar, A.; Speybroeck, N. The global burden of listeriosis: A systematic review and meta-analysis. Lancet Infect. Dis. 2014, 14, 1073–1108. [Google Scholar] [CrossRef] [Green Version]
- Gillespie, I.A.; Mook, P.; Little, C.L.; Grant, K.; Adak, G.K. Listeria monocytogenes infection in the over-60 s in England between 2005 and 2008: A retrospective case-control study utilizing market research panel data. Foodborne Pathog. Dis. 2010, 7, 1373–1379. [Google Scholar] [CrossRef] [PubMed]
- Kvistholm Jensen, A.; Ethelberg, S.; Smith, B.; Moller Nielsen, E.; Larsson, J.; Molbak, K.; Christensen, J.J.; Kemp, M. Substantial increase in listeriosis, Denmark 2009. EuroSurveill. Bull. Eur. Sur Les Mal. Transm. Eur. Commun. Dis. Bull. 2010, 15, 19522. [Google Scholar]
- Siegman-Igra, Y.; Levin, R.; Weinberger, M.; Golan, Y.; Schwartz, D.; Samra, Z.; Konigsberger, H.; Yinnon, A.; Rahav, G.; Keller, N.; et al. Listeria monocytogenes infection in Israel and review of cases worldwide. Emerg. Infect. Dis. 2002, 8, 305–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goulet, V.; Hedberg, C.; Le Monnier, A.; de Valk, H. Increasing incidence of listeriosis in France and other European countries. Emerg. Infect. Dis. 2008, 14, 734–740. [Google Scholar] [CrossRef] [PubMed]
- EFSA and ECDC. The European Union One Health 2019 zoonoses report. EFSA J. 2021, 19. [Google Scholar] [CrossRef]
- Mammina, C.; Parisi, A.; Guaita, A.; Aleo, A.; Bonura, C.; Nastasi, A.; Pontello, M. Enhanced surveillance of invasive listeriosis in the Lombardy region, Italy, in the years 2006–2010 reveals major clones and an increase in serotype 1/2a. BMC Infect. Dis. 2013, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desai, R.W.; Smith, M.A. Pregnancy-related Listeriosis. Birth Defects Res. 2017, 109, 324–335. [Google Scholar] [CrossRef] [Green Version]
- RASFF. The Rapid Alert Systems for Food and Feed. In Annual Report 2014; European Commission—Health and Food Safety: Brussels, Belgium, 2015. [Google Scholar]
- RASFF. The Rapid Alert Systems for Food and Feed. In Annual Report 2019; European Commission—Healt and Food Safety: Luxembourg, 2020. [Google Scholar]
- EFSA. Analysis of the baseline survey on the prevalence of Listeria monocytogenes in certain ready-to-eat foods in the EU, 2010-2011; Part A: Listeria monocytogenes prevalence estimates. EFSA J. 2013, 11. [Google Scholar] [CrossRef]
- Nielsen, E.M.; Björkman, J.T.; Kiil, K.; Grant, K.; Dallman, T.; Painset, A.; Amar, C.; Roussel, S.; Guillier, L.; Felix, B.; et al. Closing Gaps for Performing a Risk Assessment on Listeria Monocytogenes in Ready-to-Eat (RTE) Foods: Activity 3, the Comparison of Isolates from Different Compartments Along the Food Chain, and from Humans Using Whole Genome Sequencing (WGS) Analysis. EFSA Supporting Publ. 2017, 170. [Google Scholar] [CrossRef] [Green Version]
- Lunestad, B.T.; Truong, T.T.T.; Lindstedt, B.A. A multiple-locus variable-number tandem repeat analysis (MLVA) of Listeria monocytogenes isolated from Norwegian salmon-processing factories and from listeriosis patients. Epidemiol. Infect. 2013, 141, 2101–2110. [Google Scholar] [CrossRef] [PubMed]
- Rørvik, L.M.; Skjerve, E.; Knudsen, B.R.; Yndestad, M. Risk factors for contamination of smoked salmon with Listeria monocytogenes during processing. Int. J. Food Microbiol. 1997, 37, 215–219. [Google Scholar] [CrossRef]
- Løvdal, T.; Giske, L.A.L.; Bjørlykhaug, E.; Eri, I.B.; Mork, O.J. Hygienic standards and practices in Norwegian salmon processing plants. J. Hyg. Eng. Des. 2017, 20, 3–11. [Google Scholar]
- Rotariu, O.; Thomas, D.J.I.; Goodburn, K.E.; Hutchison, M.L.; Strachan, N.J.C. Smoked salmon industry practices and their association with Listeria monocytogenes. Food Control 2014, 35, 284–292. [Google Scholar] [CrossRef]
- Djordjevic, V.; Trbovic, D.; Lakicevic, B.; Nastasijevic, I.; Jankovic, V.; Baltic, T.; Dimitrijevic, M. Microbiological safety and quality of salmon: Health benefits and risk. Meat Technol. 2016, 57, 120–125. [Google Scholar]
- Dass, S.C.; Abu-Ghannam, N.; Antony-Babu, S.; Cummins, E.J. Ecology and molecular typing of L. monocytogenes in a processing plant for cold-smoked salmon in the Republic of Ireland. Food Res. Int. 2010, 43, 1529–1536. [Google Scholar] [CrossRef]
- Rørvik, L.M. Listeria monocytogenes in the smoked salmon industry. Int. J. Food Microbiol. 2000, 62, 183–190. [Google Scholar] [CrossRef]
- Løvdal, T. The microbiology of cold smoked salmon. Food Control 2015, 54, 360–373. [Google Scholar] [CrossRef]
- Ben Embarek, P.K. Presence, detection and growth of Listeria monocytogenes in seafoods—A review. Int. J. Food Microbiol. 1994, 23, 17–34. [Google Scholar] [CrossRef]
- Hofshagen, M.; Nygård, K.; Kruse, H. (Eds.) Listeriose; Norsk Zoonosesenter: Oslo, Norway, 2004. [Google Scholar]
- Schjørring, S.; Lassen, S.G.; Jensen, T.; Moura, A.; Kjeldgaard, J.S.; Mueller, L.; Thielke, S.; Leclercq, A.; Maury, M.M.; Tourdjman, M.; et al. Cross-border outbreak of listeriosis caused by cold-smoked salmon, revealed by integrated surveillance and whole genome sequencing (WGS), Denmark and France, 2015 to 2017. Eurosurveillance 2017, 22, 8–12. [Google Scholar] [CrossRef]
- Gillesberg Lassen, S.; Ethelberg, S.; Björkman, J.T.; Jensen, T.; Sørensen, G.; Kvistholm Jensen, A.; Müller, L.; Nielsen, E.M.; Mølbak, K. Two listeria outbreaks caused by smoked fish consumption—Using whole-genome sequencing for outbreak investigations. Clin. Microbiol. Infect. 2016, 22, 620–624. [Google Scholar] [CrossRef] [Green Version]
- EFSA; ECDC. Multi-Country Outbreak of Listeria Monocytogenes Clonal Complex 8 Infections Linked to Consumption of Cold-Smoked Fish Products; EFSA (European Food Safety Authority) and ECDC (European Centre for Disesase Prevention and Control): Stockholm, Sweden; Parma, Italy, 2019; p. 18. [Google Scholar]
- EFSA; ECDC. Multi-Country Outbreak of Listeria Monocytogenes Sequence Type 8 Infections Linked to Consumption of Salmon Products; EFSA (European Food Safety Authority) and ECDC (European Centre for Disesase Prevention and Control): Stockholm, Sweden; Parma, Italy, 2018; p. 16. [Google Scholar]
- Skjerdal, T.; Eckner, K.; Kapperud, G.; Lassen, J.; Grahek-Ogden, D.; Narvhus, J.; Nesbakken, T.; Robertson, L.; Rosnes, J.T.; Skjerve, E.; et al. Listeria Monocytogenes—Vurdering av Helseråd til Gravide og Andre Utsatte Grupper; Uttalelse fra Faggruppe for Hygiene og Smittestoffer i Vitenskapskomiteen for Mat og Miljø: Oslo, Norway, 2018. [Google Scholar]
- Jackson, B.R.; Tarr, C.; Strain, E.; Jackson, K.A.; Conrad, A.; Carleton, H.; Katz, L.S.; Stroika, S.; Gould, L.H.; Mody, R.K.; et al. Implementation of nationwide real-time whole-genome sequencing to enhance listeriosis outbreak detection and investigation. Clin. Infect. Dis. 2016, 63, 380–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chenal-Francisque, V.; Diancourt, L.; Cantinelli, T.; Passet, V.; Tran-Hykes, C.; Bracq-Dieye, H.; Leclercq, A.; Pourcel, C.; Lecuit, M.; Brisse, S. Optimized Multilocus Variable-Number Tandem-Repeat Analysis Assay and Its Complementarity with Pulsed-Field Gel Electrophoresis and Multilocus Sequence Typing for Listeria monocytogenes Clone Identification and Surveillance. J. Clin. Microbiol. 2013, 51, 1868–1880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmid, D.; Allerberger, F.; Huhulescu, S.; Pietzka, A.; Amar, C.; Kleta, S.; Prager, R.; Preussel, K.; Aichinger, E.; Mellmann, A. Whole genome sequencing as a tool to investigate a cluster of seven cases of listeriosis in Austria and Germany, 2011–2013. Clin. Microbiol. Infect. 2014, 20, 431–436. [Google Scholar] [CrossRef] [Green Version]
- Lindqvist, R.; Westöö, A. Quantitative risk assessment for Listeria monocytogenes in smoked or gravad salmon and rainbow trout in Sweden. Int. J. Food Microbiol. 2000, 58, 181–196. [Google Scholar] [CrossRef]
- Moura, A.; Criscuolo, A.; Pouseele, H.; Maury, M.M.; Leclercq, A.; Tarr, C.; Björkman, J.T.; Dallman, T.; Reimer, A.; Enouf, V.; et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2016, 2, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lindstedt, B.A.; Tham, W.; Danielsson-Tham, M.L.; Vardund, T.; Helmersson, S.; Kapperud, G. Multiple-locus variable-number tandem-repeats analysis of Listeria monocytogenes using multicolour capillary electrophoresis and comparison with pulsed-field gel electrophoresis typing. J. Microbiol. Methods 2008, 72, 141–148. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Bushnell, B. BBMap Short Read Aligner. Available online: https://sourceforge.net/projects/bbmap/ (accessed on 13 October 2020).
- Ruppitsch, W.; Pietzka, A.; Prior, K.; Bletz, S.; Fernandez, H.L.; Allerberger, F.; Harmsen, D.; Mellmann, A. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J. Clin. Microbiol. 2015, 53, 2869–2876. [Google Scholar] [CrossRef] [Green Version]
- Hyden, P.; Pietzka, A.; Lennkh, A.; Murer, A.; Springer, B.; Blaschitz, M.; Indra, A.; Huhulescu, S.; Allerberger, F.; Ruppitsch, W.; et al. Whole genome sequence-based serogrouping of Listeria monocytogenes isolates. J. Biotechnol. 2016, 235, 181–186. [Google Scholar] [CrossRef] [Green Version]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [Green Version]
- Montero, D.; Bodero, M.; Riveros, G.; Lapierre, L.; Gaggero, A.; Vidal, R.M.; Vidal, M. Molecular epidemiology and genetic diversity of Listeria monocytogenes isolates from a wide variety of ready-to-eat foods and their relationship to clinical strains from listeriosis outbreaks in Chile. Front. Microbiol. 2015, 6, 8. [Google Scholar] [CrossRef] [Green Version]
- Lambertz, S.T.; Ivarsson, S.; Lopez-Valladares, G.; Sidstedt, M.; Lindqvist, R. Subtyping of Listeria monocytogenes isolates recovered from retail ready-to-eat foods, processing plants and listeriosis patients in Sweden 2010. Int. J. Food Microbiol. 2013, 166, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Martinez, I.; Rørvik, L.M.; Brox, V.; Lassen, J.; Seppola, M.; Gram, L.; Fonnesbech-Vogel, B. Genetic variability among isolates of Listeria monocytogenes from food products, clinical samples and processing environments, estimated by RAPD typing. Int. J. Food Microbiol. 2003, 84, 285–297. [Google Scholar] [CrossRef]
- Rocourt, J.; Jacquet, C.; Reilly, A. Epidemiology of human listeriosis and seafoods. Int. J. Food Microbiol. 2000, 62, 197–209. [Google Scholar] [CrossRef]
- Wulff, G.; Gram, L.; Ahrens, P.; Vogel, B.F. One group of genetically similar Listeria monocytogenes strains frequently dominates and persists in several fish slaughter- and smokehouses. Appl. Environ. Microbiol. 2006, 72, 4313–4322. [Google Scholar] [CrossRef] [Green Version]
- Holch, A.; Webb, K.; Lukjancenko, O.; Ussery, D.; Rosenthal, B.M.; Gram, L. Genome sequencing identifies two nearly unchanged strains of persistent Listeria monocytogenes isolated at two different fish processing plants sampled 6 years apart. Appl. Environ. Microbiol. 2013, 79, 2944–2951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyoui, D.; Takahashi, H.; Miya, S.; Kuda, T.; Igimi, S.; Kimura, B. Genetic distance in the whole-genome perspective on Listeria monocytogenes strains F2-382 and NIHS-28 that show similar subtyping results. BMC Microbiol. 2014, 14, 10. [Google Scholar] [CrossRef] [Green Version]
- Hoffman, A.D.; Gall, K.L.; Norton, D.M.; Wiedmann, M. Listeria monocytogenes contamination patterns for the smoked fish processing environment and for raw fish. J. Food Prot. 2003, 66, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Rørvik, L.M.; Aase, B.; Alvestad, T.; Caugant, D.A. Molecular epidemiological survey of Listeria monocytogenes in seafoods and seafood-processing plants. Appl. Environ. Microbiol. 2000, 66, 4779–4784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Autio, T.; Hielm, S.; Miettinen, M.; Sjoberg, A.M.; Aarnisalo, K.; Bjorkroth, J.; Mattila-Sandholm, T.; Korkeala, H. Sources of Listeria monocytogenes contamination in a cold-smoked rainbow trout processing plant detected by pulsed-field gel electrophoresis typing. Appl. Environ. Microbiol. 1999, 65, 150–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rørvik, L.M.; Caugant, D.A.; Yndestad, M. Contamination pattern of Listeria monocytogenes and other Listeria spp in a salmon slaughterhouse and smoked salmon processing plant. Int. J. Food Microbiol. 1995, 25, 19–27. [Google Scholar] [CrossRef]
- Porsby, C.H.; Vogel, B.F.; Mohr, M.; Gram, L. Influence of processing steps in cold-smoked salmon production on survival and growth of persistent and presumed non-persistent Listeria monocytogenes. Int. J. Food Microbiol. 2008, 122, 287–295. [Google Scholar] [CrossRef]
- Sikorski, Z.E.; Kolodziejska, I. Microbial risks in mild hot smoking of fish. Crit. Rev. Food Sci. Nutr. 2002, 42, 35–51. [Google Scholar] [CrossRef]
- Vogel, B.F.; Huss, H.H.; Ojeniyi, B.; Ahrens, P.; Gram, L. Elucidation of Listeria monocytogenes contamination routes in cold-smoked salmon processing plants detected by DNA-based typing methods. Appl. Environ. Microbiol. 2001, 67, 2586–2595. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, V.; Wiedmann, M.; Teixeira, P.; Stasiewicz, M.J. Listeria monocytogenes persistence in food-associated environments: Epidemiology, strain characteristics, and implications for public health. J. Food Prot. 2014, 77, 150–170. [Google Scholar] [CrossRef]
- Nakari, U.-M.; Rantala, L.; Pihlajasaari, A.; Toikkanen, S.; Johansson, T.; Hellsten, C.; Raulo, S.M.; Kuusi, M.; Siitonen, A.; Rimhanen-Finne, R. Investigation of increased listeriosis revealed two fishery production plants with persistent Listeria contamination in Finland 2010. Epidemiol. Infect. 2014, 142, 2261–2269. [Google Scholar] [CrossRef] [PubMed]
- Palma, F.; Pasquali, F.; Lucchi, A.; De Cesare, A.; Manfreda, G. Whole genome sequencing for typing and characterisation of Listeria monocytogenes isolated in a rabbit meat processing plant. Ital. J. Food Saf. 2017, 6, 125–130. [Google Scholar] [CrossRef] [Green Version]
- Chenal-Francisque, V.; Lopez, J.; Cantinelli, T.; Caro, V.; Tran, C.; Leclercq, A.; Lecuit, M.; Brisse, S. Worldwide distribution of major clones of Listeria monocytogenes. Emerg. Infect. Dis. 2011, 17, 1110–1112. [Google Scholar] [CrossRef] [PubMed]
- Painset, A.; Bjorkman, J.T.; Kiil, K.; Guillier, L.; Mariet, J.F.; Felix, B.; Amar, C.; Rotariu, O.; Roussel, S.; Perez-Reche, F.; et al. LiSEQ—Whole-genome sequencing of a cross-sectional survey of Listeria monocytogenes in ready-to-eat foods and human clinical cases in Europe. Microb. Genom. 2019, 5, 11. [Google Scholar] [CrossRef]
- Maury, M.M.; Tsai, Y.H.; Charlier, C.; Touchon, M.; Chenal-Francisque, V.; Leclercq, A.; Criscuolo, A.; Gaultier, C.; Roussel, S.; Brisabois, A.; et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 2016, 48, 308–313. [Google Scholar] [CrossRef] [Green Version]
- Kuenne, C.; Billion, A.; Abu Mraheil, M.; Strittmatter, A.; Daniel, R.; Goesmann, A.; Barbuddhe, S.; Hain, T.; Chakraborty, T. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genom. 2013, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pightling, A.W.; Pettengill, J.B.; Luo, Y.; Baugher, J.D.; Rand, H.; Strain, E. Interpreting Whole-Genome Sequence Analyses of Foodborne Bacteria for Regulatory Applications and Outbreak Investigations. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Pettengill, J.B.; Pightling, A.W.; Timme, R.; Allard, M.; Strain, E.; Rand, H. Genetic Diversity of Salmonella and Listeria Isolates from Food Facilities. J. Food Prot. 2018, 81, 2082–2089. [Google Scholar] [CrossRef] [PubMed]
- Rychli, K.; Mueller, A.; Zaiser, A.; Schoder, D.; Allerberger, F.; Wagner, M.; Schmitz-Esser, S. Genome sequencing of Listeria monocytogenes “Quargel” listeriosis outbreak strains reveals two different strains with distinct in vitro virulence potential. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [Green Version]
| Key | Source of Isolation | Year of Isolation | MLVA | MLST ST | CC | cgMLST CT | Predicted PCR Serogroup * | Predicted Lineage |
|---|---|---|---|---|---|---|---|---|
| S12 | Environmental—Water tank | 2015 | 6-0-14-6-9 | 1 | 1 | 3063 | IVb (2) | I |
| S10 | Environmental—Floor | 2015 | 7-7-19-6-10 | 2 | 2 | 4059 | IVb (3) | |
| 1106-0479 | Clinical—Unknown | 2006 | 4065 | |||||
| S8 | Environmental—Floor | 2015 | 7-7-22-6-10 | 4059 | ||||
| 16EP000085 | Clinical—Blood | 2016 | 1127 | |||||
| 3BS29 | Environmental—Drain | 2007 | 9-4-18-6-9 | 6 | 6 | 4064 | IVb (13) | |
| 1108-0593 | Clinical—Unknown | 2008 | ||||||
| 1108-3537 | Clinical—Blood | 2008 | 4039 | |||||
| MB5 | Environmental—Gutting machine | 2015 | 7-7-10-10-6 | 7 | 7 | 4060 | IIa (4) | II |
| MB2 | Environmental—Gutting machine | 2015 | ||||||
| 15EP000821 | Clinical—Blood | 2015 | 665 | |||||
| 1BR16 | Environmental—Workers apron and hands | 2007 | 2657 | |||||
| C7 | Environmental—Gutting machine | 2015 | 6-9-26-18-6 | 8 | 8 | 4057 | IIa (2) | |
| C3 | Environmental—Floor | 2015 | ||||||
| 16EP000110 | Clinical—Blood | 2016 | 4042 | |||||
| CH3K- | Environmental—Gutting machine | 2015 | unassigned | 4058 | ||||
| 2BR21 | Environmental—Swab of forceps | 2007 | 5-8-15-10-6 | 14 | 14 | 112 | ||
| MB4 | Environmental—Conveyor belt | 2015 | ||||||
| 1BR5 | Environmental—Ungutted salmon | 2007 | 5-8-14-10-6 | |||||
| 1112-0170 | Clinical—CSF | 2012 | 847 | |||||
| 3BS27 | Environmental—Gloves, forceps and knives | 2007 | 8-8-17-19-6 | 18 | 18 | 2806 | ||
| 2BR25 | Environmental—Raw salmon fillet | 2007 | ||||||
| 14EP001609 | Clinical—Blood | 2014 | 3562 | |||||
| 3BS28 | Environmental—Run-off water | 2007 | 6-10-5-16-6 | 18 | 18 | 2806 | ||
| 3BS44 | Environmental—Gutted salmon | 2007 | 20 | 20 | 4061 | IIa (10) | ||
| 1110-0057 | Clinical—Unknown | 2010 | ||||||
| 3BS5 | Environmental—Slaughtering table | 2007 | 6-8-14-18-6 | 31 | 31 | 4063 | ||
| 1109-1422 | Clinical—Unknown | 2009 | 4062 | |||||
| 2HF15 | Environmental—Conveyor belt | 2007 | 6-9-4-10-6 | 101 | 101 | 3358 | IIa (14) | |
| 2HF33 | Environmental—Drain | 2007 | 6-7-14-10-6 | 121 | 121 | 2278 | IIa (5) | |
| 14EP001215 | Clinical—CSF | 2014 | ||||||
| 3BS90 | Environmental—Floor | 2007 | 6-10-17-21-6 | 398 | 398 | 3510 | IIa (1) | |
| 1106-3928 | Clinical—Unknown | 2006 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Løvdal, T.; Brandal, L.T.; Sundaram, A.Y.M.; Naseer, U.; Roth, B.; Lunestad, B.T. Small-Scale Comparative Genomic Analysis of Listeria monocytogenes Isolated from Environments of Salmon Processing Plants and Human Cases in Norway. Hygiene 2021, 1, 43-55. https://doi.org/10.3390/hygiene1010005
Løvdal T, Brandal LT, Sundaram AYM, Naseer U, Roth B, Lunestad BT. Small-Scale Comparative Genomic Analysis of Listeria monocytogenes Isolated from Environments of Salmon Processing Plants and Human Cases in Norway. Hygiene. 2021; 1(1):43-55. https://doi.org/10.3390/hygiene1010005
Chicago/Turabian StyleLøvdal, Trond, Lin T. Brandal, Arvind Y. M. Sundaram, Umaer Naseer, Bjørn Roth, and Bjørn Tore Lunestad. 2021. "Small-Scale Comparative Genomic Analysis of Listeria monocytogenes Isolated from Environments of Salmon Processing Plants and Human Cases in Norway" Hygiene 1, no. 1: 43-55. https://doi.org/10.3390/hygiene1010005
APA StyleLøvdal, T., Brandal, L. T., Sundaram, A. Y. M., Naseer, U., Roth, B., & Lunestad, B. T. (2021). Small-Scale Comparative Genomic Analysis of Listeria monocytogenes Isolated from Environments of Salmon Processing Plants and Human Cases in Norway. Hygiene, 1(1), 43-55. https://doi.org/10.3390/hygiene1010005






