Comprehensive Polygenic Score Profiling Reveals Autism Spectrum Disorder Subgroups with Different Genetic Predisposition Related to High-Density Lipoprotein Cholesterol, Urea, and Body Mass Index
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
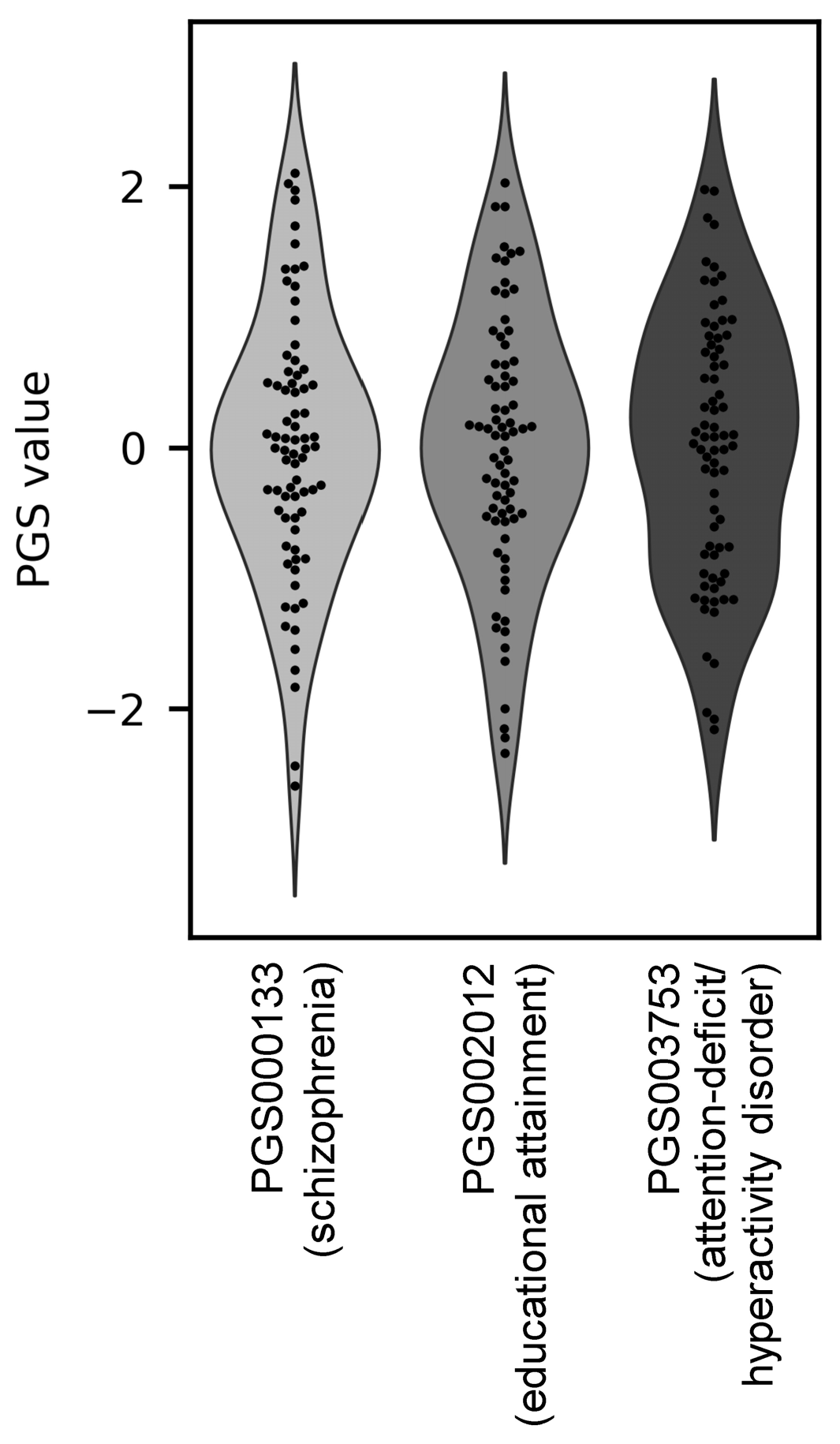
2.2. Polygenic Scores (PGS)
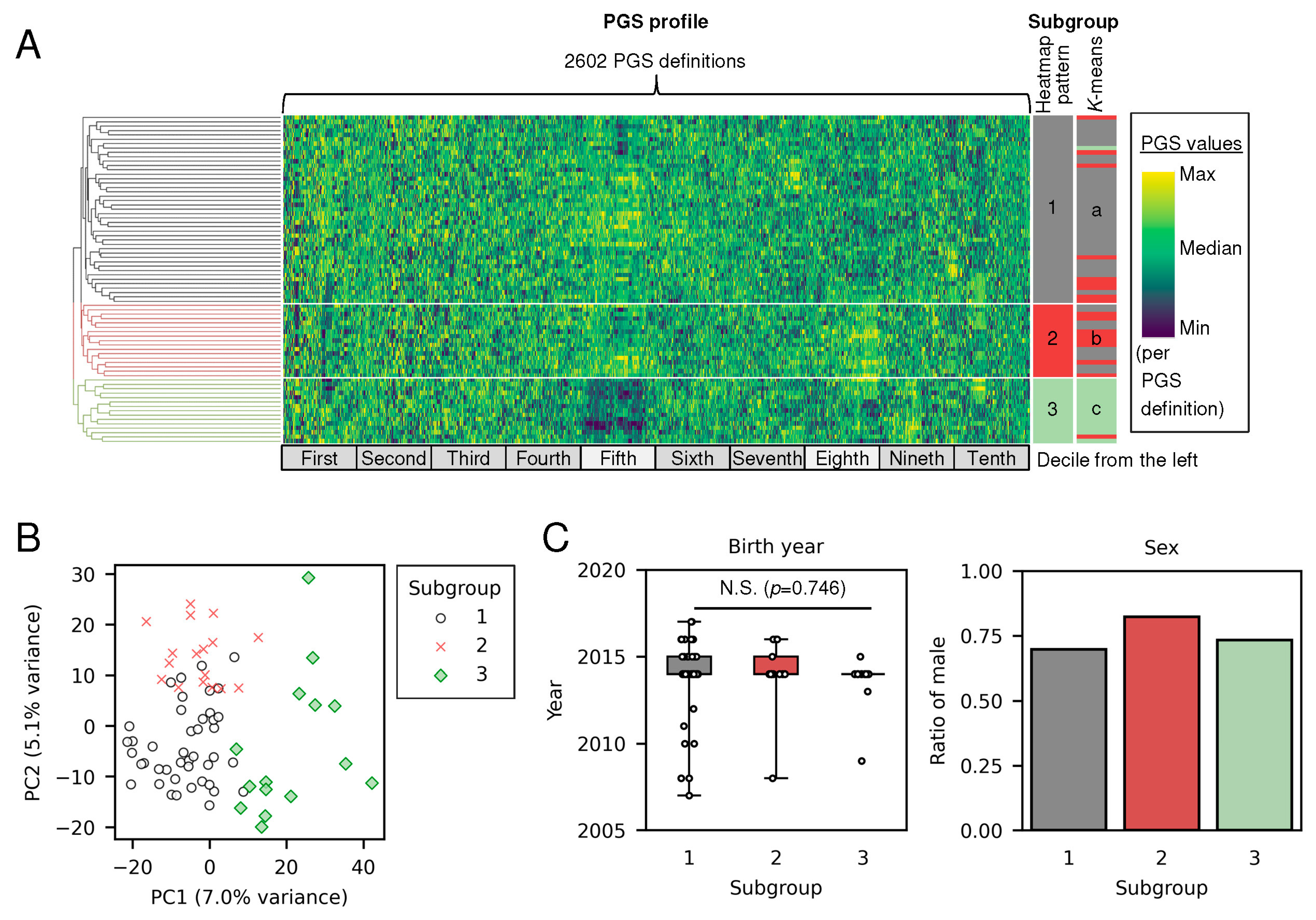
2.3. Stratification of Individuals with Autism Spectrum Disorder (ASD) Using PGS Profiles
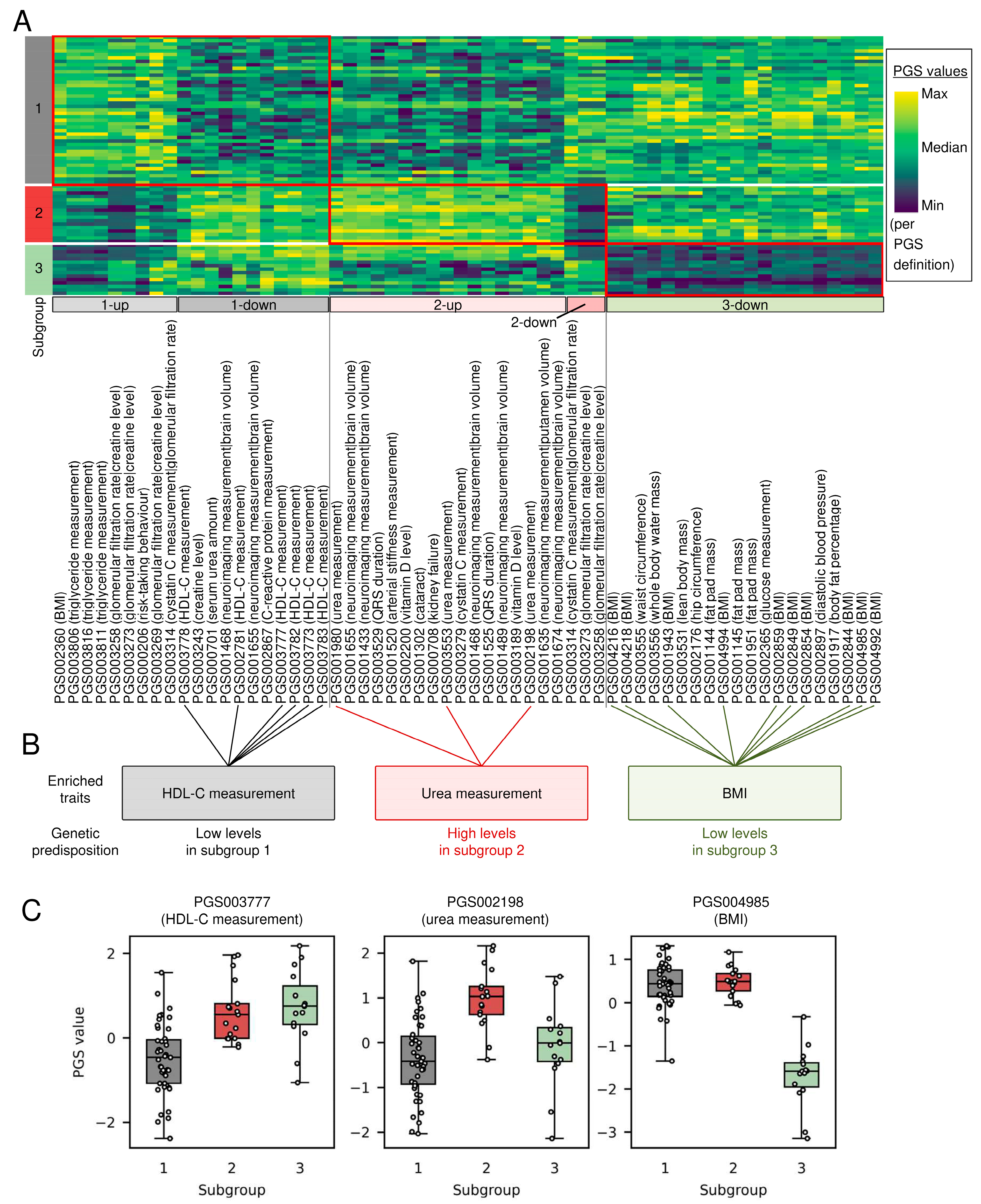
2.4. Enrichment Analysis for Distinctive PGSs in Each Subgroup
3. Results
3.1. Polygenic Score Profiles Revealed Three Subgroups of Individuals with ASD
3.2. Distinctive PGSs in Each Subgroup Enriched High-Density Lipoprotein Cholesterol (HDL-C) Measurements, Urea Measurement, and Body Mass Index (BMI)
4. Discussion
4.1. Polygenic Scores Are a Potential Biomarker to Stratify Individuals with ASD into Subgroups of Different Genetic Predispositions
4.2. ASD Subgroups with Different Genetic Predisposition Toward Obesity May Warrant Investigation of Obesity-Targeted Interventions
4.3. ASD Subgroup with Different Genetic Predisposition Toward Dyslipidemia May Warrant Investigation of Lipid Metabolism-Targeted Interventions
4.4. ASD Subgroup with Different Genetic Predisposition Toward Impaired Renal Function Provide Insights into Potential Therapeutic Targets Common to Both Kidney Diseases and ASD
4.5. This Study Has Strengths and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ASD | Autism spectrum disorder |
| BMI | Body mass index |
| DNA | Deoxyribonucleic Acid |
| FDR | False discovery rate |
| GWAS | Genome-wide association study |
| HDL-C | High-density lipoprotein cholesterol |
| PC | Principal component |
| PCA | Principal component analysis |
| PGS | Polygenic score |
| RNA | Ribonucleic Acid |
| SNP | Single nucleotide polymorphism |
| TMM BirThree | Tohoku Medical Megabank Birth and Three-generation |
| ToMMo | Tohoku Medical Megabank Organization |
References
- Zeidan, J.; Fombonne, E.; Scorah, J.; Ibrahim, A.; Durkin, M.S.; Saxena, S.; Yusuf, A.; Shih, A.; Elsabbagh, M. Global Prevalence of Autism: A Systematic Review Update. Autism Res. 2022, 15, 778–790. [Google Scholar] [CrossRef]
- Hirota, T.; King, B.H. Autism Spectrum Disorder: A Review. JAMA 2023, 329, 157–168. [Google Scholar] [CrossRef] [PubMed]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, Text Revision (DSM-V-TR), 5th ed.; American Psychiatric Association: Washington, DC, USA, 2022; pp. 56–68. [Google Scholar]
- Masi, A.; DeMayo, M.M.; Glozier, N.; Guastella, A.J. An Overview of Autism Spectrum Disorder, Heterogeneity and Treatment Options. Neurosci. Bull. 2017, 33, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Kereszturi, É. Diversity and Classification of Genetic Variations in Autism Spectrum Disorder. Int. J. Mol. Sci. 2023, 24, 16768. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, H.; Liang, Z.; Ma, G.; Qureshi, A.; Ran, X.; Feng, C.; Liu, X.; Yan, X.; Shen, L. Autism Spectrum Disorder: Pathogenesis, Biomarker, and Intervention Therapy. MedComm 2024, 5, e497. [Google Scholar] [CrossRef]
- Dell’Osso, L.; Bonelli, C.; Giovannoni, F.; Poli, F.; Anastasio, L.; Cerofolini, G.; Nardi, B.; Cremone, I.M.; Pini, S.; Carpita, B. Available Treatments for Autism Spectrum Disorder: From Old Strategies to New Options. Pharmaceuticals 2025, 18, 324. [Google Scholar] [CrossRef]
- Loth, E.; Spooren, W.; Ham, L.M.; Isaac, M.B.; Auriche-Benichou, C.; Banaschewski, T.; Baron-Cohen, S.; Broich, K.; Bölte, S.; Bourgeron, T.; et al. Identification and Validation of Biomarkers for Autism Spectrum Disorders. Nat. Rev. Drug Discov. 2016, 15, 70–73. [Google Scholar] [CrossRef]
- Pérez-Cano, L.; Azidane Chenlo, S.; Sabido-Vera, R.; Sirci, F.; Durham, L.; Guney, E. Translating Precision Medicine for Autism Spectrum Disorder: A Pressing Need. Drug Discov. Today 2023, 28, 103486. [Google Scholar] [CrossRef]
- Loth, E.; Murphy, D.G.; Spooren, W. Defining Precision Medicine Approaches to Autism Spectrum Disorders: Concepts and Challenges. Front. Psychiatry 2016, 7, 188. [Google Scholar] [CrossRef]
- El-Ahmad, P.; Mendes-Silva, A.P.; Diniz, B.S. Liquid Biopsy in Neuropsychiatric Disorders: A Step Closer to Precision Medicine. Mol. Neurobiol. 2025, 62, 3462–3479. [Google Scholar] [CrossRef]
- Pichitpunpong, C.; Thongkorn, S.; Kanlayaprasit, S.; Yuwattana, W.; Plaingam, W.; Sangsuthum, S.; Aizat, W.M.; Baharum, S.N.; Tencomnao, T.; Hu, V.W.; et al. Phenotypic Subgrouping and Multi-Omics Analyses Reveal Reduced Diazepam-Binding Inhibitor (DBI) Protein Levels in Autism Spectrum Disorder with Severe Language Impairment. PLoS ONE 2019, 14, e0214198. [Google Scholar] [CrossRef]
- Lin, P.I.; Moni, M.A.; Gau, S.S.F.; Eapen, V. Identifying Subgroups of Patients with Autism by Gene Expression Profiles Using Machine Learning Algorithms. Front. Psychiatry 2021, 12, 637022. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.G.; Kohane, I.S.; Kong, S.W. Pathway-Based Outlier Method Reveals Heterogeneous Genomic Structure of Autism in Blood Transcriptome. BMC Med. Genom. 2013, 6, 34. [Google Scholar] [CrossRef] [PubMed]
- Rifai, N.; Gillette, M.A.; Carr, S.A. Protein Biomarker Discovery and Validation: The Long and Uncertain Path to Clinical Utility. Nat. Biotechnol. 2006, 24, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.N.; Masud, M.K.; Haque, M.H.; Hossain, M.S.A.; Yamauchi, Y.; Nguyen, N.-T.; Shiddiky, M.J.A. RNA Biomarkers: Diagnostic and Prognostic Potentials and Recent Developments of Electrochemical Biosensors. Small Methods 2017, 1, 1700131. [Google Scholar] [CrossRef]
- Leek, J.T.; Scharpf, R.B.; Bravo, H.C.; Simcha, D.; Langmead, B.; Johnson, W.E.; Geman, D.; Baggerly, K.; Irizarry, R.A. Tackling the Widespread and Critical Impact of Batch Effects in High-Throughput Data. Nat. Rev. Genet. 2010, 11, 733–739. [Google Scholar] [CrossRef]
- Choi, S.W.; Mak, T.S.-H.; O’Reilly, P.F. Tutorial: A Guide to Performing Polygenic Risk Score Analyses. Nat. Protoc. 2020, 15, 2759–2772. [Google Scholar] [CrossRef]
- Chapman, C.R. Ethical, Legal, and Social Implications of Genetic Risk Prediction for Multifactorial Disease: A Narrative Review Identifying Concerns about Interpretation and Use of Polygenic Scores. J. Community Genet. 2023, 14, 441–452. [Google Scholar] [CrossRef]
- Lambert, S.A.; Gil, L.; Jupp, S.; Ritchie, S.C.; Xu, Y.; Buniello, A.; McMahon, A.; Abraham, G.; Chapman, M.; Parkinson, H.; et al. The Polygenic Score Catalog as an Open Database for Reproducibility and Systematic Evaluation. Nat. Genet. 2021, 53, 420–425. [Google Scholar] [CrossRef]
- Lambert, S.A.; Wingfield, B.; Gibson, J.T.; Gil, L.; Ramachandran, S.; Yvon, F.; Saverimuttu, S.; Tinsley, E.; Lewis, E.; Ritchie, S.C.; et al. Enhancing the Polygenic Score Catalog with Tools for Score Calculation and Ancestry Normalization. Nat. Genet. 2024, 56, 1989–1994. [Google Scholar] [CrossRef]
- Haworth, C.M.A.; Davis, O.S.P. From Observational to Dynamic Genetics. Front. Genet. 2014, 5, 6. [Google Scholar] [CrossRef]
- Matange, K.; Tuck, J.M.; Keung, A.J. DNA Stability: A Central Design Consideration for DNA Data Storage Systems. Nat. Commun. 2021, 12, 1358. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Xu, L.; Liu, J.; Jones, W.D.; Su, Z.; Ning, B.; Perkins, R.; Ge, W.; Miclaus, K.; Zhang, L.; et al. Technical Reproducibility of Genotyping SNP Arrays Used in Genome-Wide Association Studies. PLoS ONE 2012, 7, e44483. [Google Scholar] [CrossRef] [PubMed]
- Warrier, V.; Zhang, X.; Reed, P.; Havdahl, A.; Moore, T.M.; Cliquet, F.; Leblond, C.S.; Rolland, T.; Rosengren, A.; EU-AIMS LEAP; et al. Genetic Correlates of Phenotypic Heterogeneity in Autism. Nat. Genet. 2022, 54, 1293–1304. [Google Scholar] [CrossRef] [PubMed]
- Antaki, D.; Guevara, J.; Maihofer, A.X.; Klein, M.; Gujral, M.; Grove, J.; Carey, C.E.; Hong, O.; Arranz, M.J.; Hervas, A.; et al. A Phenotypic Spectrum of Autism Is Attributable to the Combined Effects of Rare Variants, Polygenic Risk and Sex. Nat. Genet. 2022, 54, 1284–1292. [Google Scholar] [CrossRef]
- Klein, L.; D’Urso, S.; Eapen, V.; Hwang, L.-D.; Lin, P.-I. Exploring Polygenic Contributors to Subgroups of Comorbid Conditions in Autism Spectrum Disorder. Sci. Rep. 2022, 12, 3416. [Google Scholar] [CrossRef]
- Albiñana, C.; Zhu, Z.; Schork, A.J.; Ingason, A.; Aschard, H.; Brikell, I.; Bulik, C.M.; Petersen, L.V.; Agerbo, E.; Grove, J.; et al. Multi-PGS Enhances Polygenic Prediction by Combining 937 Polygenic Scores. Nat. Commun. 2023, 14, 4702. [Google Scholar] [CrossRef]
- Kuriyama, S.; Metoki, H.; Kikuya, M.; Obara, T.; Ishikuro, M.; Yamanaka, C.; Nagai, M.; Matsubara, H.; Kobayashi, T.; Sugawara, J.; et al. Cohort Profile: Tohoku Medical Megabank Project Birth and Three-Generation Cohort Study (TMM BirThree Cohort Study): Rationale, Progress and Perspective. Int. J. Epidemiol. 2020, 49, 18–19m. [Google Scholar] [CrossRef]
- Ogishima, S.; Nagaie, S.; Mizuno, S.; Ishiwata, R.; Iida, K.; Shimokawa, K.; Takai-Igarashi, T.; Nakamura, N.; Nagase, S.; Nakamura, T.; et al. dbTMM: An Integrated Database of Large-Scale Cohort, Genome and Clinical Data for the Tohoku Medical Megabank Project. Hum. Genome Var. 2021, 8, 44. [Google Scholar] [CrossRef]
- Nagasaki, M.; Yasuda, J.; Katsuoka, F.; Nariai, N.; Kojima, K.; Kawai, Y.; Yamaguchi-Kabata, Y.; Yokozawa, J.; Danjoh, I.; Saito, S.; et al. Rare Variant Discovery by Deep Whole-Genome Sequencing of 1,070 Japanese Individuals. Nat. Commun. 2015, 6, 8018. [Google Scholar] [CrossRef]
- Abraham, G.; Havulinna, A.S.; Bhalala, O.G.; Byars, S.G.; De Livera, A.M.; Yetukuri, L.; Tikkanen, E.; Perola, M.; Schunkert, H.; Sijbrands, E.J.; et al. Genomic Prediction of Coronary Heart Disease. Eur. Heart J. 2016, 37, 3267–3278. [Google Scholar] [CrossRef] [PubMed]
- Abraham, G.; Malik, R.; Yonova-Doing, E.; Salim, A.; Wang, T.; Danesh, J.; Butterworth, A.S.; Howson, J.M.M.; Inouye, M.; Dichgans, M. Genomic Risk Score Offers Predictive Performance Comparable to Clinical Risk Factors for Ischaemic Stroke. Nat. Commun. 2019, 10, 5819. [Google Scholar] [CrossRef] [PubMed]
- Abraham, G.; Rohmer, A.; Tye-Din, J.A.; Inouye, M. Genomic Prediction of Celiac Disease Targeting HLA-Positive Individuals. Genome Med. 2015, 7, 72. [Google Scholar] [CrossRef] [PubMed]
- Abraham, G.; Tye-Din, J.A.; Bhalala, O.G.; Kowalczyk, A.; Zobel, J.; Inouye, M. Accurate and Robust Genomic Prediction of Celiac Disease Using Statistical Learning. PLoS Genet. 2014, 10, e1004137. [Google Scholar] [CrossRef]
- Agrawal, S.; Wang, M.; Klarqvist, M.D.R.; Smith, K.; Shin, J.; Dashti, H.; Diamant, N.; Choi, S.H.; Jurgens, S.J.; Ellinor, P.T.; et al. Inherited Basis of Visceral, Abdominal Subcutaneous and Gluteofemoral Fat Depots. Nat. Commun. 2022, 13, 3771. [Google Scholar] [CrossRef]
- Ahmed, R.A.; Shi, Z.; Rifkin, A.S.; Wei, J.; Lilly Zheng, S.; Helfand, B.T.; Hulick, P.J.; Woo, J.S.H.; Qamar, A.; Davidson, D.J.; et al. Reclassification of Coronary Artery Disease Risk Using Genetic Risk Score among Subjects with Borderline or Intermediate Clinical Risk. Int. J. Cardiol. Heart Vasc. 2022, 43, 101136. [Google Scholar] [CrossRef]
- Al-Janabi, A.; Eyre, S.; Foulkes, A.C.; Khan, A.R.; Dand, N.; Burova, E.; DeSilva, B.; Makrygeorgou, A.; Davies, E.; Smith, C.H.; et al. Atopic Polygenic Risk Score Is Associated with Paradoxical Eczema Developing in Patients with Psoriasis Treated with Biologics. J. Investig. Dermatol. 2023, 143, 1470–1478.e1. [Google Scholar] [CrossRef]
- Al-Janabi, A.; Martin, P.; Khan, A.R.; Foulkes, A.C.; Smith, C.H.; Griffiths, C.E.M.; Morris, A.P.; Eyre, S.; Warren, R.B. BSTOP Study Group and the BADBIR Study Group Integrated Proteomics and Genomics Analysis of Paradoxical Eczema in Psoriasis Patients Treated with Biologics. J. Allergy Clin. Immunol. 2023, 152, 1237–1246. [Google Scholar] [CrossRef]
- Archambault, A.N.; Jeon, J.; Lin, Y.; Thomas, M.; Harrison, T.A.; Bishop, D.T.; Brenner, H.; Casey, G.; Chan, A.T.; Chang-Claude, J.; et al. Risk Stratification for Early-Onset Colorectal Cancer Using a Combination of Genetic and Environmental Risk Scores: An International Multi-Center Study. J. Natl. Cancer Inst. 2022, 114, 528–539. [Google Scholar] [CrossRef]
- Archambault, A.N.; Su, Y.-R.; Jeon, J.; Thomas, M.; Lin, Y.; Conti, D.V.; Win, A.K.; Sakoda, L.C.; Lansdorp-Vogelaar, I.; Peterse, E.F.P.; et al. Cumulative Burden of Colorectal Cancer-Associated Genetic Variants Is More Strongly Associated with Early-Onset vs Late-Onset Cancer. Gastroenterology 2020, 158, 1274–1286.e12. [Google Scholar] [CrossRef]
- Bakker, M.K.; Kanning, J.P.; Abraham, G.; Martinsen, A.E.; Winsvold, B.S.; Zwart, J.-A.; Bourcier, R.; Sawada, T.; Koido, M.; Kamatani, Y.; et al. Genetic Risk Score for Intracranial Aneurysms: Prediction of Subarachnoid Hemorrhage and Role in Clinical Heterogeneity. Stroke 2023, 54, 810–818. [Google Scholar] [CrossRef]
- Barr, P.B.; Ksinan, A.; Su, J.; Johnson, E.C.; Meyers, J.L.; Wetherill, L.; Latvala, A.; Aliev, F.; Chan, G.; Kuperman, S.; et al. Using Polygenic Scores for Identifying Individuals at Increased Risk of Substance Use Disorders in Clinical and Population Samples. Transl. Psychiatry 2020, 10, 196. [Google Scholar] [CrossRef] [PubMed]
- Bellenguez, C.; Küçükali, F.; Jansen, I.E.; Kleineidam, L.; Moreno-Grau, S.; Amin, N.; Naj, A.C.; Campos-Martin, R.; Grenier-Boley, B.; Andrade, V.; et al. New Insights into the Genetic Etiology of Alzheimer’s Disease and Related Dementias. Nat. Genet. 2022, 54, 412–436. [Google Scholar] [CrossRef] [PubMed]
- Bianco, C.; Jamialahmadi, O.; Pelusi, S.; Baselli, G.; Dongiovanni, P.; Zanoni, I.; Santoro, L.; Maier, S.; Liguori, A.; Meroni, M.; et al. Non-Invasive Stratification of Hepatocellular Carcinoma Risk in Non-Alcoholic Fatty Liver Using Polygenic Risk Scores. J. Hepatol. 2021, 74, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Bobbili, D.R.; Banda, P.; Krüger, R.; May, P. Excess of Singleton Loss-of-Function Variants in Parkinson’s Disease Contributes to Genetic Risk. J. Med. Genet. 2020, 57, 617–623. [Google Scholar] [CrossRef]
- Bonfiglio, F.; Liu, X.; Smillie, C.; Pandit, A.; Kurilshikov, A.; Bacigalupe, R.; Zheng, T.; Nim, H.; Garcia-Etxebarria, K.; Bujanda, L.; et al. GWAS of Stool Frequency Provides Insights into Gastrointestinal Motility and Irritable Bowel Syndrome. Cell Genom. 2021, 1, 100069. [Google Scholar] [CrossRef]
- Boumtje, V.; Manikpurage, H.D.; Li, Z.; Gaudreault, N.; Armero, V.S.; Boudreau, D.K.; Renaut, S.; Henry, C.; Racine, C.; Eslami, A.; et al. Polygenic Inheritance and Its Interplay with Smoking History in Predicting Lung Cancer Diagnosis: A French-Canadian Case-Control Cohort. EBioMedicine 2024, 106, 105234. [Google Scholar] [CrossRef]
- Brentnall, A.R.; van Veen, E.M.; Harkness, E.F.; Rafiq, S.; Byers, H.; Astley, S.M.; Sampson, S.; Howell, A.; Newman, W.G.; Cuzick, J.; et al. A Case-Control Evaluation of 143 Single Nucleotide Polymorphisms for Breast Cancer Risk Stratification with Classical Factors and Mammographic Density. Int. J. Cancer 2020, 146, 2122–2129. [Google Scholar] [CrossRef]
- Campos, A.I.; Mulcahy, A.; Thorp, J.G.; Wray, N.R.; Byrne, E.M.; Lind, P.A.; Medland, S.E.; Martin, N.G.; Hickie, I.B.; Rentería, M.E. Understanding Genetic Risk Factors for Common Side Effects of Antidepressant Medications. Commun. Med. 2021, 1, 45. [Google Scholar] [CrossRef]
- Cánovas, R.; Cobb, J.; Brozynska, M.; Bowes, J.; Li, Y.R.; Smith, S.L.; Hakonarson, H.; Thomson, W.; Ellis, J.A.; Abraham, G.; et al. Genomic Risk Scores for Juvenile Idiopathic Arthritis and Its Subtypes. Ann. Rheum. Dis. 2020, 79, 1572–1579. [Google Scholar] [CrossRef]
- Canzian, F.; Piredda, C.; Macauda, A.; Zawirska, D.; Andersen, N.F.; Nagler, A.; Zaucha, J.M.; Mazur, G.; Dumontet, C.; Wątek, M.; et al. A Polygenic Risk Score for Multiple Myeloma Risk Prediction. Eur. J. Hum. Genet. 2022, 30, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Spracklen, C.N.; Marenne, G.; Varshney, A.; Corbin, L.J.; Luan, J.; Willems, S.M.; Wu, Y.; Zhang, X.; Horikoshi, M.; et al. The Trans-Ancestral Genomic Architecture of Glycemic Traits. Nat. Genet. 2021, 53, 840–860. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, Y.-F.; Liu, L.; Bielowka, A.; Ahmed, R.; Zhang, H.; Tombleson, P.; Roberts, A.L.; Odhams, C.A.; Cunninghame Graham, D.S.; et al. Genome-Wide Assessment of Genetic Risk for Systemic Lupus Erythematosus and Disease Severity. Hum. Mol. Genet. 2020, 29, 1745–1756. [Google Scholar] [CrossRef] [PubMed]
- Cherny, S.S.; Livshits, G.; Wells, H.R.R.; Freidin, M.B.; Malkin, I.; Dawson, S.J.; Williams, F.M.K. Self-Reported Hearing Loss Questions Provide a Good Measure for Genetic Studies: A Polygenic Risk Score Analysis from UK Biobank. Eur. J. Hum. Genet. 2020, 28, 1056–1065. [Google Scholar] [CrossRef]
- Chikowore, T.; Läll, K.; Micklesfield, L.K.; Lombard, Z.; Goedecke, J.H.; Fatumo, S.; Norris, S.A.; Magi, R.; Ramsay, M.; Franks, P.W.; et al. Variability of Polygenic Prediction for Body Mass Index in Africa. Genome Med. 2024, 16, 74. [Google Scholar] [CrossRef]
- China Kadoorie Biobank Collaborative Group. Joint Impact of Polygenic Risk Score and Lifestyles on Early-and Late-Onset Cardiovascular Diseases. Nat. Hum. Behav. 2024, 8, 1810–1818. [Google Scholar] [CrossRef]
- Choi, J.; Jia, G.; Wen, W.; Long, J.; Zheng, W. Evaluating Polygenic Risk Scores in Assessing Risk of Nine Solid and Hematologic Cancers in European Descendants. Int. J. Cancer 2020, 147, 3416–3423. [Google Scholar] [CrossRef]
- Christiansen, M.R.; Kilpeläinen, T.O.; McCaffery, J.M. Abdominal Obesity Genetic Variants Predict Waist Circumference Regain After Weight Loss. Diabetes 2023, 72, 1424–1432. [Google Scholar] [CrossRef]
- Clark, R.; Lee, S.S.-Y.; Du, R.; Wang, Y.; Kneepkens, S.C.M.; Charng, J.; Huang, Y.; Hunter, M.L.; Jiang, C.; Tideman, J.W.L.; et al. A New Polygenic Score for Refractive Error Improves Detection of Children at Risk of High Myopia but Not the Prediction of Those at Risk of Myopic Macular Degeneration. EBioMedicine 2023, 91, 104551. [Google Scholar] [CrossRef]
- Dareng, E.O.; Tyrer, J.P.; Barnes, D.R.; Jones, M.R.; Yang, X.; Aben, K.K.H.; Adank, M.A.; Agata, S.; Andrulis, I.L.; Anton-Culver, H.; et al. Polygenic Risk Modeling for Prediction of Epithelial Ovarian Cancer Risk. Eur. J. Hum. Genet. 2022, 30, 349–362. [Google Scholar] [CrossRef]
- de Rojas, I.; Moreno-Grau, S.; Tesi, N.; Grenier-Boley, B.; Andrade, V.; Jansen, I.E.; Pedersen, N.L.; Stringa, N.; Zettergren, A.; Hernández, I.; et al. Common Variants in Alzheimer’s Disease and Risk Stratification by Polygenic Risk Scores. Nat. Commun. 2021, 12, 3417. [Google Scholar] [CrossRef]
- Deng, W.Q.; Belisario, K.; Gray, J.C.; Levitt, E.E.; Mohammadi-Shemirani, P.; Singh, D.; Pare, G.; MacKillop, J. Leveraging Related Health Phenotypes for Polygenic Prediction of Impulsive Choice, Impulsive Action, and Impulsive Personality Traits in 1534 European Ancestry Community Adults. Genes Brain Behav. 2023, 22, e12848. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Hou, K.; Xu, Z.; Pimplaskar, A.; Petter, E.; Boulier, K.; Privé, F.; Vilhjálmsson, B.J.; Olde Loohuis, L.M.; Pasaniuc, B. Polygenic Scoring Accuracy Varies across the Genetic Ancestry Continuum. Nature 2023, 618, 774–781. [Google Scholar] [CrossRef] [PubMed]
- Dongiovanni, P.; Stender, S.; Pietrelli, A.; Mancina, R.M.; Cespiati, A.; Petta, S.; Pelusi, S.; Pingitore, P.; Badiali, S.; Maggioni, M.; et al. Causal Relationship of Hepatic Fat with Liver Damage and Insulin Resistance in Nonalcoholic Fatty Liver. J. Intern. Med. 2018, 283, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Downie, M.L.; Gupta, S.; Chan, M.M.Y.; Sadeghi-Alavijeh, O.; Cao, J.; Parekh, R.S.; Diz, C.B.; Bierzynska, A.; Levine, A.P.; Pepper, R.J.; et al. Shared Genetic Risk across Different Presentations of Gene Test-Negative Idiopathic Nephrotic Syndrome. Pediatr. Nephrol. 2023, 38, 1793–1800. [Google Scholar] [CrossRef]
- Dron, J.S.; Wang, M.; Patel, A.P.; Kartoun, U.; Ng, K.; Hegele, R.A.; Khera, A.V. Genetic Predictor to Identify Individuals with High Lipoprotein(a) Concentrations. Circ. Genom. Precis. Med. 2021, 14, e003182. [Google Scholar] [CrossRef]
- Ebenau, J.L.; van der Lee, S.J.; Hulsman, M.; Tesi, N.; Jansen, I.E.; Verberk, I.M.W.; van Leeuwenstijn, M.; Teunissen, C.E.; Barkhof, F.; Prins, N.D.; et al. Risk of Dementia in APOE Ε4 Carriers Is Mitigated by a Polygenic Risk Score. Alzheimers Dement 2021, 13, e12229. [Google Scholar] [CrossRef]
- El-Boraie, A.; Chenoweth, M.J.; Pouget, J.G.; Benowitz, N.L.; Fukunaga, K.; Mushiroda, T.; Kubo, M.; Nollen, N.L.; Sanderson Cox, L.; Lerman, C.; et al. Transferability of Ancestry-Specific and Cross-Ancestry CYP2A6 Activity Genetic Risk Scores in African and European Populations. Clin. Pharmacol. Ther. 2021, 110, 975–985. [Google Scholar] [CrossRef]
- El-Boraie, A.; Taghavi, T.; Chenoweth, M.J.; Fukunaga, K.; Mushiroda, T.; Kubo, M.; Lerman, C.; Nollen, N.L.; Benowitz, N.L.; Tyndale, R.F. Evaluation of a Weighted Genetic Risk Score for the Prediction of Biomarkers of CYP2A6 Activity. Addict. Biol. 2020, 25, e12741. [Google Scholar] [CrossRef]
- Elliott, J.; Bodinier, B.; Bond, T.A.; Chadeau-Hyam, M.; Evangelou, E.; Moons, K.G.M.; Dehghan, A.; Muller, D.C.; Elliott, P.; Tzoulaki, I. Predictive Accuracy of a Polygenic Risk Score-Enhanced Prediction Model vs a Clinical Risk Score for Coronary Artery Disease. JAMA 2020, 323, 636–645. [Google Scholar] [CrossRef]
- Emdin, C.A.; Haas, M.; Ajmera, V.; Simon, T.G.; Homburger, J.; Neben, C.; Jiang, L.; Wei, W.-Q.; Feng, Q.; Zhou, A.; et al. Association of Genetic Variation with Cirrhosis: A Multi-Trait Genome-Wide Association and Gene-Environment Interaction Study. Gastroenterology 2021, 160, 1620–1633.e13. [Google Scholar] [CrossRef] [PubMed]
- Flynn, E.; Tanigawa, Y.; Rodriguez, F.; Altman, R.B.; Sinnott-Armstrong, N.; Rivas, M.A. Sex-Specific Genetic Effects across Biomarkers. Eur. J. Hum. Genet. 2021, 29, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Folkersen, L.; Gustafsson, S.; Wang, Q.; Hansen, D.H.; Hedman, Å.K.; Schork, A.; Page, K.; Zhernakova, D.V.; Wu, Y.; Peters, J.; et al. Genomic and Drug Target Evaluation of 90 Cardiovascular Proteins in 30,931 Individuals. Nat. Metab. 2020, 2, 1135–1148. [Google Scholar] [CrossRef] [PubMed]
- Folsom, A.R.; Tang, W.; Hong, C.-P.; Rosamond, W.D.; Lane, J.A.; Cushman, M.; Pankratz, N. Prediction of Venous Thromboembolism Incidence in the General Adult Population Using Two Published Genetic Risk Scores. PLoS ONE 2023, 18, e0280657. [Google Scholar] [CrossRef]
- Fontanillas, P.; Alipanahi, B.; Furlotte, N.A.; Johnson, M.; Wilson, C.H.; 23andMe Research Team; Pitts, S.J.; Gentleman, R.; Auton, A. Disease Risk Scores for Skin Cancers. Nat. Commun. 2021, 12, 160. [Google Scholar] [CrossRef]
- Gao, C.; Polley, E.C.; Hart, S.N.; Huang, H.; Hu, C.; Gnanaolivu, R.; Lilyquist, J.; Boddicker, N.J.; Na, J.; Ambrosone, C.B.; et al. Risk of Breast Cancer Among Carriers of Pathogenic Variants in Breast Cancer Predisposition Genes Varies by Polygenic Risk Score. J. Clin. Oncol. 2021, 39, 2564–2573. [Google Scholar] [CrossRef]
- García-González, P.; de Rojas, I.; Moreno-Grau, S.; Montrreal, L.; Puerta, R.; Alarcón-Martín, E.; Quintela, I.; Orellana, A.; Andrade, V.; Adami, P.V.M.; et al. Mendelian Randomisation Confirms the Role of Y-Chromosome Loss in Alzheimer’s Disease Aetiopathogenesis in Men. Int. J. Mol. Sci. 2023, 24, 898. [Google Scholar] [CrossRef]
- Ge, T.; Irvin, M.R.; Patki, A.; Srinivasasainagendra, V.; Lin, Y.-F.; Tiwari, H.K.; Armstrong, N.D.; Benoit, B.; Chen, C.-Y.; Choi, K.W.; et al. Development and Validation of a Trans-Ancestry Polygenic Risk Score for Type 2 Diabetes in Diverse Populations. Genome Med. 2022, 14, 70. [Google Scholar] [CrossRef]
- Gibson, M.J.; Lawlor, D.A.; Millard, L.A.C. Identifying the Potential Causal Role of Insomnia Symptoms on 11,409 Health-Related Outcomes: A Phenome-Wide Mendelian Randomisation Analysis in UK Biobank. BMC Med. 2023, 21, 128. [Google Scholar] [CrossRef]
- Gorman, B.R.; Voloudakis, G.; Igo, R.P.; Kinzy, T.; Halladay, C.W.; Bigdeli, T.B.; Zeng, B.; Venkatesh, S.; Cooke Bailey, J.N.; Crawford, D.C.; et al. Genome-Wide Association Analyses Identify Distinct Genetic Architectures for Age-Related Macular Degeneration across Ancestries. Nat. Genet. 2024, 56, 2659–2671. [Google Scholar] [CrossRef]
- Gunn, S.; Wang, X.; Posner, D.C.; Cho, K.; Huffman, J.E.; Gaziano, M.; Wilson, P.W.; Sun, Y.V.; Peloso, G.; Lunetta, K.L. Comparison of Methods for Building Polygenic Scores for Diverse Populations. HGG Adv. 2025, 6, 100355. [Google Scholar] [CrossRef] [PubMed]
- Haas, M.E.; Pirruccello, J.P.; Friedman, S.N.; Wang, M.; Emdin, C.A.; Ajmera, V.H.; Simon, T.G.; Homburger, J.R.; Guo, X.; Budoff, M.; et al. Machine Learning Enables New Insights into Genetic Contributions to Liver Fat Accumulation. Cell Genom. 2021, 1, 100066. [Google Scholar] [CrossRef] [PubMed]
- Harper, A.R.; Goel, A.; Grace, C.; Thomson, K.L.; Petersen, S.E.; Xu, X.; Waring, A.; Ormondroyd, E.; Kramer, C.M.; Ho, C.Y.; et al. Common Genetic Variants and Modifiable Risk Factors Underpin Hypertrophic Cardiomyopathy Susceptibility and Expressivity. Nat. Genet. 2021, 53, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Hassanin, E.; Lee, K.-H.; Hsieh, T.-C.; Aldisi, R.; Lee, Y.-L.; Bobbili, D.; Krawitz, P.; May, P.; Chen, C.-Y.; Maj, C. Trans-Ancestry Polygenic Models for the Prediction of LDL Blood Levels: An Analysis of the UK Biobank and Taiwan Biobank. medRxiv 2023, preprint. [Google Scholar] [CrossRef]
- Honda, S.; Ikari, K.; Yano, K.; Terao, C.; Tanaka, E.; Harigai, M.; Kochi, Y. Association of Polygenic Risk Scores with Radiographic Progression in Patients with Rheumatoid Arthritis. Arthritis Rheumatol. 2022, 74, 791–800. [Google Scholar] [CrossRef]
- Huynh-Le, M.-P.; Fan, C.C.; Karunamuni, R.; Thompson, W.K.; Martinez, M.E.; Eeles, R.A.; Kote-Jarai, Z.; Muir, K.; Schleutker, J.; Pashayan, N.; et al. Polygenic Hazard Score Is Associated with Prostate Cancer in Multi-Ethnic Populations. Nat. Commun. 2021, 12, 1236. [Google Scholar] [CrossRef]
- Huynh-Le, M.-P.; Karunamuni, R.; Fan, C.C.; Asona, L.; Thompson, W.K.; Martinez, M.E.; Eeles, R.A.; Kote-Jarai, Z.; Muir, K.R.; Lophatananon, A.; et al. Prostate Cancer Risk Stratification Improvement across Multiple Ancestries with New Polygenic Hazard Score. Prostate Cancer Prostatic Dis. 2022, 25, 755–761. [Google Scholar] [CrossRef]
- Ibanez, L.; Dube, U.; Saef, B.; Budde, J.; Black, K.; Medvedeva, A.; Del-Aguila, J.L.; Davis, A.A.; Perlmutter, J.S.; Harari, O.; et al. Parkinson Disease Polygenic Risk Score Is Associated with Parkinson Disease Status and Age at Onset but Not with Alpha-Synuclein Cerebrospinal Fluid Levels. BMC Neurol. 2017, 17, 198. [Google Scholar] [CrossRef]
- Ibáñez-Sanz, G.; Díez-Villanueva, A.; Alonso, M.H.; Rodríguez-Moranta, F.; Pérez-Gómez, B.; Bustamante, M.; Martin, V.; Llorca, J.; Amiano, P.; Ardanaz, E.; et al. Risk Model for Colorectal Cancer in Spanish Population Using Environmental and Genetic Factors: Results from the MCC-Spain Study. Sci. Rep. 2017, 7, 43263. [Google Scholar] [CrossRef]
- Inouye, M.; Abraham, G.; Nelson, C.P.; Wood, A.M.; Sweeting, M.J.; Dudbridge, F.; Lai, F.Y.; Kaptoge, S.; Brozynska, M.; Wang, T.; et al. Genomic Risk Prediction of Coronary Artery Disease in 480,000 Adults: Implications for Primary Prevention. J. Am. Coll. Cardiol. 2018, 72, 1883–1893. [Google Scholar] [CrossRef]
- Ishigaki, K.; Sakaue, S.; Terao, C.; Luo, Y.; Sonehara, K.; Yamaguchi, K.; Amariuta, T.; Too, C.L.; Laufer, V.A.; Scott, I.C.; et al. Multi-Ancestry Genome-Wide Association Analyses Identify Novel Genetic Mechanisms in Rheumatoid Arthritis. Nat. Genet. 2022, 54, 1640–1651. [Google Scholar] [CrossRef] [PubMed]
- Jee, Y.H.; Ho, W.-K.; Park, S.; Easton, D.F.; Teo, S.-H.; Jung, K.J.; Kraft, P. Polygenic Risk Scores for Prediction of Breast Cancer in Korean Women. Int. J. Epidemiol. 2023, 52, 796–805. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Lu, Y.; Wen, W.; Long, J.; Liu, Y.; Tao, R.; Li, B.; Denny, J.C.; Shu, X.-O.; Zheng, W. Evaluating the Utility of Polygenic Risk Scores in Identifying High-Risk Individuals for Eight Common Cancers. JNCI Cancer Spectr. 2020, 4, pkaa021. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Ping, J.; Guo, X.; Yang, Y.; Tao, R.; Li, B.; Ambs, S.; Barnard, M.E.; Chen, Y.; Garcia-Closas, M.; et al. Genome-Wide Association Analyses of Breast Cancer in Women of African Ancestry Identify New Susceptibility Loci and Improve Risk Prediction. Nat. Genet. 2024, 56, 819–826. [Google Scholar] [CrossRef]
- Jones, A.C.; Patki, A.; Srinivasasainagendra, V.; Tiwari, H.K.; Armstrong, N.D.; Chaudhary, N.S.; Limdi, N.A.; Hidalgo, B.A.; Davis, B.; Cimino, J.J.; et al. Single-Ancestry versus Multi-Ancestry Polygenic Risk Scores for CKD in Black American Populations. J. Am. Soc. Nephrol. 2024, 35, 1558–1569. [Google Scholar] [CrossRef]
- Jung, H.; Jung, H.-U.; Baek, E.J.; Kwon, S.Y.; Kang, J.-O.; Lim, J.E.; Oh, B. Integration of Risk Factor Polygenic Risk Score with Disease Polygenic Risk Score for Disease Prediction. Commun. Biol. 2024, 7, 180. [Google Scholar] [CrossRef]
- Jung, S.-H.; Kim, H.-R.; Chun, M.Y.; Jang, H.; Cho, M.; Kim, B.; Kim, S.; Jeong, J.H.; Yoon, S.J.; Park, K.W.; et al. Transferability of Alzheimer Disease Polygenic Risk Score Across Populations and Its Association with Alzheimer Disease-Related Phenotypes. JAMA Netw. Open 2022, 5, e2247162. [Google Scholar] [CrossRef]
- Kanoni, S.; Graham, S.E.; Wang, Y.; Surakka, I.; Ramdas, S.; Zhu, X.; Clarke, S.L.; Bhatti, K.F.; Vedantam, S.; Winkler, T.W.; et al. Implicating Genes, Pleiotropy, and Sexual Dimorphism at Blood Lipid Loci through Multi-Ancestry Meta-Analysis. Genome Biol. 2022, 23, 268. [Google Scholar] [CrossRef]
- Karunamuni, R.A.; Huynh-Le, M.-P.; Fan, C.C.; Thompson, W.; Eeles, R.A.; Kote-Jarai, Z.; Muir, K.; UKGPCS Collaborators; Lophatananon, A.; Tangen, C.M.; et al. African-Specific Improvement of a Polygenic Hazard Score for Age at Diagnosis of Prostate Cancer. Int. J. Cancer 2021, 148, 99–105. [Google Scholar] [CrossRef]
- Karunamuni, R.A.; Huynh-Le, M.-P.; Fan, C.C.; Thompson, W.; Eeles, R.A.; Kote-Jarai, Z.; Muir, K.; Lophatananon, A.; UKGPCS collaborators; Schleutker, J.; et al. Additional SNPs Improve Risk Stratification of a Polygenic Hazard Score for Prostate Cancer. Prostate Cancer Prostatic Dis. 2021, 24, 532–541. [Google Scholar] [CrossRef]
- Kawai, V.K.; Shi, M.; Liu, G.; Feng, Q.; Wei, W.; Chung, C.P.; Walunas, T.L.; Gordon, A.S.; Linneman, J.G.; Hebbring, S.J.; et al. Pleiotropy of Systemic Lupus Erythematosus Risk Alleles and Cardiometabolic Disorders: A Phenome-Wide Association Study and Inverse-Variance Weighted Meta-Analysis. Lupus 2021, 30, 1264–1272. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Turchin, M.C.; Patki, A.; Srinivasasainagendra, V.; Shang, N.; Nadukuru, R.; Jones, A.C.; Malolepsza, E.; Dikilitas, O.; Kullo, I.J.; et al. Genome-Wide Polygenic Score to Predict Chronic Kidney Disease across Ancestries. Nat. Med. 2022, 28, 1412–1420. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yuan, C.; Babic, A.; Bao, Y.; Clish, C.B.; Pollak, M.N.; Amundadottir, L.T.; Klein, A.P.; Stolzenberg-Solomon, R.Z.; Pandharipande, P.V.; et al. Genetic and Circulating Biomarker Data Improve Risk Prediction for Pancreatic Cancer in the General Population. Cancer Epidemiol. Biomarkers Prev. 2020, 29, 999–1008. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Jang, H.; Wang, M.; Shi, Q.; Strain, T.; Sharp, S.J.; Yeung, S.L.A.; Luo, S.; Griffin, S.; Wareham, N.J.; et al. Replacing Device-Measured Sedentary Time with Physical Activity Is Associated with Lower Risk of Coronary Heart Disease Regardless of Genetic Risk. J. Intern. Med. 2024, 295, 38–50. [Google Scholar] [CrossRef]
- Kloeve-Mogensen, K.; Rohde, P.D.; Twisttmann, S.; Nygaard, M.; Koldby, K.M.; Steffensen, R.; Dahl, C.M.; Rytter, D.; Overgaard, M.T.; Forman, A.; et al. Polygenic Risk Score Prediction for Endometriosis. Front. Reprod. Health 2021, 3, 793226. [Google Scholar] [CrossRef]
- Kloosterman, M.; Santema, B.T.; Roselli, C.; Nelson, C.P.; Koekemoer, A.; Romaine, S.P.R.; Van Gelder, I.C.; Lam, C.S.P.; Artola, V.A.; Lang, C.C.; et al. Genetic Risk and Atrial Fibrillation in Patients with Heart Failure. Eur. J. Heart Fail. 2020, 22, 519–527. [Google Scholar] [CrossRef]
- Knevel, R.; le Cessie, S.; Terao, C.C.; Slowikowski, K.; Cui, J.; Huizinga, T.W.J.; Costenbader, K.H.; Liao, K.P.; Karlson, E.W.; Raychaudhuri, S. Using Genetics to Prioritize Diagnoses for Rheumatology Outpatients with Inflammatory Arthritis. Sci. Transl. Med. 2020, 12, eaay1548. [Google Scholar] [CrossRef]
- Ko, C.-L.; Lin, W.-Z.; Lee, M.-T.; Chang, Y.-T.; Lin, H.-C.; Wu, Y.-S.; Lin, J.-F.; Pan, K.-T.; Chang, Y.-C.; Lee, K.-H.; et al. Genome-Wide Association Study Reveals Ethnicity-Specific SNPs Associated with Ankylosing Spondylitis in the Taiwanese Population. J. Transl. Med. 2022, 20, 589. [Google Scholar] [CrossRef]
- Kolin, D.A.; Kulm, S.; Elemento, O. Prediction of Primary Venous Thromboembolism Based on Clinical and Genetic Factors within the U.K. Biobank. Sci. Rep. 2021, 11, 21340. [Google Scholar] [CrossRef]
- Kothalawala, D.M.; Kadalayil, L.; Curtin, J.A.; Murray, C.S.; Simpson, A.; Custovic, A.; Tapper, W.J.; Arshad, S.H.; Rezwan, F.I.; Holloway, J.W.; et al. Integration of Genomic Risk Scores to Improve the Prediction of Childhood Asthma Diagnosis. J. Pers. Med. 2022, 12, 75. [Google Scholar] [CrossRef]
- Koyama, S.; Ito, K.; Terao, C.; Akiyama, M.; Horikoshi, M.; Momozawa, Y.; Matsunaga, H.; Ieki, H.; Ozaki, K.; Onouchi, Y.; et al. Population-Specific and Trans-Ancestry Genome-Wide Analyses Identify Distinct and Shared Genetic Risk Loci for Coronary Artery Disease. Nat. Genet. 2020, 52, 1169–1177. [Google Scholar] [CrossRef] [PubMed]
- Krohn, L.; Heilbron, K.; Blauwendraat, C.; Reynolds, R.H.; Yu, E.; Senkevich, K.; Rudakou, U.; Estiar, M.A.; Gustavsson, E.K.; Brolin, K.; et al. Genome-Wide Association Study of REM Sleep Behavior Disorder Identifies Polygenic Risk and Brain Expression Effects. Nat. Commun. 2022, 13, 7496. [Google Scholar] [CrossRef] [PubMed]
- Kuchenbaecker, K.; Telkar, N.; Reiker, T.; Walters, R.G.; Lin, K.; Eriksson, A.; Gurdasani, D.; Gilly, A.; Southam, L.; Tsafantakis, E.; et al. The Transferability of Lipid Loci across African, Asian and European Cohorts. Nat. Commun. 2019, 10, 4330. [Google Scholar] [CrossRef] [PubMed]
- Kurniansyah, N.; Goodman, M.O.; Kelly, T.N.; Elfassy, T.; Wiggins, K.L.; Bis, J.C.; Guo, X.; Palmas, W.; Taylor, K.D.; Lin, H.J.; et al. A Multi-Ethnic Polygenic Risk Score Is Associated with Hypertension Prevalence and Progression throughout Adulthood. Nat. Commun. 2022, 13, 3549. [Google Scholar] [CrossRef]
- Kurniansyah, N.; Goodman, M.O.; Khan, A.T.; Wang, J.; Feofanova, E.; Bis, J.C.; Wiggins, K.L.; Huffman, J.E.; Kelly, T.; Elfassy, T.; et al. Evaluating the Use of Blood Pressure Polygenic Risk Scores across Race/Ethnic Background Groups. Nat. Commun. 2023, 14, 3202. [Google Scholar] [CrossRef]
- Lai, D.; Johnson, E.C.; Colbert, S.; Pandey, G.; Chan, G.; Bauer, L.; Francis, M.W.; Hesselbrock, V.; Kamarajan, C.; Kramer, J.; et al. Evaluating Risk for Alcohol Use Disorder: Polygenic Risk Scores and Family History. Alcohol. Clin. Exp. Res. 2022, 46, 374–383. [Google Scholar] [CrossRef]
- Lai, D.; Schwantes-An, T.-H.; Abreu, M.; Chan, G.; Hesselbrock, V.; Kamarajan, C.; Liu, Y.; Meyers, J.L.; Nurnberger, J.I.; Plawecki, M.H.; et al. Gene-Based Polygenic Risk Scores Analysis of Alcohol Use Disorder in African Americans. Transl. Psychiatry 2022, 12, 266. [Google Scholar] [CrossRef]
- Law, M.H.; Aoude, L.G.; Duffy, D.L.; Long, G.V.; Johansson, P.A.; Pritchard, A.L.; Khosrotehrani, K.; Mann, G.J.; Montgomery, G.W.; Iles, M.M.; et al. Multiplex Melanoma Families Are Enriched for Polygenic Risk. Hum. Mol. Genet. 2020, 29, 2976–2985. [Google Scholar] [CrossRef]
- Li, D.; Xie, J.; Wang, L.; Sun, Y.; Hu, Y.; Tian, Y. Genetic Susceptibility and Lifestyle Modify the Association of Long-Term Air Pollution Exposure on Major Depressive Disorder: A Prospective Study in UK Biobank. BMC Med. 2023, 21, 67. [Google Scholar] [CrossRef]
- Liu, G.; Peng, J.; Liao, Z.; Locascio, J.J.; Corvol, J.-C.; Zhu, F.; Dong, X.; Maple-Grødem, J.; Campbell, M.C.; Elbaz, A.; et al. Genome-Wide Survival Study Identifies a Novel Synaptic Locus and Polygenic Score for Cognitive Progression in Parkinson’s Disease. Nat. Genet. 2021, 53, 787–793. [Google Scholar] [CrossRef]
- Liu, W.; Wang, T.; Zhu, M.; Jin, G. Healthy Diet, Polygenic Risk Score, and Upper Gastrointestinal Cancer Risk: A Prospective Study from UK Biobank. Nutrients 2023, 15, 1344. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yan, C.; Yin, S.; Wang, T.; Zhu, M.; Liu, L.; Jin, G. Genetic Risk, Metabolic Syndrome, and Gastrointestinal Cancer Risk: A Prospective Cohort Study. Cancer Med. 2023, 12, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Lourida, I.; Hannon, E.; Littlejohns, T.J.; Langa, K.M.; Hyppönen, E.; Kuzma, E.; Llewellyn, D.J. Association of Lifestyle and Genetic Risk with Incidence of Dementia. JAMA 2019, 322, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Martucci, V.L.; Quandt, Z.; Groha, S.; Murray, M.H.; Lovly, C.M.; Rizvi, H.; Egger, J.V.; Plodkowski, A.J.; Abu-Akeel, M.; et al. Immunotherapy-Mediated Thyroid Dysfunction: Genetic Risk and Impact on Outcomes with PD-1 Blockade in Non-Small Cell Lung Cancer. Clin. Cancer Res. 2021, 27, 5131–5140. [Google Scholar] [CrossRef]
- Ma, Y.; Patil, S.; Zhou, X.; Mukherjee, B.; Fritsche, L.G. ExPRSweb: An Online Repository with Polygenic Risk Scores for Common Health-Related Exposures. Am. J. Hum. Genet. 2022, 109, 1742–1760. [Google Scholar] [CrossRef]
- Mack, S.; Coassin, S.; Rueedi, R.; Yousri, N.A.; Seppälä, I.; Gieger, C.; Schönherr, S.; Forer, L.; Erhart, G.; Marques-Vidal, P.; et al. A Genome-Wide Association Meta-Analysis on Lipoprotein (a) Concentrations Adjusted for Apolipoprotein (a) Isoforms. J. Lipid Res. 2017, 58, 1834–1844. [Google Scholar] [CrossRef]
- Mansour Aly, D.; Dwivedi, O.P.; Prasad, R.B.; Käräjämäki, A.; Hjort, R.; Thangam, M.; Åkerlund, M.; Mahajan, A.; Udler, M.S.; Florez, J.C.; et al. Genome-Wide Association Analyses Highlight Etiological Differences Underlying Newly Defined Subtypes of Diabetes. Nat. Genet. 2021, 53, 1534–1542. [Google Scholar] [CrossRef]
- Marston, N.A.; Kamanu, F.K.; Nordio, F.; Gurmu, Y.; Roselli, C.; Sever, P.S.; Pedersen, T.R.; Keech, A.C.; Wang, H.; Lira Pineda, A.; et al. Predicting Benefit from Evolocumab Therapy in Patients with Atherosclerotic Disease Using a Genetic Risk Score: Results from the FOURIER Trial. Circulation 2020, 141, 616–623, Correction in Circulation 2024, 149, e1414. [Google Scholar] [CrossRef]
- Mavaddat, N.; Michailidou, K.; Dennis, J.; Lush, M.; Fachal, L.; Lee, A.; Tyrer, J.P.; Chen, T.-H.; Wang, Q.; Bolla, M.K.; et al. Polygenic Risk Scores for Prediction of Breast Cancer and Breast Cancer Subtypes. Am. J. Hum. Genet. 2019, 104, 21–34. [Google Scholar] [CrossRef]
- Mavaddat, N.; Pharoah, P.D.P.; Michailidou, K.; Tyrer, J.; Brook, M.N.; Bolla, M.K.; Wang, Q.; Dennis, J.; Dunning, A.M.; Shah, M.; et al. Prediction of Breast Cancer Risk Based on Profiling with Common Genetic Variants. J. Natl. Cancer Inst. 2015, 107, djv036. [Google Scholar] [CrossRef]
- Mayerhofer, E.; Malik, R.; Parodi, L.; Burgess, S.; Harloff, A.; Dichgans, M.; Rosand, J.; Anderson, C.D.; Georgakis, M.K. Genetically Predicted On-Statin LDL Response Is Associated with Higher Intracerebral Haemorrhage Risk. Brain 2022, 145, 2677–2686. [Google Scholar] [CrossRef] [PubMed]
- Mayerhofer, E.; Parodi, L.; Prapiadou, S.; Malik, R.; Rosand, J.; Georgakis, M.K.; Anderson, C.D. Genetic Risk Score Improves Risk Stratification for Anticoagulation-Related Intracerebral Hemorrhage. Stroke 2023, 54, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Middha, P.; Thummalapalli, R.; Betti, M.J.; Yao, L.; Quandt, Z.; Balaratnam, K.; Bejan, C.A.; Cardenas, E.; Falcon, C.J.; Faleck, D.M.; et al. Polygenic Risk Score for Ulcerative Colitis Predicts Immune Checkpoint Inhibitor-Mediated Colitis. Nat. Commun. 2024, 15, 2568. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.P.; Mishra, B.H.; Lyytikäinen, L.-P.; Goebeler, S.; Martiskainen, M.; Hakamaa, E.; Kleber, M.E.; Delgado, G.E.; März, W.; Kähönen, M.; et al. Genetic Risk Score for Coronary Artery Calcification and Its Predictive Ability for Coronary Artery Disease. Am. J. Prev. Cardiol. 2024, 20, 100884. [Google Scholar] [CrossRef]
- Miyazawa, K.; Ito, K.; Ito, M.; Zou, Z.; Kubota, M.; Nomura, S.; Matsunaga, H.; Koyama, S.; Ieki, H.; Akiyama, M.; et al. Cross-Ancestry Genome-Wide Analysis of Atrial Fibrillation Unveils Disease Biology and Enables Cardioembolic Risk Prediction. Nat. Genet. 2023, 55, 187–197. [Google Scholar] [CrossRef]
- Moll, M.; Sakornsakolpat, P.; Shrine, N.; Hobbs, B.D.; DeMeo, D.L.; John, C.; Guyatt, A.L.; McGeachie, M.J.; Gharib, S.A.; Obeidat, M.; et al. Chronic Obstructive Pulmonary Disease and Related Phenotypes: Polygenic Risk Scores in Population-Based and Case-Control Cohorts. Lancet Respir. Med. 2020, 8, 696–708, Correction in Lancet Respir. Med. 2024, 12, E70. [Google Scholar] [CrossRef]
- Morieri, M.L.; Gao, H.; Pigeyre, M.; Shah, H.S.; Sjaarda, J.; Mendonca, C.; Hastings, T.; Buranasupkajorn, P.; Motsinger-Reif, A.A.; Rotroff, D.M.; et al. Genetic Tools for Coronary Risk Assessment in Type 2 Diabetes: A Cohort Study from the ACCORD Clinical Trial. Diabetes Care 2018, 41, 2404–2413. [Google Scholar] [CrossRef]
- Mukadam, N.; Giannakopoulou, O.; Bass, N.; Kuchenbaecker, K.; McQuillin, A. Genetic Risk Scores and Dementia Risk across Different Ethnic Groups in UK Biobank. PLoS ONE 2022, 17, e0277378. [Google Scholar] [CrossRef]
- Namba, S.; Saito, Y.; Kogure, Y.; Masuda, T.; Bondy, M.L.; Gharahkhani, P.; Gockel, I.; Heider, D.; Hillmer, A.; Jankowski, J.; et al. Common Germline Risk Variants Impact Somatic Alterations and Clinical Features across Cancers. Cancer Res. 2023, 83, 20–27. [Google Scholar] [CrossRef]
- Namjou, B.; Lape, M.; Malolepsza, E.; DeVore, S.B.; Weirauch, M.T.; Dikilitas, O.; Jarvik, G.P.; Kiryluk, K.; Kullo, I.J.; Liu, C.; et al. Multiancestral Polygenic Risk Score for Pediatric Asthma. J. Allergy Clin. Immunol. 2022, 150, 1086–1096. [Google Scholar] [CrossRef]
- Oh, J.J.; Kim, E.; Woo, E.; Song, S.H.; Kim, J.K.; Lee, H.; Lee, S.; Hong, S.K.; Byun, S.-S. Evaluation of Polygenic Risk Scores for Prediction of Prostate Cancer in Korean Men. Front. Oncol. 2020, 10, 583625. [Google Scholar] [CrossRef] [PubMed]
- Ohbe, H.; Hachiya, T.; Yamaji, T.; Nakano, S.; Miyamoto, Y.; Sutoh, Y.; Otsuka-Yamasaki, Y.; Shimizu, A.; Yasunaga, H.; Sawada, N.; et al. Development and Validation of Genome-Wide Polygenic Risk Scores for Predicting Breast Cancer Incidence in Japanese Females: A Population-Based Case-Cohort Study. Breast Cancer Res. Treat. 2023, 197, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Ohta, R.; Tanigawa, Y.; Suzuki, Y.; Kellis, M.; Morishita, S. A Polygenic Score Method Boosted by Non-Additive Models. Nat. Commun. 2024, 15, 4433. [Google Scholar] [CrossRef] [PubMed]
- Ojima, T.; Namba, S.; Suzuki, K.; Yamamoto, K.; Sonehara, K.; Narita, A.; Tohoku Medical Megabank Project Study Group; Biobank Japan Project; Kamatani, Y.; Tamiya, G.; et al. Body Mass Index Stratification Optimizes Polygenic Prediction of Type 2 Diabetes in Cross-Biobank Analyses. Nat. Genet. 2024, 56, 1100–1109. [Google Scholar] [CrossRef]
- Paquette, M.; Chong, M.; Thériault, S.; Dufour, R.; Paré, G.; Baass, A. Polygenic Risk Score Predicts Prevalence of Cardiovascular Disease in Patients with Familial Hypercholesterolemia. J. Clin. Lipidol. 2017, 11, 725–732.e5. [Google Scholar] [CrossRef]
- Parcha, V.; Pampana, A.; Shetty, N.S.; Irvin, M.R.; Natarajan, P.; Lin, H.J.; Guo, X.; Rich, S.S.; Rotter, J.I.; Li, P.; et al. Association of a Multiancestry Genome-Wide Blood Pressure Polygenic Risk Score with Adverse Cardiovascular Events. Circ. Genom. Precis. Med. 2022, 15, e003946. [Google Scholar] [CrossRef]
- Pashayan, N.; Pharoah, P.D.; Schleutker, J.; Talala, K.; Tammela, T.L.; Määttänen, L.; Harrington, P.; Tyrer, J.; Eeles, R.; Duffy, S.W.; et al. Reducing Overdiagnosis by Polygenic Risk-Stratified Screening: Findings from the Finnish Section of the ERSPC. Br. J. Cancer 2015, 113, 1086–1093. [Google Scholar] [CrossRef]
- Patel, A.P.; Wang, M.; Ruan, Y.; Koyama, S.; Clarke, S.L.; Yang, X.; Tcheandjieu, C.; Agrawal, S.; Fahed, A.C.; Ellinor, P.T.; et al. A Multi-Ancestry Polygenic Risk Score Improves Risk Prediction for Coronary Artery Disease. Nat. Med. 2023, 29, 1793–1803. [Google Scholar] [CrossRef]
- Ping, J.; Yang, Y.; Wen, W.; Kweon, S.-S.; Matsuda, K.; Jia, W.-H.; Shin, A.; Gao, Y.-T.; Matsuo, K.; Kim, J.; et al. Developing and Validating Polygenic Risk Scores for Colorectal Cancer Risk Prediction in East Asians. Int. J. Cancer 2022, 151, 1726–1736. [Google Scholar] [CrossRef]
- Pirruccello, J.P.; Chaffin, M.D.; Chou, E.L.; Fleming, S.J.; Lin, H.; Nekoui, M.; Khurshid, S.; Friedman, S.F.; Bick, A.G.; Arduini, A.; et al. Deep Learning Enables Genetic Analysis of the Human Thoracic Aorta. Nat. Genet. 2022, 54, 40–51. [Google Scholar] [CrossRef]
- Pirruccello, J.P.; Khurshid, S.; Lin, H.; Weng, L.-C.; Zamirpour, S.; Kany, S.; Raghavan, A.; Koyama, S.; Vasan, R.S.; Benjamin, E.J.; et al. The AORTA Gene Score for Detection and Risk Stratification of Ascending Aortic Dilation. Eur. Heart J. 2024, 45, 4318–4332. [Google Scholar] [CrossRef] [PubMed]
- Privé, F.; Aschard, H.; Carmi, S.; Folkersen, L.; Hoggart, C.; O’Reilly, P.F.; Vilhjálmsson, B.J. Portability of 245 Polygenic Scores When Derived from the UK Biobank and Applied to 9 Ancestry Groups from the Same Cohort. Am. J. Hum. Genet. 2022, 109, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Raben, T.G.; Lello, L.; Widen, E.; Hsu, S.D.H. Biobank-Scale Methods and Projections for Sparse Polygenic Prediction from Machine Learning. Sci. Rep. 2023, 13, 11662. [Google Scholar] [CrossRef]
- Revez, J.A.; Lin, T.; Qiao, Z.; Xue, A.; Holtz, Y.; Zhu, Z.; Zeng, J.; Wang, H.; Sidorenko, J.; Kemper, K.E.; et al. Genome-Wide Association Study Identifies 143 Loci Associated with 25 Hydroxyvitamin D Concentration. Nat. Commun. 2020, 11, 1647. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, S.C.; Lambert, S.A.; Arnold, M.; Teo, S.M.; Lim, S.; Scepanovic, P.; Marten, J.; Zahid, S.; Chaffin, M.; Liu, Y.; et al. Integrative Analysis of the Plasma Proteome and Polygenic Risk of Cardiometabolic Diseases. Nat. Metab. 2021, 3, 1476–1483. [Google Scholar] [CrossRef]
- Ritchie, S.C.; Taylor, H.J.; Liang, Y.; Manikpurage, H.D.; Pennells, L.; Foguet, C.; Abraham, G.; Gibson, J.T.; Jiang, X.; Liu, Y.; et al. Integrated Clinical Risk Prediction of Type 2 Diabetes with a Multifactorial Polygenic Risk Score. medRxiv 2024. [Google Scholar] [CrossRef]
- Robinson, J.R.; Carroll, R.J.; Bastarache, L.; Chen, Q.; Pirruccello, J.; Mou, Z.; Wei, W.-Q.; Connolly, J.; Mentch, F.; Crane, P.K.; et al. Quantifying the Phenome-Wide Disease Burden of Obesity Using Electronic Health Records and Genomics. Obesity 2022, 30, 2477–2488. [Google Scholar] [CrossRef]
- Ruan, X.; Huang, D.; Huang, J.; Xu, D.; Na, R. Application of European-Specific Polygenic Risk Scores for Predicting Prostate Cancer Risk in Different Ancestry Populations. Prostate 2023, 83, 30–38. [Google Scholar] [CrossRef]
- Ruan, X.; Huang, D.; Huang, J.; Huang, J.; Zhan, Y.; Wu, Y.; Ding, Q.; Xu, D.; Jiang, H.; Xue, W.; et al. The Combined Effect of Polygenic Risk Score and Prostate Health Index in Chinese Men Undergoing Prostate Biopsy. J. Clin. Med. 2023, 12, 1343. [Google Scholar] [CrossRef]
- Sapkota, Y.; Qiu, W.; Dixon, S.B.; Wilson, C.L.; Wang, Z.; Zhang, J.; Leisenring, W.; Chow, E.J.; Bhatia, S.; Armstrong, G.T.; et al. Genetic Risk Score Enhances the Risk Prediction of Severe Obesity in Adult Survivors of Childhood Cancer. Nat. Med. 2022, 28, 1590–1598. [Google Scholar] [CrossRef]
- Sato, J.R.; Biazoli, C.E.; Bueno, A.P.A.; Caye, A.; Pan, P.M.; Santoro, M.; Honorato-Mauer, J.; Salum, G.A.; Hoexter, M.Q.; Bressan, R.A.; et al. Polygenic Risk Score for Attention-Deficit/Hyperactivity Disorder and Brain Functional Networks Segregation in a Community-Based Sample. Genes. Brain Behav. 2023, 22, e12838. [Google Scholar] [CrossRef] [PubMed]
- Schoepf, I.C.; Thorball, C.W.; Ledergerber, B.; Engel, T.; Raffenberg, M.; Kootstra, N.A.; Reiss, P.; Hasse, B.; Marzolini, C.; Thurnheer, C.; et al. Coronary Artery Disease-Associated and Longevity-Associated Polygenic Risk Scores for Prediction of Coronary Artery Disease Events in Persons Living With Human Immunodeficiency Virus: The Swiss HIV Cohort Study. Clin. Infect. Dis. 2021, 73, 1597–1604. [Google Scholar] [CrossRef] [PubMed]
- Shams, H.; Shao, X.; Santaniello, A.; Kirkish, G.; Harroud, A.; Ma, Q.; Isobe, N.; University of California San Francisco MS-EPIC Team; Schaefer, C.A.; McCauley, J.L.; et al. Polygenic Risk Score Association with Multiple Sclerosis Susceptibility and Phenotype in Europeans. Brain 2023, 146, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Shetty, N.S.; Pampana, A.; Patel, N.; Li, P.; Yerabolu, K.; Gaonkar, M.; Arora, G.; Arora, P. Sex Differences in the Association of Genome-Wide Systolic Blood Pressure Polygenic Risk Score with Hypertension. Circ. Genom. Precis. Med. 2023, 16, e004259. [Google Scholar] [CrossRef]
- Shi, M.; Shelley, J.P.; Schaffer, K.R.; Tosoian, J.J.; Bagheri, M.; Witte, J.S.; Kachuri, L.; Mosley, J.D. Clinical Consequences of a Genetic Predisposition toward Higher Benign Prostate-Specific Antigen Levels. EBioMedicine 2023, 97, 104838. [Google Scholar] [CrossRef]
- Shi, Z.; Yu, H.; Wu, Y.; Lin, X.; Bao, Q.; Jia, H.; Perschon, C.; Duggan, D.; Helfand, B.T.; Zheng, S.L.; et al. Systematic Evaluation of Cancer-Specific Genetic Risk Score for 11 Types of Cancer in The Cancer Genome Atlas and Electronic Medical Records and Genomics Cohorts. Cancer Med. 2019, 8, 3196–3205. [Google Scholar] [CrossRef]
- Shi, Z.; Zhan, J.; Wei, J.; Ladson-Gary, S.; Wang, C.-H.; Hulick, P.J.; Zheng, S.L.; Cooney, K.A.; Isaacs, W.B.; Helfand, B.T.; et al. Reliability of Ancestry-Specific Prostate Cancer Genetic Risk Score in Four Racial and Ethnic Populations. Eur. Urol. Open Sci. 2022, 45, 23–30. [Google Scholar] [CrossRef]
- Shrine, N.; Izquierdo, A.G.; Chen, J.; Packer, R.; Hall, R.J.; Guyatt, A.L.; Batini, C.; Thompson, R.J.; Pavuluri, C.; Malik, V.; et al. Multi-Ancestry Genome-Wide Association Analyses Improve Resolution of Genes and Pathways Influencing Lung Function and Chronic Obstructive Pulmonary Disease Risk. Nat. Genet. 2023, 55, 410–422. [Google Scholar] [CrossRef]
- Sinnott-Armstrong, N.; Tanigawa, Y.; Amar, D.; Mars, N.; Benner, C.; Aguirre, M.; Venkataraman, G.R.; Wainberg, M.; Ollila, H.M.; Kiiskinen, T.; et al. Genetics of 35 Blood and Urine Biomarkers in the UK Biobank. Nat. Genet. 2021, 53, 185–194. [Google Scholar] [CrossRef]
- Sofer, T.; Kurniansyah, N.; Granot-Hershkovitz, E.; Goodman, M.O.; Tarraf, W.; Broce, I.; Lipton, R.B.; Daviglus, M.; Lamar, M.; Wassertheil-Smoller, S.; et al. A Polygenic Risk Score for Alzheimer’s Disease Constructed Using APOE-Region Variants Has Stronger Association than APOE Alleles with Mild Cognitive Impairment in Hispanic/Latino Adults in the U.S. Alzheimers Res. Ther. 2023, 15, 146. [Google Scholar] [CrossRef]
- Sofer, T.; Kurniansyah, N.; Murray, M.; Ho, Y.-L.; Abner, E.; Esko, T.; Estonian Biobank Research Team; Huffman, J.E.; Cho, K.; Wilson, P.W.F.; et al. Genome-Wide Association Study of Obstructive Sleep Apnoea in the Million Veteran Program Uncovers Genetic Heterogeneity by Sex. EBioMedicine 2023, 90, 104536. [Google Scholar] [CrossRef]
- Song, S.H.; Kim, E.; Woo, E.; Kwon, E.; Yoon, S.; Kim, J.K.; Lee, H.; Oh, J.J.; Lee, S.; Hong, S.K.; et al. Prediction of Clinically Significant Prostate Cancer Using Polygenic Risk Models in Asians. Investig. Clin. Urol. 2022, 63, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, J.; Iles, M.M.; Lee, J.Y.; Wang, X.; Law, M.H.; Smit, A.K.; Nguyen-Dumont, T.; Giles, G.G.; Southey, M.C.; Milne, R.L.; et al. Independent Evaluation of Melanoma Polygenic Risk Scores in UK and Australian Prospective Cohorts. Br. J. Dermatol. 2022, 186, 823–834. [Google Scholar] [CrossRef]
- Sumpter, N.A.; Takei, R.; Cadzow, M.; Topless, R.K.G.; Phipps-Green, A.J.; Murphy, R.; de Zoysa, J.; Watson, H.; Qasim, M.; Lupi, A.S.; et al. Association of Gout Polygenic Risk Score with Age at Disease Onset and Tophaceous Disease in European and Polynesian Men with Gout. Arthritis Rheumatol. 2023, 75, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Verma, S.P.; Jia, G.; Wang, X.; Ping, J.; Guo, X.; Shu, X.-O.; Chen, J.; Derkach, A.; Cai, Q.; et al. Case-Case Genome-Wide Analyses Identify Subtype-Informative Variants That Confer Risk for Breast Cancer. Cancer Res. 2024, 84, 2533–2548. [Google Scholar] [CrossRef]
- Tamlander, M.; Jermy, B.; Seppälä, T.T.; Färkkilä, M.; Gen, F.; Widén, E.; Ripatti, S.; Mars, N. Genome-Wide Polygenic Risk Scores for Colorectal Cancer Have Implications for Risk-Based Screening. Br. J. Cancer 2024, 130, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Tanigawa, Y.; Qian, J.; Venkataraman, G.; Justesen, J.M.; Li, R.; Tibshirani, R.; Hastie, T.; Rivas, M.A. Significant Sparse Polygenic Risk Scores across 813 Traits in UK Biobank. PLoS Genet. 2022, 18, e1010105. [Google Scholar] [CrossRef]
- Testori, A.; Vaksman, Z.; Diskin, S.J.; Hakonarson, H.; Capasso, M.; Iolascon, A.; Maris, J.M.; Devoto, M. Genetic Analysis in African American Children Supports Ancestry-Specific Neuroblastoma Susceptibility. Cancer Epidemiol. Biomarkers Prev. 2022, 31, 870–875. [Google Scholar] [CrossRef]
- Trinder, M.; Uddin, M.M.; Finneran, P.; Aragam, K.G.; Natarajan, P. Clinical Utility of Lipoprotein(a) and LPA Genetic Risk Score in Risk Prediction of Incident Atherosclerotic Cardiovascular Disease. JAMA Cardiol. 2021, 6, 287–295. [Google Scholar] [CrossRef]
- Valenti, L.; Tripodi, A.; La Mura, V.; Pelusi, S.; Bianco, C.; Scalambrino, E.; Margarita, S.; Malvestiti, F.; Ronzoni, L.; Clerici, M.; et al. Clinical and Genetic Determinants of the Fatty Liver-Coagulation Balance Interplay in Individuals with Metabolic Dysfunction. JHEP Rep. 2022, 4, 100598. [Google Scholar] [CrossRef]
- Vaura, F.; Kauko, A.; Suvila, K.; Havulinna, A.S.; Mars, N.; Salomaa, V.; Gen, F.; Cheng, S.; Niiranen, T. Polygenic Risk Scores Predict Hypertension Onset and Cardiovascular Risk. Hypertension 2021, 77, 1119–1127. [Google Scholar] [CrossRef]
- Wang, J.; Dron, J.S.; Ban, M.R.; Robinson, J.F.; McIntyre, A.D.; Alazzam, M.; Zhao, P.J.; Dilliott, A.A.; Cao, H.; Huff, M.W.; et al. Polygenic Versus Monogenic Causes of Hypercholesterolemia Ascertained Clinically. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 2439–2445. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Namba, S.; Lopera, E.; Kerminen, S.; Tsuo, K.; Läll, K.; Kanai, M.; Zhou, W.; Wu, K.-H.; Favé, M.-J.; et al. Global Biobank Analyses Provide Lessons for Developing Polygenic Risk Scores across Diverse Cohorts. Cell Genom. 2023, 3, 100241. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-F.; Zhang, Y.; Lin, Z.; Zhang, H.; Wang, T.-Y.; Cao, Y.; Morris, D.L.; Sheng, Y.; Yin, X.; Zhong, S.-L.; et al. Identification of 38 Novel Loci for Systemic Lupus Erythematosus and Genetic Heterogeneity between Ancestral Groups. Nat. Commun. 2021, 12, 772. [Google Scholar] [CrossRef] [PubMed]
- Weissbrod, O.; Kanai, M.; Shi, H.; Gazal, S.; Peyrot, W.J.; Khera, A.V.; Okada, Y.; Biobank Japan Project; Martin, A.R.; Finucane, H.K.; et al. Leveraging Fine-Mapping and Multipopulation Training Data to Improve Cross-Population Polygenic Risk Scores. Nat. Genet. 2022, 54, 450–458. [Google Scholar] [CrossRef]
- Xicota, L.; Gyorgy, B.; Grenier-Boley, B.; Lecoeur, A.; Fontaine, G.; Danjou, F.; Gonzalez, J.S.; Colliot, O.; Amouyel, P.; Martin, G.; et al. Association of APOE-Independent Alzheimer Disease Polygenic Risk Score with Brain Amyloid Deposition in Asymptomatic Older Adults. Neurology 2022, 99, e462–e475. [Google Scholar] [CrossRef]
- Xu, J.; Isaacs, W.B.; Mamawala, M.; Shi, Z.; Landis, P.; Petkewicz, J.; Wei, J.; Wang, C.-H.; Resurreccion, W.K.; Na, R.; et al. Association of Prostate Cancer Polygenic Risk Score with Number and Laterality of Tumor Cores in Active Surveillance Patients. Prostate 2021, 81, 703–709. [Google Scholar] [CrossRef]
- Xu, Y.; Vuckovic, D.; Ritchie, S.C.; Akbari, P.; Jiang, T.; Grealey, J.; Butterworth, A.S.; Ouwehand, W.H.; Roberts, D.J.; Di Angelantonio, E.; et al. Machine Learning Optimized Polygenic Scores for Blood Cell Traits Identify Sex-Specific Trajectories and Genetic Correlations with Disease. Cell Genom. 2022, 2, 100086. [Google Scholar] [CrossRef]
- Yang, Y.; Tao, R.; Shu, X.; Cai, Q.; Wen, W.; Gu, K.; Gao, Y.-T.; Zheng, Y.; Kweon, S.-S.; Shin, M.-H.; et al. Incorporating Polygenic Risk Scores and Nongenetic Risk Factors for Breast Cancer Risk Prediction Among Asian Women. JAMA Netw. Open 2022, 5, e2149030. [Google Scholar] [CrossRef]
- Yu, Z.; Jin, J.; Tin, A.; Köttgen, A.; Yu, B.; Chen, J.; Surapaneni, A.; Zhou, L.; Ballantyne, C.M.; Hoogeveen, R.C.; et al. Polygenic Risk Scores for Kidney Function and Their Associations with Circulating Proteome, and Incident Kidney Diseases. J. Am. Soc. Nephrol. 2021, 32, 3161–3173. [Google Scholar] [CrossRef]
- Zhang, H.; Ahearn, T.U.; Lecarpentier, J.; Barnes, D.; Beesley, J.; Qi, G.; Jiang, X.; O’Mara, T.A.; Zhao, N.; Bolla, M.K.; et al. Genome-Wide Association Study Identifies 32 Novel Breast Cancer Susceptibility Loci from Overall and Subtype-Specific Analyses. Nat. Genet. 2020, 52, 572–581. [Google Scholar] [CrossRef]
- Zhang, H.; Zhan, J.; Jin, J.; Zhang, J.; Lu, W.; Zhao, R.; Ahearn, T.U.; Yu, Z.; O’Connell, J.; Jiang, Y.; et al. A New Method for Multiancestry Polygenic Prediction Improves Performance across Diverse Populations. Nat. Genet. 2023, 55, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Elgart, M.; Kurniansyah, N.; Spitzer, B.W.; Wang, H.; Kim, D.; Shah, N.; Daviglus, M.; Zee, P.C.; Cai, J.; et al. Genetic Determinants of Cardiometabolic and Pulmonary Phenotypes and Obstructive Sleep Apnoea in HCHS/SOL. EBioMedicine 2022, 84, 104288. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.L.; Henry, A.; Cannie, D.; Lee, M.; Miller, D.; McGurk, K.A.; Bond, I.; Xu, X.; Issa, H.; Francis, C.; et al. Genome-Wide Association Analysis Provides Insights into the Molecular Etiology of Dilated Cardiomyopathy. Nat. Genet. 2024, 56, 2646–2658. [Google Scholar] [CrossRef] [PubMed]
- Zheutlin, A.B.; Dennis, J.; Karlsson Linnér, R.; Moscati, A.; Restrepo, N.; Straub, P.; Ruderfer, D.; Castro, V.M.; Chen, C.-Y.; Ge, T.; et al. Penetrance and Pleiotropy of Polygenic Risk Scores for Schizophrenia in 106,160 Patients Across Four Health Care Systems. Am. J. Psychiatry 2019, 176, 846–855. [Google Scholar] [CrossRef]
- Zubair, N.; Conomos, M.P.; Hood, L.; Omenn, G.S.; Price, N.D.; Spring, B.J.; Magis, A.T.; Lovejoy, J.C. Genetic Predisposition Impacts Clinical Changes in a Lifestyle Coaching Program. Sci. Rep. 2019, 9, 6805. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Tadaka, S.; Katsuoka, F.; Ueki, M.; Kojima, K.; Makino, S.; Saito, S.; Otsuki, A.; Gocho, C.; Sakurai-Yageta, M.; Danjoh, I.; et al. 3.5KJPNv2: An Allele Frequency Panel of 3552 Japanese Individuals Including the X Chromosome. Hum. Genome Var. 2019, 6, 28. [Google Scholar] [CrossRef]
- Miyano, T.; Mikkaichi, T.; Nakamura, K.; Yoshigae, Y.; Abernathy, K.; Ogura, Y.; Kiyosawa, N. Circulating microRNA Profiles Identify a Patient Subgroup with High Inflammation and Severe Symptoms in Schizophrenia Experiencing Acute Psychosis. Int. J. Mol. Sci. 2024, 25, 4291. [Google Scholar] [CrossRef]
- Miyano, T.; Hirouchi, M.; Yoshimura, N.; Hattori, K.; Mikkaichi, T.; Kiyosawa, N. Plasma microRNAs Associate Positive, Negative, and Cognitive Symptoms with Inflammation in Schizophrenia. Int. J. Mol. Sci. 2024, 25, 13522. [Google Scholar] [CrossRef]
- Malone, J.; Holloway, E.; Adamusiak, T.; Kapushesky, M.; Zheng, J.; Kolesnikov, N.; Zhukova, A.; Brazma, A.; Parkinson, H. Modeling Sample Variables with an Experimental Factor Ontology. Bioinformatics 2010, 26, 1112–1118. [Google Scholar] [CrossRef] [PubMed]
- Pain, O.; Gillett, A.C.; Austin, J.C.; Folkersen, L.; Lewis, C.M. A Tool for Translating Polygenic Scores onto the Absolute Scale Using Summary Statistics. Eur. J. Hum. Genet. 2022, 30, 339–348. [Google Scholar] [CrossRef]
- Sandling, J.K.; Pucholt, P.; Hultin Rosenberg, L.; Farias, F.H.G.; Kozyrev, S.V.; Eloranta, M.-L.; Alexsson, A.; Bianchi, M.; Padyukov, L.; Bengtsson, C.; et al. Molecular Pathways in Patients with Systemic Lupus Erythematosus Revealed by Gene-Centred DNA Sequencing. Ann. Rheum. Dis. 2021, 80, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Sharew, N.T.; Clark, S.R.; Schubert, K.O.; Amare, A.T. Pharmacogenomic Scores in Psychiatry: Systematic Review of Current Evidence. Transl. Psychiatry 2024, 14, 322. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.-K.; Su, Y.; Zhang, Y.-Y.-N.; Yu, H.; Lu, Z.; Li, W.-Q.; Yang, Y.-F.; Xiao, X.; Yan, H.; Lu, T.-L.; et al. Prediction of Treatment Response to Antipsychotic Drugs for Precision Medicine Approach to Schizophrenia: Randomized Trials and Multiomics Analysis. Mil. Med. Res. 2023, 10, 24. [Google Scholar] [CrossRef]
- Werner, M.C.F.; Wirgenes, K.V.; Haram, M.; Bettella, F.; Lunding, S.H.; Rødevand, L.; Hjell, G.; Agartz, I.; Djurovic, S.; Melle, I.; et al. Indicated Association between Polygenic Risk Score and Treatment-Resistance in a Naturalistic Sample of Patients with Schizophrenia Spectrum Disorders. Schizophr. Res. 2020, 218, 55–62. [Google Scholar] [CrossRef]
- Amare, A.T.; Schubert, K.O.; Hou, L.; Clark, S.R.; Papiol, S.; Cearns, M.; Heilbronner, U.; Degenhardt, F.; Tekola-Ayele, F.; Hsu, Y.-H.; et al. Association of Polygenic Score for Major Depression with Response to Lithium in Patients with Bipolar Disorder. Mol. Psychiatry 2021, 26, 2457–2470. [Google Scholar] [CrossRef]
- International Consortium on Lithium Genetics (ConLi+Gen); Amare, A.T.; Schubert, K.O.; Hou, L.; Clark, S.R.; Papiol, S.; Heilbronner, U.; Degenhardt, F.; Tekola-Ayele, F.; Hsu, Y.-H.; et al. Association of Polygenic Score for Schizophrenia and HLA Antigen and Inflammation Genes With Response to Lithium in Bipolar Affective Disorder: A Genome-Wide Association Study. JAMA Psychiatry 2018, 75, 65–74. [Google Scholar] [CrossRef]
- Nuttall, F.Q. Body Mass Index: Obesity, BMI, and Health: A Critical Review. Nutr. Today 2015, 50, 117–128. [Google Scholar] [CrossRef]
- Zheng, Z.; Zhang, L.; Li, S.; Zhao, F.; Wang, Y.; Huang, L.; Huang, J.; Zou, R.; Qu, Y.; Mu, D. Association among Obesity, Overweight and Autism Spectrum Disorder: A Systematic Review and Meta-Analysis. Sci. Rep. 2017, 7, 11697. [Google Scholar] [CrossRef]
- Sammels, O.; Karjalainen, L.; Dahlgren, J.; Wentz, E. Autism Spectrum Disorder and Obesity in Children: A Systematic Review and Meta-Analysis. Obes. Facts 2022, 15, 305–320. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.E.; Pinto-Martin, J.A.; Bradley, C.B.; Chittams, J.; Johnson, S.L.; Pandey, J.; Pomykacz, A.; Ramirez, A.; Reynolds, A.; Rubenstein, E.; et al. Relationship of Weight Outcomes, Co-Occurring Conditions, and Severity of Autism Spectrum Disorder in the Study to Explore Early Development. J. Pediatr. 2019, 205, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Dhaliwal, K.K.; Orsso, C.E.; Richard, C.; Haqq, A.M.; Zwaigenbaum, L. Risk Factors for Unhealthy Weight Gain and Obesity among Children with Autism Spectrum Disorder. Int. J. Mol. Sci. 2019, 20, 3285. [Google Scholar] [CrossRef] [PubMed]
- Must, A.; Curtin, C.; Hubbard, K.; Sikich, L.; Bedford, J.; Bandini, L. Obesity Prevention for Children with Developmental Disabilities. Curr. Obes. Rep. 2014, 3, 156–170. [Google Scholar] [CrossRef]
- Makin, L.; Meyer, A.; Zesch, E.; Mondelli, V.; Tchanturia, K. Autism, ADHD, and Their Traits in Adults with Obesity: A Scoping Review. Nutrients 2025, 17, 787. [Google Scholar] [CrossRef]
- Crowley, B.; Howe, Y.J.; McDougle, C.J. Topiramate for Weight Loss in Two Young Adult Women with Autism Spectrum Disorder. J. Child. Adolesc. Psychopharmacol. 2015, 25, 183–185. [Google Scholar] [CrossRef]
- Järvinen, A.; Laine, M.K.; Tikkanen, R.; Castrén, M.L. Beneficial Effects of GLP-1 Agonist in a Male with Compulsive Food-Related Behavior Associated with Autism. Front. Psychiatry 2019, 10, 97. [Google Scholar] [CrossRef]
- Nakashima, R.; Ikeda, S.; Shinohara, K.; Matsumoto, S.; Yoshida, D.; Ono, Y.; Nakashima, H.; Miyamoto, R.; Matsushima, S.; Kishimoto, J.; et al. Triglyceride/High Density Lipoprotein Cholesterol Index and Future Cardiovascular Events in Diabetic Patients without Known Cardiovascular Disease. Sci. Rep. 2025, 15, 9217. [Google Scholar] [CrossRef]
- Dhanasekara, C.S.; Ancona, D.; Cortes, L.; Hu, A.; Rimu, A.H.; Robohm-Leavitt, C.; Payne, D.; Wakefield, S.M.; Mastergeorge, A.M.; Kahathuduwa, C.N. Association Between Autism Spectrum Disorders and Cardiometabolic Diseases: A Systematic Review and Meta-Analysis. JAMA Pediatr. 2023, 177, 248–257. [Google Scholar] [CrossRef]
- Luçardo, J.d.C.; Monk, G.F.; Dias, M.d.S.; Martins-Silva, T.; Fernandes, M.P.; Maia, J.C.; Valle, S.C.; Vaz, J.D.S. Interest in Food and Triglyceride Concentrations in Children and Adolescents with Autistic Spectrum Disorder. J. Pediatr. 2021, 97, 103–108. [Google Scholar] [CrossRef]
- Doaei, S.; Bourbour, F.; Teymoori, Z.; Jafari, F.; Kalantari, N.; Abbas Torki, S.; Ashoori, N.; Nemat Gorgani, S.; Gholamalizadeh, M. The Effect of Omega-3 Fatty Acids Supplementation on Social and Behavioral Disorders of Children with Autism: A Randomized Clinical Trial. Pediatr. Endocrinol. Diabetes Metab. 2021, 27, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Al-Beltagi, M.; Saeed, N.K.; Bediwy, A.S.; Elbeltagi, R. Metabolomic Changes in Children with Autism. World J. Clin. Pediatr. 2024, 13, 92737. [Google Scholar] [CrossRef] [PubMed]
- Salazar, J.H. Overview of Urea and Creatinine. Lab. Med. 2014, 45, e19–e20. [Google Scholar] [CrossRef]
- Clothier, J.; Absoud, M. Autism Spectrum Disorder and Kidney Disease. Pediatr. Nephrol. 2021, 36, 2987–2995. [Google Scholar] [CrossRef]
- Capal, J.K.; Williams, M.E.; Pearson, D.A.; Kissinger, R.; Horn, P.S.; Murray, D.; Currans, K.; Kent, B.; Bebin, M.; Northrup, H.; et al. Profile of Autism Spectrum Disorder in Tuberous Sclerosis Complex: Results from a Longitudinal, Prospective, Multisite Study. Ann. Neurol. 2021, 90, 874–886. [Google Scholar] [CrossRef]
- Kingswood, J.C.; Belousova, E.; Benedik, M.P.; Carter, T.; Cottin, V.; Curatolo, P.; Dahlin, M.; D’ Amato, L.; d’Augères, G.B.; de Vries, P.J.; et al. Renal Angiomyolipoma in Patients with Tuberous Sclerosis Complex: Findings from the TuberOus SClerosis Registry to Increase Disease Awareness. Nephrol. Dial. Transplant. 2019, 34, 502–508. [Google Scholar] [CrossRef]
- Ewalt, D.H.; Sheffield, E.; Sparagana, S.P.; Delgado, M.R.; Roach, E.S. Renal Lesion Growth in Children with Tuberous Sclerosis Complex. J. Urol. 1998, 160, 141–145. [Google Scholar] [CrossRef]
- Bissler, J.J.; Budde, K.; Sauter, M.; Franz, D.N.; Zonnenberg, B.A.; Frost, M.D.; Belousova, E.; Berkowitz, N.; Ridolfi, A.; Christopher Kingswood, J. Effect of Everolimus on Renal Function in Patients with Tuberous Sclerosis Complex: Evidence from EXIST-1 and EXIST-2. Nephrol. Dial. Transplant. 2019, 34, 1000–1008. [Google Scholar] [CrossRef]
- Mizuguchi, M.; Ikeda, H.; Kagitani-Shimono, K.; Yoshinaga, H.; Suzuki, Y.; Aoki, M.; Endo, M.; Yonemura, M.; Kubota, M. Everolimus for Epilepsy and Autism Spectrum Disorder in Tuberous Sclerosis Complex: EXIST-3 Substudy in Japan. Brain Dev. 2019, 41, 1–10. [Google Scholar] [CrossRef]
- Lo, Y.-C.; Tian, H.; Chan, T.F.; Jeon, S.; Alatorre, K.; Dinh, B.L.; Maskarinec, G.; Taparra, K.; Nakatsuka, N.; Yu, M.; et al. The Accuracy of Polygenic Score Models for BMI and Type II Diabetes in the Native Hawaiian Population. Commun. Biol. 2025, 8, 651. [Google Scholar] [CrossRef]
- Chang, X.; Shih, C.C.; Chen, J.; Lee, A.S.; Tan, P.; Wang, L.; Liu, J.; Li, J.; Yuan, J.-M.; Khor, C.C.; et al. Predictive Capabilities of Polygenic Scores in an East-Asian Population-Based Cohort: The Singapore Chinese Health Study. Commun. Biol. 2025, 8, 1228. [Google Scholar] [CrossRef]
| Characteristics | n = 75 |
|---|---|
| Birth year, mean ± S.D. | 2013.9 ± 2.0 |
| Male, n (%) | 55 (73.3) |
| Ethnicity: Japanese, n (%) | 75 (100) |
| PGS Set | Enriched Trait | Nmapped/Nall | q-Value |
|---|---|---|---|
| Twenty distinctive PGSs in subgroup 1 | HDL-C measurement | 6/35 | 8.62 × 10−5 |
| Twenty distinctive PGSs in subgroup 2 | Urea measurement | 3/4 | 6.93 × 10−4 |
| Twenty distinctive PGSs in subgroup 3 | BMI | 10/69 | 1.78 × 10−8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miyano, T.; Mikkaichi, T. Comprehensive Polygenic Score Profiling Reveals Autism Spectrum Disorder Subgroups with Different Genetic Predisposition Related to High-Density Lipoprotein Cholesterol, Urea, and Body Mass Index. Int. J. Transl. Med. 2025, 5, 57. https://doi.org/10.3390/ijtm5040057
Miyano T, Mikkaichi T. Comprehensive Polygenic Score Profiling Reveals Autism Spectrum Disorder Subgroups with Different Genetic Predisposition Related to High-Density Lipoprotein Cholesterol, Urea, and Body Mass Index. International Journal of Translational Medicine. 2025; 5(4):57. https://doi.org/10.3390/ijtm5040057
Chicago/Turabian StyleMiyano, Takuya, and Tsuyoshi Mikkaichi. 2025. "Comprehensive Polygenic Score Profiling Reveals Autism Spectrum Disorder Subgroups with Different Genetic Predisposition Related to High-Density Lipoprotein Cholesterol, Urea, and Body Mass Index" International Journal of Translational Medicine 5, no. 4: 57. https://doi.org/10.3390/ijtm5040057
APA StyleMiyano, T., & Mikkaichi, T. (2025). Comprehensive Polygenic Score Profiling Reveals Autism Spectrum Disorder Subgroups with Different Genetic Predisposition Related to High-Density Lipoprotein Cholesterol, Urea, and Body Mass Index. International Journal of Translational Medicine, 5(4), 57. https://doi.org/10.3390/ijtm5040057






