Sirtuins: Emergent Players in Tissue and Organ Regeneration
Abstract
1. Introduction
2. Sirtuins Regulate Different Cellular Processes
2.1. Epithelial–Mesenchymal Transition and Cellular Migration
2.2. Cellular Proliferation and Cell Cycle
2.3. Osteogenic Chondrogenic and Adipogenic Differentiation
2.4. Connection of Regeneration and Cancer
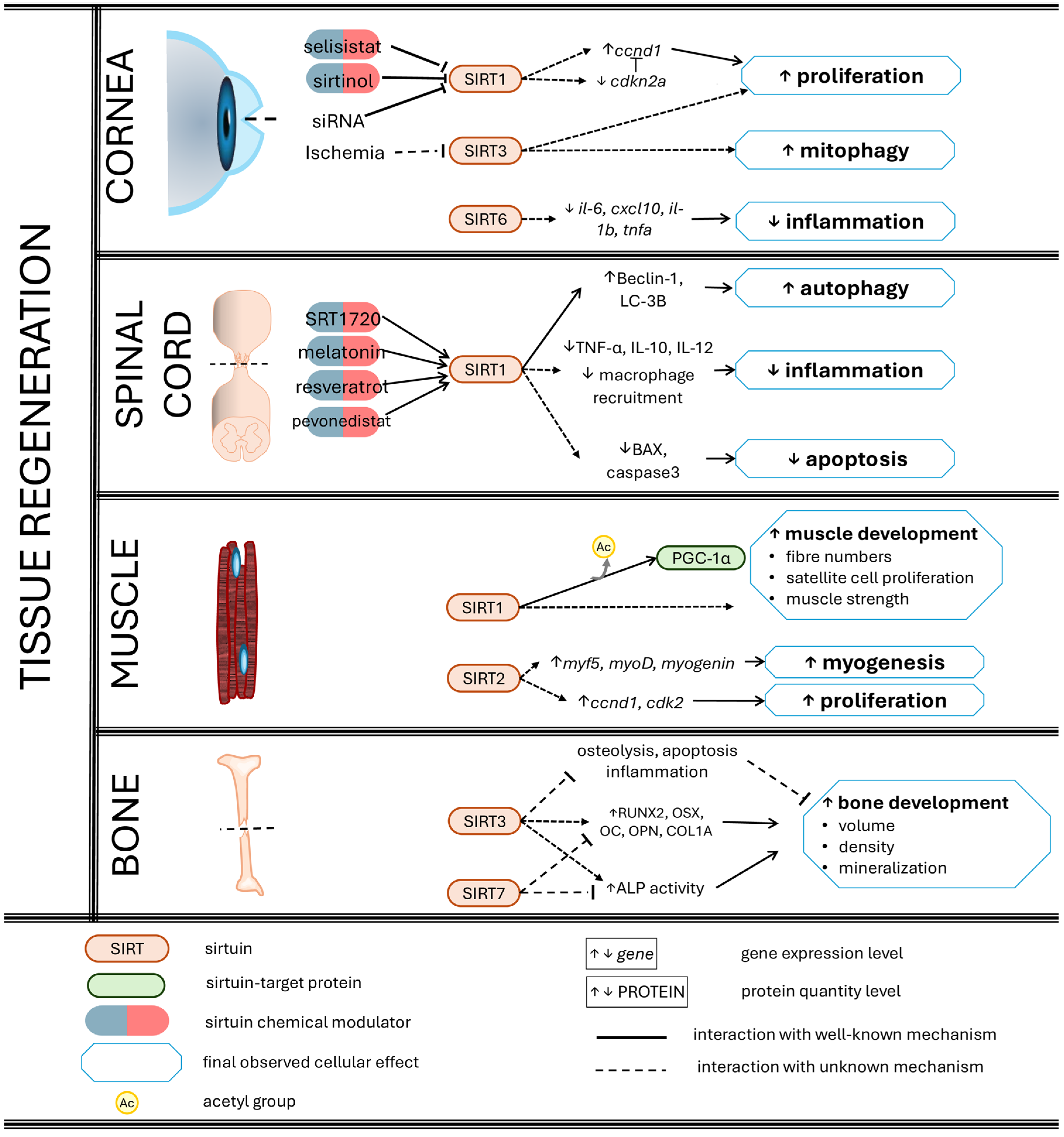
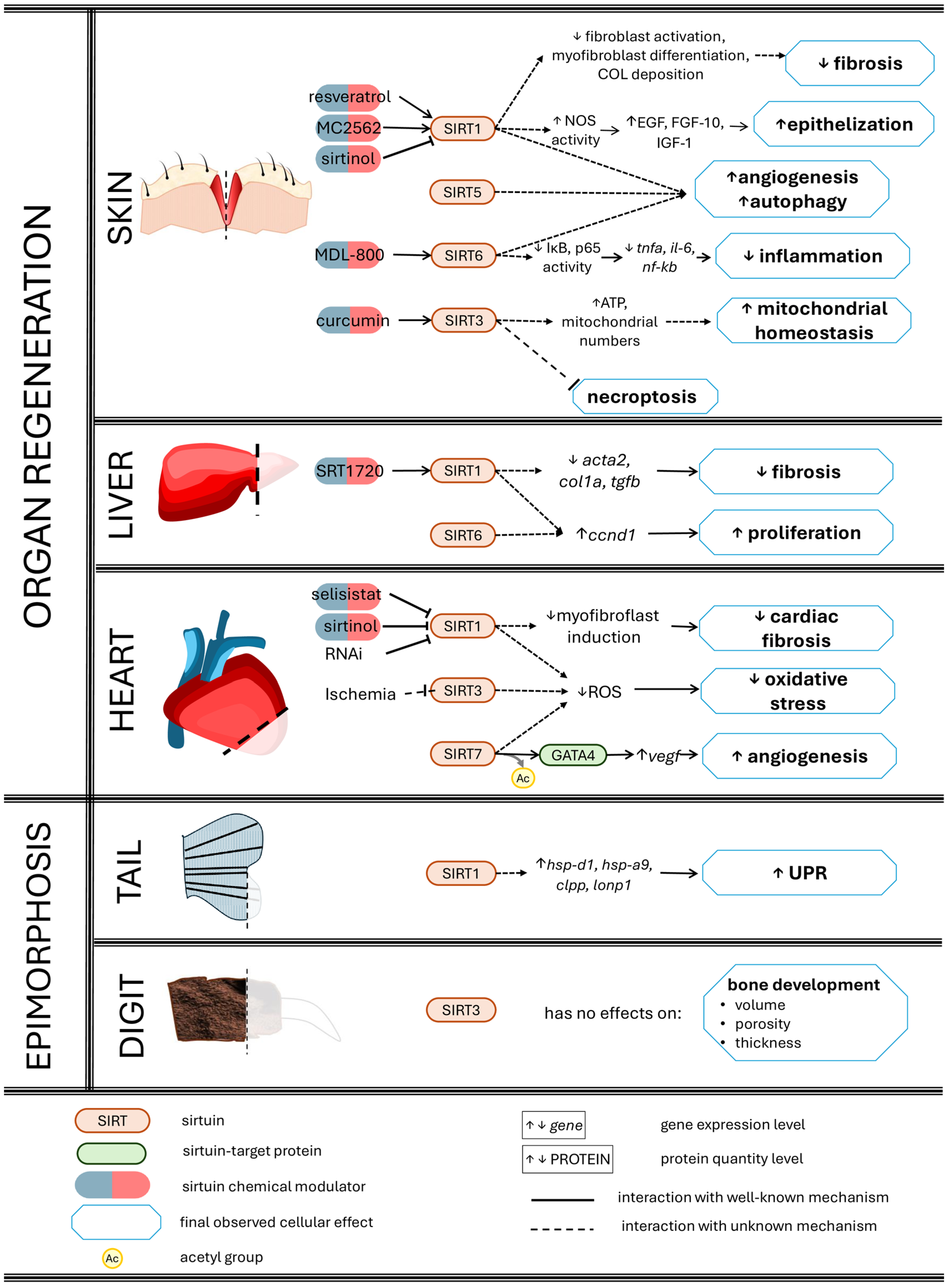
3. Sirtuins Promote Tissue and Organ Regeneration
3.1. Skin Wound Healing
3.2. Corneal Re-Epithelialization
3.3. Liver Regeneration
3.4. Ischaemia-Injured Hearts
3.5. Spinal Cord Regeneration and Motor Restoration
3.6. Bone Regeneration
3.7. Muscle Repair
3.8. Sirtuins as Potential Keys in Epimorphosis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dai, H.; Sinclair, D.A.; Ellis, J.L.; Steegborn, C. Sirtuin Activators and Inhibitors: Promises, Achievements, and Challenges. Pharmacol. Ther. 2018, 188, 140–154. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Liu, H. Sirtuins—Novel Regulators of Epigenetic Alterations in Airway Inflammation. Front. Genet. 2022, 13, 862577. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Kang, W.; Wang, Y.; Ge, J.; Huang, J.; Yang, J.; Yang, W.; Tang, X.; Xie, S. SIRT1 Promotes Osteogenic Differentiation in Human Dental Pulp Stem Cells through Counteracting the Activation of STAT3. Coatings 2021, 11, 1353. [Google Scholar] [CrossRef]
- Deng, Z.; Sun, M.; Wu, J.; Fang, H.; Cai, S.; An, S.; Huang, Q.; Chen, Z.; Wu, C.; Zhou, Z.; et al. SIRT1 Attenuates Sepsis-Induced Acute Kidney Injury via Beclin1 Deacetylation-Mediated Autophagy Activation. Cell Death Dis. 2021, 12, 217. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.; Xiong, H.; Zhan, T.; Cheng, G.; Jia, H.; Ye, Y.; Su, Z.; Chen, H.; Lin, H.; Lai, L.; et al. Sirtuin 1 and Autophagy Attenuate Cisplatin-Induced Hair Cell Death in the Mouse Cochlea and Zebrafish Lateral Line. Front. Cell. Neurosci. 2019, 12, 515. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Shao, Y.; Wu, C.; Ma, X.; Lv, C.; Wang, Q. Metformin Alleviates Oxidative Stress and Enhances Autophagy in Diabetic Kidney Disease via AMPK/SIRT1-FoxO1 Pathway. Mol. Cell. Endocrinol. 2020, 500, 110628. [Google Scholar] [CrossRef]
- Ben Salem, I.; Boussabbeh, M.; Da Silva, J.P.; Guilbert, A.; Bacha, H.; Abid-Essefi, S.; Lemaire, C. SIRT1 Protects Cardiac Cells against Apoptosis Induced by Zearalenone or Its Metabolites α- and β-Zearalenol through an Autophagy-Dependent Pathway. Toxicol. Appl. Pharmacol. 2017, 314, 82–90. [Google Scholar] [CrossRef]
- Luo, G.; Jian, Z.; Zhu, Y.; Zhu, Y.; Chen, B.; Ma, R.; Tang, F.; Xiao, Y. Sirt1 Promotes Autophagy and Inhibits Apoptosis to Protect Cardiomyocytes from Hypoxic Stress. Int. J. Mol. Med. 2019, 43, 2033–2043. [Google Scholar] [CrossRef] [PubMed]
- Gillum, M.P.; Kotas, M.E.; Erion, D.M.; Kursawe, R.; Chatterjee, P.; Nead, K.T.; Muise, E.S.; Hsiao, J.J.; Frederick, D.W.; Yonemitsu, S.; et al. SirT1 Regulates Adipose Tissue Inflammation. Diabetes 2011, 60, 3235–3245. [Google Scholar] [CrossRef]
- Wang, F.; Nguyen, M.; Qin, F.X.F.; Tong, Q. SIRT2 Deacetylates FOXO3a in Response to Oxidative Stress and Caloric Restriction. Aging Cell 2007, 6, 505–514. [Google Scholar] [CrossRef]
- Liu, S.; Gao, X.; Fan, Z.; Wang, Q. SIRT2 Affects Cell Proliferation and Apoptosis by Suppressing the Level of Autophagy in Renal Podocytes. Dis. Markers 2022, 2022, 4586198. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Nakayama, Y.; Li, Y.; Matsumori, H.; Takahashi, H.; Kojima, H.; Wanibuchi, H.; Katoh, M.; Oshimura, M. SIRT2 Knockdown Increases Basal Autophagy and Prevents Postslippage Death by Abnormally Prolonging the Mitotic Arrest That Is Induced by Microtubule Inhibitors. FEBS J. 2014, 281, 2623–2637. [Google Scholar] [CrossRef] [PubMed]
- Gal, J.; Bang, Y.; Choi, H.J. SIRT2 Interferes with Autophagy-Mediated Degradation of Protein Aggregates in Neuronal Cells under Proteasome Inhibition. Neurochem. Int. 2012, 61, 992–1000. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Cao, J.; Hu, K.; He, X.; Yun, D.; Tong, T.; Han, L. Sirtuins and Their Biological Relevance in Aging and Age-Related Diseases. Aging Dis. 2020, 11, 927–945. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Li, Y.; Song, S.; Zhang, Y.; Wang, Y.; Wang, H.; Yang, Z.; Wang, Y. The Dual Role of Sirtuins in Cancer: Biological Functions and Implications. Front. Oncol. 2024, 14, 1384928. [Google Scholar] [CrossRef]
- Bhalla, S.; Gordon, L.I. Functional Characterization of NAD Dependent De-Acetylases SIRT1 and SIRT2 in B-Cell Chronic Lymphocytic Leukemia (CLL). Cancer Biol. Ther. 2016, 17, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.R.; Sanstrum, B.J.; Liu, Y.; Kwon, S.H. Distinct Role of Sirtuin 1 (SIRT1) and Sirtuin 2 (SIRT2) in Inhibiting Cargo-Loading and Release of Extracellular Vesicles. Sci. Rep. 2019, 9, 20049. [Google Scholar] [CrossRef]
- Kabiljo, J.; Murko, C.; Pusch, O.; Zupkovitz, G. Spatio-Temporal Expression Profile of Sirtuins during Aging of the Annual Fish Nothobranchius Furzeri. Gene Expr. Patterns 2019, 33, 11–19. [Google Scholar] [CrossRef]
- Pereira, T.C.B.; Rico, E.P.; Rosemberg, D.B.; Schirmer, H.; Dias, R.D.; Souto, A.A.; Bonan, C.D.; Bogo, M.R. Zebrafish as a Model Organism to Evaluate Drugs Potentially Able to Modulate Sirtuin Expression. Zebrafish 2011, 8, 9–16. [Google Scholar] [CrossRef]
- Sidorova-Darmos, E.; Wither, R.G.; Shulyakova, N.; Fisher, C.; Ratnam, M.; Aarts, M.; Lilge, L.; Monnier, P.P.; Eubanks, J.H. Differential Expression of Sirtuin Family Members in the Developing, Adult, and Aged Rat Brain. Front. Aging Neurosci. 2014, 6, 333. [Google Scholar] [CrossRef]
- Luo, H.; Zhou, M.; Ji, K.; Zhuang, J.; Dang, W.; Fu, S.; Sun, T.; Zhang, X. Expression of Sirtuins in the Retinal Neurons of Mice, Rats, and Humans. Front. Aging Neurosci. 2017, 9, 366. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, C.M.; Ye, Z.C.; Huang, J.; Li, Y.; Lai, W.; Peng, H.; Lou, T. qi Sirt3 Modulates Fatty Acid Oxidation and Attenuates Cisplatin-Induced AKI in Mice. J. Cell. Mol. Med. 2020, 24, 5109–5121. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Quan, Y.; Xia, W. SIRT3 Inhibits Prostate Cancer Metastasis through Regulation of FOXO3A by Suppressing Wnt/β-Catenin Pathway. Exp. Cell Res. 2018, 364, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Ahmedy, O.A.; Abdelghany, T.M.; El-Shamarka, M.E.A.; Khattab, M.A.; El-Tanbouly, D.M. Apigenin Attenuates LPS-Induced Neurotoxicity and Cognitive Impairment in Mice via Promoting Mitochondrial Fusion/Mitophagy: Role of SIRT3/PINK1/Parkin Pathway. Psychopharmacology 2022, 239, 3903–3917. [Google Scholar] [CrossRef] [PubMed]
- D’onofrio, N.; Martino, E.; Mele, L.; Colloca, A.; Maione, M.; Cautela, D.; Castaldo, D.; Balestrieri, M.L. Colorectal Cancer Apoptosis Induced by Dietary δ-Valerobetaine Involves Pink1/Parkin Dependent-Mitophagy and Sirt3. Int. J. Mol. Sci. 2021, 22, 8117. [Google Scholar] [CrossRef]
- Laurent, G.; de Boer, V.C.J.; Finley, L.W.S.; Sweeney, M.; Lu, H.; Schug, T.T.; Cen, Y.; Jeong, S.M.; Li, X.; Sauve, A.A.; et al. SIRT4 Represses Peroxisome Proliferator-Activated Receptor α Activity To Suppress Hepatic Fat Oxidation. Mol. Cell. Biol. 2013, 33, 4552–4561. [Google Scholar] [CrossRef]
- Nasrin, N.; Wu, X.; Fortier, E.; Feng, Y.; Baré, O.C.; Chen, S.; Ren, X.; Wu, Z.; Streeper, R.S.; Bordone, L. SIRT4 Regulates Fatty Acid Oxidation and Mitochondrial Gene Expression in Liver and Muscle Cells. J. Biol. Chem. 2010, 285, 31995–32002. [Google Scholar] [CrossRef]
- Lang, A.; Piekorz, R.P. Novel Role of the SIRT4-OPA1 Axis in Mitochondrial Quality Control. Cell Stress 2018, 2, 1–3. [Google Scholar] [CrossRef]
- Lang, A.; Anand, R.; Altinoluk-Hambüchen, S.; Ezzahoini, H.; Stefanski, A.; Iram, A.; Bergmann, L.; Urbach, J.; Böhler, P.; Hänsel, J.; et al. SIRT4 Interacts with OPA1 and Regulates Mitochondrial Quality Control and Mitophagy. Aging 2017, 9, 2160–2186. [Google Scholar] [CrossRef]
- Jung, Y.H.; Chae, C.W.; Chang, H.S.; Choi, G.E.; Lee, H.J.; Han, H.J. Silencing SIRT5 Induces the Senescence of UCB-MSCs Exposed to TNF-α by Reduction of Fatty Acid β-Oxidation and Anti-Oxidation. Free Radic. Biol. Med. 2022, 192, 1–12. [Google Scholar] [CrossRef]
- Rardin, M.J.; He, W.; Nishida, Y.; Newman, J.C.; Carrico, C.; Danielson, S.R.; Guo, A.; Gut, P.; Sahu, A.K.; Li, B.; et al. SIRT5 Regulates the Mitochondrial Lysine Succinylome and Metabolic Networks. Cell Metab. 2013, 18, 920–933. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bharathi, S.S.; Rardin, M.J.; Lu, J.; Maringer, K.V.; Sims-Lucas, S.; Prochownik, E.V.; Gibson, B.W.; Goetzman, E.S. Lysine Desuccinylase SIRT5 Binds to Cardiolipin and Regulates the Electron Transport Chain. J. Biol. Chem. 2017, 292, 10239–10249. [Google Scholar] [CrossRef] [PubMed]
- Cardus, A.; Uryga, A.K.; Walters, G.; Erusalimsky, J.D. SIRT6 Protects Human Endothelial Cells from DNA Damage, Telomere Dysfunction, and Senescence. Cardiovasc. Res. 2013, 97, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Cea, M.; Cagnetta, A.; Adamia, S.; Acharya, C.; Tai, Y.T.; Fulciniti, M.; Ohguchi, H.; Munshi, A.; Acharya, P.; Bhasin, M.K.; et al. Evidence for a Role of the Histone Deacetylase SIRT6 in DNA Damage Response of Multiple Myeloma Cells. Blood 2016, 127, 1138–1150. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, J.; Sun, X.; Yu, J.; Qian, Z.; Wu, L.; Xu, X.; Wan, X.; Jiang, Y.; Zhang, J.; et al. The SIRT6 Activator MDL-800 Improves Genomic Stability and Pluripotency of Old Murine-Derived IPS Cells. Aging Cell 2020, 19, e13185. [Google Scholar] [CrossRef]
- Geng, A.; Tang, H.; Huang, J.; Qian, Z.; Qin, N.; Yao, Y.; Xu, Z.; Chen, H.; Lan, L.; Xie, H.; et al. The Deacetylase SIRT6 Promotes the Repair of UV-Induced DNA Damage by Targeting DDB2. Nucleic Acids Res. 2020, 48, 9181–9194. [Google Scholar] [CrossRef]
- Gao, Y.; Tan, J.; Jin, J.; Ma, H.; Chen, X.; Leger, B.; Xu, J.; Spagnol, S.T.; Dahl, K.N.; Levine, A.S.; et al. SIRT6 Facilitates Directional Telomere Movement upon Oxidative Damage. Sci. Rep. 2018, 8, 5407. [Google Scholar] [CrossRef]
- Nagai, K.; Matsushita, T.; Matsuzaki, T.; Takayama, K.; Matsumoto, T.; Kuroda, R.; Kurosaka, M. Depletion of SIRT6 Causes Cellular Senescence, DNA Damage, and Telomere Dysfunction in Human Chondrocytes. Osteoarthr. Cartil. 2015, 23, 1412–1420. [Google Scholar] [CrossRef]
- Tang, M.; Li, Z.; Zhang, C.; Lu, X.; Tu, B.; Cao, Z.; Li, Y.; Chen, Y.; Jiang, L.; Wang, H.; et al. SIRT7-Mediated ATM Deacetylation Is Essential for Its Deactivation and DNA Damage Repair. Sci. Adv. 2019, 5, eaav1118. [Google Scholar] [CrossRef]
- Su, Y.; Wu, C.; Chang, Y.; Li, L.; Chen, Y.; Jia, X.; Wang, X.; Lv, Y.; Yu, B.; Yuan, J. USP17L2-SIRT7 Axis Regulates DNA Damage Repair and Chemoresistance in Breast Cancer Cells. Breast Cancer Res. Treat. 2022, 196, 31–44. [Google Scholar] [CrossRef]
- Carlson, B.M. Principles of Regenerative Biology, 1st ed.; Academic Press: Cambridge, MA, USA, 2007; ISBN 9780123694393. [Google Scholar]
- Zeisberg, M.; Neilson, E.G. Biomarkers for Epithelial-Mesenchymal Transitions. J. Clin. Invest. 2009, 119, 1429–1437. [Google Scholar] [CrossRef] [PubMed]
- Loh, C.Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The E-Cadherin and n-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, K.; He, S.; Terasaki, H.; Nazari, H.; Zhang, H.; Spee, C.; Kannan, R.; Hinton, D.R. Resveratrol Inhibits Epithelial-Mesenchymal Transition of Retinal Pigment Epithelium and Development of Proliferative Vitreoretinopathy. Sci. Rep. 2015, 5, 16386. [Google Scholar] [CrossRef]
- Watanabe, M.; Masuyama, N.; Fukuda, M.; Nishida, E. Regulation of Intracellular Dynamics of Smad4 by Its Leucine-Rich Nuclear Export Signal. EMBO Rep. 2000, 1, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Simic, P.; Williams, E.O.; Bell, E.L.; Gong, J.J.; Bonkowski, M.; Guarente, L. SIRT1 Suppresses the Epithelial-to-Mesenchymal Transition in Cancer Metastasis and Organ Fibrosis. Cell Rep. 2013, 3, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Gamart, J.; Barozzi, I.; Laurent, F.; Reinhardt, R.; Martins, L.R.; Oberholzer, T.; Visel, A.; Zeller, R.; Zuniga, A. SMAD4 Target Genes Are Part of a Transcriptional Network That Integrates the Response to BMP and SHH Signaling during Early Limb Bud Patterning. Development 2021, 148, dev200182. [Google Scholar] [CrossRef] [PubMed]
- Sarvagalla, S.; Kolapalli, S.P.; Vallabhapurapu, S. The Two Sides of YY1 in Cancer: A Friend and a Foe. Front. Oncol. 2019, 9, 1230. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Qian, X.; Li, Y.; Li, X.-Z.; He, L.-L.; Xu, L.; Liu, Y.-Q.; Li, C.-C.; Ma, P.; Shu, F.-L.; et al. Sirt1 Inhibits Renal Tubular Cell Epithelial–mesenchymal Transition through YY1 Deacetylation in Diabetic Nephropathy. Acta Pharmacol. Sin. 2021, 42, 242–251. [Google Scholar] [CrossRef]
- Chen, F.; Zhou, J.; Li, Y.; Zhao, Y.; Yuan, J.; Cao, Y.; Wang, L.; Zhang, Z.; Zhang, B.; Wang, C.C.; et al. YY 1 Regulates Skeletal Muscle Regeneration through Controlling Metabolic Reprogramming of Satellite Cells. EMBO J. 2019, 38, e99727. [Google Scholar] [CrossRef]
- Lin, Y.; Li, L.; Liu, J.; Zhao, X.; Ye, J.; Reinach, P.S.; Qu, J.; Yan, D. SIRT1 Deletion Impairs Retinal Endothelial Cell Migration through Downregulation of VEGF-A/VEGFR-2 and MMP14. Investig. Ophthalmol. Vis. Sci. 2018, 59, 5431–5440. [Google Scholar] [CrossRef]
- Kunimoto, R.; Jimbow, K.; Tanimura, A.; Sato, M.; Horimoto, K.; Hayashi, T.; Hisahara, S.; Sugino, T.; Hirobe, T.; Yamashita, T.; et al. SIRT1 Regulates Lamellipodium Extension and Migration of Melanoma Cells. J. Invest. Dermatol. 2014, 134, 1693–1700. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, X.; Cui, Y.; Wei, Q.; Chen, S.; Wang, X. Effects of SIRT1 Silencing on Viability, Invasion and Metastasis of Human Glioma Cell Lines. Oncol. Lett. 2019, 17, 3701–3708. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Jiao, L.; Wang, Y.; Yu, Y.; Ming, L. SIRT1 Induces Epithelial-Mesenchymal Transition by Promoting Autophagic Degradation of E-Cadherin in Melanoma Cells Article. Cell Death Dis. 2018, 9, 136. [Google Scholar] [CrossRef] [PubMed]
- Han, L.L.; Jia, L.; Wu, F.; Huang, C. Sirtuin6 (SIRT6) Promotes the EMT of Hepatocellular Carcinoma by Stimulating Autophagic Degradation of e-Cadherin. Mol. Cancer Res. 2019, 17, 2267–2280. [Google Scholar] [CrossRef] [PubMed]
- Zeng, K.; Feng, Q.G.; Lin, B.T.; Ma, D.H.; Liu, C.M. Effects of MicroRNA-211 on Proliferation and Apoptosis of Lens Epithelial Cells by Targeting SIRT1 Gene in Diabetic Cataract Mice. Biosci. Rep. 2017, 37, BSR20170695. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhang, Y.; You, S.; Tian, Y.; Lu, S.; Cao, L.; Sun, Y. Septin4 Prevents PDGF-BB-Induced HAVSMC Phenotypic Transformation, Proliferation and Migration by Promoting SIRT1-STAT3 Deacetylation and Dephosphorylation. Int. J. Biol. Sci. 2020, 16, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.H.; Xiao, Z.Q.; Zhou, J.D.; Yin, C.Q.; Chen, Z.Z.; Tang, F.J.; Wang, S.H. MiR-199a-5p Represses the Stemness of Cutaneous Squamous Cell Carcinoma Stem Cells by Targeting Sirt1 and CD44ICD Cleavage Signaling. Cell Cycle 2020, 19, 1–14. [Google Scholar] [CrossRef]
- Deng, Y.W.; Shu, Y.G.; Sun, S.L. MiR-376a Inhibits Glioma Proliferation and Angiogenesis by Regulating YAP1/VEGF Signalling via Targeting of SIRT1. Transl. Oncol. 2022, 15, 101270. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Li, M.; Li, X.; Li, H.; Lai, Y.; Huang, S.; He, X.; Si, X.; Zheng, H.; Liao, W.; et al. Sirt1-Inducible Deacetylation of P21 Promotes Cardiomyocyte Proliferation. Aging 2019, 11, 12546–12567. [Google Scholar] [CrossRef]
- Wang, F.; Li, Z.; Zhou, J.; Wang, G.; Zhang, W.; Xu, J.; Liang, A. SIRT1 Regulates the Phosphorylation and Degradation of P27 by Deacetylating CDK2 to Promote T-Cell Acute Lymphoblastic Leukemia Progression. J. Exp. Clin. Cancer Res. 2021, 40, 259. [Google Scholar] [CrossRef]
- Mateo, F.; Vidal-laliena, M.; Canela, N.; Zecchin, A.; Martínez-balbás, M.; Agell, N.; Giacca, M.; Pujol, M.J.; Bachs, O. The Transcriptional Co-Activator PCAF Regulates Cdk2 Activity. Nucleic Acids Res. 2009, 37, 7072–7084. [Google Scholar] [CrossRef] [PubMed]
- Sheaff, R.J.; Groudine, M.; Gordon, M.; Roberts, J.M.; Clurman, B.E. Cyclin E-CDK2 Is a Regulator of P27(Kip1). Genes Dev. 1997, 11, 1464–1478. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cui, W.; Song, B.; Ye, X.; Li, Z.; Lu, C. Autophagy-Sirtuin1(SIRT1) Alleviated the Coronary Atherosclerosis (AS) in Mice through Regulating the Proliferation and Migration of Endothelial Progenitor Cells (EPCs) via Wnt/β-Catenin/GSK3β Signaling Pathway. J. Nutr. Health Aging 2022, 26, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, X.; Wan, L.; Han, B.; Ma, S.; Pan, H.; Wei, J.; Cui, X. Metformin Suppresses Cardiac Fibroblast Proliferation under High-Glucose Conditions via Regulating the Mitochondrial Complex I Protein Grim-19 Involved in the Sirt1/Stat3 Signaling Pathway. Free Radic. Biol. Med. 2023, 206, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sherry, M.M.; Reeves, A.; Wu, J.K.; Cochran, B.H. STAT3 Is Required for Proliferation and Maintenance of Multipotency in Glioblastoma Stem Cells. Stem Cells 2009, 27, 2383–2392. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Xiao, F.; Wang, G.; Wei, X.; Jiang, L.; Chen, Y.; Zhu, L.; Wang, H.; Diao, Y.; Wang, H.; et al. STAT3 Regulates Self-Renewal of Adult Muscle Satellite Cells during Injury-Induced Muscle Regeneration. Cell Rep. 2016, 16, 2102–2115. [Google Scholar] [CrossRef]
- Liang, J.; Wang, D.; Renaud, G.; Wolfsberg, T.G.; Wilson, A.F.; Burgess, S.M. The Stat3/Socs3a Pathway Is a Key Regulator of Hair Cell Regeneration in Zebrafish Stat3/Socs3a Pathway: Regulator of Hair Cell Regeneration. J. Neurosci. 2012, 32, 10662–10673. [Google Scholar] [CrossRef]
- Fang, Y.; Gupta, V.; Karra, R.; Holdway, J.E.; Kikuchi, K.; Poss, K.D. Translational Profiling of Cardiomyocytes Identifies an Early Jak1/Stat3 Injury Response Required for Zebrafish Heart Regeneration. Proc. Natl. Acad. Sci. USA 2013, 110, 13416–13421. [Google Scholar] [CrossRef]
- Sánchez Alvarado, A.; Yamanaka, S. Rethinking Differentiation: Stem Cells, Regeneration, and Plasticity. Cell 2014, 157, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.; Nejak-Bowen, K.; Monga, S.P.S. Crosstalk of the Wnt Signaling Pathway. In Targeting the Wnt Pathway in Cancer; Springer: New York, NY, USA, 2011; pp. 51–80. ISBN 9781441980229. [Google Scholar]
- Lu, Y.; Ma, Z.X.; Deng, R.; Jiang, H.T.; Chu, L.; Deng, Z.L. The SIRT1 Activator SRT2104 Promotes BMP9-Induced Osteogenic and Angiogenic Differentiation in Mesenchymal Stem Cells. Mech. Ageing Dev. 2022, 207, 111724. [Google Scholar] [CrossRef]
- Borojević, A.; Jauković, A.; Kukolj, T.; Mojsilović, S.; Obradović, H.; Trivanović, D.; Živanović, M.; Zečević, Ž.; Simić, M.; Gobeljić, B.; et al. Vitamin D3 Stimulates Proliferation Capacity, Expression of Pluripotency Markers, and Osteogenesis of Human Bone Marrow Mesenchymal Stromal/Stem Cells, Partly through SIRT1 Signaling. Biomolecules 2022, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Song, C.Y.; Guo, Y.; Chen, F.Y.; Liu, W.G. Resveratrol Promotes Osteogenic Differentiation of Bone Marrow-Derived Mesenchymal Stem Cells Through MiR-193a/SIRT7 Axis. Calcif. Tissue Int. 2022, 110, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, X.; Liu, W.; Lu, W. Thrombin-Activated Platelet-Rich Plasma Enhances Osteogenic Differentiation of Human Periodontal Ligament Stem Cells by Activating SIRT1-Mediated Autophagy. Eur. J. Med. Res. 2021, 26, 105. [Google Scholar] [CrossRef]
- Gao, L.; Gong, F.-Z.; Ma, L.-Y.; Yang, J.-H. Uncarboxylated Osteocalcin Promotes Osteogenesis and Inhibits Adipogenesis of Mouse Bone Marrow-derived Mesenchymal Stem Cells via the PKA-AMPK-SIRT1 Axis. Exp. Ther. Med. 2021, 22, 880. [Google Scholar] [CrossRef] [PubMed]
- Luo, B.; Yang, J.F.; Wang, Y.H.; Qu, G.B.; Hao, P.D.; Zeng, Z.J.; Yuan, J.; Yang, R.; Yuan, Y. MicroRNA-579-3P Promotes the Progression of Osteoporosis by Inhibiting Osteogenic Differentiation of Mesenchymal Stem Cells through Regulating Sirt1. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 6791–6799. [Google Scholar] [CrossRef]
- Lin, C.H.; Li, N.T.; Cheng, H.S.; Yen, M.L. Oxidative Stress Induces Imbalance of Adipogenic/Osteoblastic Lineage Commitment in Mesenchymal Stem Cells through Decreasing SIRT1 Functions. J. Cell. Mol. Med. 2018, 22, 786–796. [Google Scholar] [CrossRef]
- Zhu, C.; Ding, H.; Shi, L.; Zhang, S.; Tong, X.; Huang, M.; Liu, L.; Guan, X.; Zou, J.; Yuan, Y.; et al. Exercise Improved Bone Health in Aging Mice: A Role of SIRT1 in Regulating Autophagy and Osteogenic Differentiation of BMSCs. Front. Endocrinol. 2023, 14, 1156637. [Google Scholar] [CrossRef]
- Ouyang, X.; Ding, Y.; Yu, L.; Xin, F.; Yang, X. LncRNA TUG Regulates Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells via MiRNA-204/SIRT 1. J. Musculoskelet. Neuronal Interact. 2022, 22, 401–410. [Google Scholar]
- Li, M.; Yan, J.; Chen, X.; Tam, W.; Zhou, L.; Liu, T.; Pan, G.; Lin, J.; Yang, H.; Pei, M.; et al. Spontaneous Up-Regulation of SIRT1 during Osteogenesis Contributes to Stem Cells’ Resistance to Oxidative Stress. J. Cell. Biochem. 2018, 119, 4928–4944. [Google Scholar] [CrossRef]
- Qu, B.; Gong, K.; Yang, H.S.; Li, Y.G.; Jiang, T.; Zeng, Z.M.; Cao, Z.R.; Pan, X.M. MiR-449 Overexpression Inhibits Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells via Suppressing Sirt1/Fra-1 Pathway in High Glucose and Free Fatty Acids Microenvironment. Biochem. Biophys. Res. Commun. 2018, 496, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Qu, B.; He, J.; Zeng, Z.; Yang, H.; Liu, Z.; Cao, Z.; Yu, H.; Zhao, W.; Pan, X. MiR-155 Inhibition Alleviates Suppression of Osteoblastic Differentiation by High Glucose and Free Fatty Acids in Human Bone Marrow Stromal Cells by Upregulating SIRT1. Pflugers Arch. Eur. J. Physiol. 2020, 472, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Qu, H.; Li, T.; Jin, H.; Zhang, S.; He, B. Silent Mating Type Information Regulation 2 Homolog (SIRT1) Influences Osteogenic Proliferation and Differentiation of MC3T3-E1 Cells via Regulation of MiR-132-3p. Med. Sci. Monit. 2019, 25, 2289–2295. [Google Scholar] [CrossRef] [PubMed]
- Tseng, P.C.; Hou, S.M.; Chen, R.J.; Peng, H.W.; Hsieh, C.F.; Kuo, M.L.; Yen, M.L. Resveratrol Promotes Osteogenesis of Human Mesenchymal Stem Cells by Upregulating RUNX2 Gene Expression via the SIRT1/FOXO3A Axis. J. Bone Miner. Res. 2011, 26, 2552–2563. [Google Scholar] [CrossRef]
- Smith, C.A.; Humphreys, P.A.; Bates, N.; Naven, M.A.; Cain, S.A.; Dvir-Ginzberg, M.; Kimber, S.J. SIRT1 Activity Orchestrates ECM Expression during HESC-Chondrogenic Differentiation. FASEB J. 2022, 36, e22314. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhou, L.; Wang, L.; He, S.; Ren, H.; Zhou, N.; Hu, Z. The Role of SIRT1 in BMP2-Induced Chondrogenic Differentiation and Cartilage Maintenance under Oxidative Stress. Aging 2020, 12, 9000–9013. [Google Scholar] [CrossRef]
- Gao, J.; Feng, Z.; Wang, X.; Zeng, M.; Liu, J.; Han, S.; Xu, J.; Chen, L.; Cao, K.; Long, J.; et al. SIRT3/SOD2 Maintains Osteoblast Differentiation and Bone Formation by Regulating Mitochondrial Stress. Cell Death Differ. 2018, 25, 229–240. [Google Scholar] [CrossRef]
- Zheng, K.; Bai, J.; Li, N.; Li, M.; Sun, H.; Zhang, W.; Ge, G.; Liang, X.; Tao, H.; Xue, Y.; et al. Protective Effects of Sirtuin 3 on Titanium Particle-Induced Osteogenic Inhibition by Regulating the NLRP3 Inflammasome via the GSK-3β/β-Catenin Signalling Pathway. Bioact. Mater. 2021, 6, 3343–3357. [Google Scholar] [CrossRef]
- Zaganjor, E.; Yoon, H.; Spinelli, J.B.; Nunn, E.R.; Laurent, G.; Keskinidis, P.; Sivaloganathan, S.; Joshi, S.; Notarangelo, G.; Mulei, S.; et al. SIRT4 Is an Early Regulator of Branched-Chain Amino Acid Catabolism That Promotes Adipogenesis. Cell Rep. 2021, 36, 109345. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Li, S.; Wang, X.; Mei, C.; Zan, L. Study of Expression Analysis of SIRT4 and the Coordinate Regulation of Bovine Adipocyte Differentiation by SIRT4 and Its Transcription Factors. Biosci. Rep. 2018, 38, BSR20181705. [Google Scholar] [CrossRef]
- Singh, C.K.; Chhabra, G.; Ndiaye, M.A.; Garcia-Peterson, L.M.; MacK, N.J.; Ahmad, N. The Role of Sirtuins in Antioxidant and Redox Signaling. Antioxid. Redox Signal. 2018, 28, 643–661. [Google Scholar] [CrossRef]
- Shuai, L.; Zhang, L.N.; Li, B.H.; Tang, C.L.; Wu, L.Y.; Li, J.; Li, J.Y. SIRT5 Regulates Brown Adipocyte Differentiation and Browning of Subcutaneous White Adipose Tissue. Diabetes 2019, 68, 1449–1461. [Google Scholar] [CrossRef] [PubMed]
- Molinari, F.; Feraco, A.; Mirabilii, S.; Saladini, S.; Sansone, L.; Vernucci, E.; Tomaselli, G.; Marzolla, V.; Rotili, D.; Russo, M.A.; et al. Sirt5 Inhibition Induces Brown Fat-like Phenotype in 3t3-L1 Preadipocytes. Cells 2021, 10, 1126. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Chen, J.; Wang, Q.; Sun, X.; Han, J.; Guastaldi, F.; Xiang, S.; Ye, Q.; He, Y. SIRT6 Promotes Osteogenic Differentiation of Adipose-Derived Mesenchymal Stem Cells Through Antagonizing DNMT1. Front. Cell Dev. Biol. 2021, 9, 648627. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, S.; Zhang, W.; Ni, L.; Hu, Z.; Sheng, Z.; Yin, B. MIR-128 Inhibits the Osteogenic Differentiation in Osteoporosis by down-Regulating SIRT6 Expression. Biosci. Rep. 2019, 39, BSR20191405. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Qin, S.; Li, W.; Yao, L.; Huang, P.; Liao, J.; Liu, J.; Li, S. Osteogenic Differentiation of Rat Bone Mesenchymal Stem Cells Modulated by MiR-186 via SIRT6. Life Sci. 2020, 253, 117660. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Zhou, Y.; Liu, Y.; Xie, M.; Guo, G. Inhibitory Effect of Sirtuin6 (SIRT6) on Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells. Med. Sci. Monit. 2019, 25, 8412–8421. [Google Scholar] [CrossRef]
- Piao, J.; Tsuji, K.; Ochi, H.; Iwata, M.; Koga, D.; Okawa, A.; Morita, S.; Takeda, S.; Asou, Y. Sirt6 Regulates Postnatal Growth Plate Differentiation and Proliferation via Ihh Signaling. Sci. Rep. 2013, 3, 3022. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Guo, S.; Tong, S.; Sun, X. Exosomal MiR-130a-3p Regulates Osteogenic Differentiation of Human Adipose-Derived Stem Cells through Mediating SIRT7/Wnt/β-Catenin Axis. Cell Prolif. 2020, 53, e12890. [Google Scholar] [CrossRef]
- Chen, E.E.M.; Zhang, W.; Ye, C.C.Y.; Gao, X.; Jiang, L.L.J.; Zhao, T.T.F.; Pan, Z.Z.J.; Xue, D.D.T. Knockdown of SIRT7 Enhances the Osteogenic Differentiation of Human Bone Marrow Mesenchymal Stem Cells Partly via Activation of the Wnt/β-Catenin Signaling Pathway. Cell Death Dis. 2017, 8, e3042. [Google Scholar] [CrossRef]
- Martinez-Pastor, B.; Mostoslavsky, R. Sirtuins, Metabolism, and Cancer. Front. Pharmacol. 2012, 3 FEB, 22. [Google Scholar] [CrossRef]
- German, N.J.; Haigis, M.C. Sirtuins and the Metabolic Hurdles in Cancer. Curr. Biol. 2015, 25, R569–R583. [Google Scholar] [CrossRef] [PubMed]
- Zhao, E.; Hou, J.; Ke, X.; Abbas, M.N.; Kausar, S.; Zhang, L.; Cui, H. The Roles of Sirtuin Family Proteins in Cancer Progression. Cancers 2019, 11, 1949. [Google Scholar] [CrossRef] [PubMed]
- Gantwerker, E.A.; Hom, D.B. Skin: Histology and Physiology of Wound Healing. Clin. Plast. Surg. 2011, 19, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Spallotta, F.; Cencioni, C.; Straino, S.; Nanni, S.; Rosati, J.; Artuso, S.; Manni, I.; Colussi, C.; Piaggio, G.; Martelli, F.; et al. A Nitric Oxide-Dependent Cross-Talk between Class i and III Histone Deacetylases Accelerates Skin Repair. J. Biol. Chem. 2013, 288, 11004–11012. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shen, K.; Wang, J.; Liu, K.; Wu, G.; Li, Y.; Luo, L.; Zheng, Z.; Hu, D. Hypoxic Preconditioning Combined with Curcumin Promotes Cell Survival and Mitochondrial Quality of Bone Marrow Mesenchymal Stem Cells, and Accelerates Cutaneous Wound Healing via PGC-1α/SIRT3/HIF-1α Signaling. Free Radic. Biol. Med. 2020, 159, 164–176. [Google Scholar] [CrossRef]
- Jiang, X.; Yao, Z.; Wang, K.; Lou, L.; Xue, K.; Chen, J.; Zhang, G.; Zhang, Y.; Du, J.; Lin, C.; et al. MDL-800, the SIRT6 Activator, Suppresses Inflammation via the NF- κ B Pathway and Promotes Angiogenesis to Accelerate Cutaneous Wound Healing in Mice. Oxid. Med. Cell. Longev. 2022, 2022, 1619651. [Google Scholar] [CrossRef]
- Lei, M.; Lien, W.H.; Li, J. Editorial: Inflammation, Stem Cells and Wound Healing in Skin Aging. Front. Cell Dev. Biol. 2022, 10, 1046022. [Google Scholar] [CrossRef]
- Xiu, Y.; Su, Y.; Gao, L.; Yuan, H.; Xu, S.; Liu, Y.; Qiu, Y.; Liu, Z.; Li, Y. Corylin Accelerated Wound Healing through SIRT1 and PI3K/AKT Signaling: A Candidate Remedy for Chronic Non-Healing Wounds. Front. Pharmacol. 2023, 14, 1153810. [Google Scholar] [CrossRef]
- Shi, R.; Jin, Y.; Hu, W.; Lian, W.; Cao, C.; Han, S.; Zhao, S.; Yuan, H.; Yang, X.; Shi, J.; et al. Exosomes Derived from Mmu_circ_0000250-Modified Adipose-Derived Mesenchymal Stem Cells Promote Wound Healing in Diabetic Mice by Inducing MiR-128-3p/SIRT1-Mediated Autophagy. Am. J. Physiol. Cell Physiol. 2020, 318, C848–C856. [Google Scholar] [CrossRef]
- Shang, B.; Xu, T.; Hu, N.; Mao, Y.; Du, X. Circ-Klhl8 Overexpression Increased the Therapeutic Effect of EPCs in Diabetic Wound Healing via the MiR-212-3p/SIRT5 Axis. J. Diabetes Complicat. 2021, 35, 108020. [Google Scholar] [CrossRef]
- Thandavarayan, R.A.; Garikipati, V.N.S.; Joladarashi, D.; Suresh Babu, S.; Jeyabal, P.; Verma, S.K.; Mackie, A.R.; Khan, M.; Arumugam, S.; Watanabe, K.; et al. Sirtuin-6 Deficiency Exacerbates Diabetes-Induced Impairment of Wound Healing. Exp. Dermatol. 2015, 24, 773–778. [Google Scholar] [CrossRef] [PubMed]
- Wahedi, H.M.; Chae, J.K.; Subedi, L.; Kang, M.C.; Cho, H.; Kim, S.; Kim, S.Y. NED416, a Novel Synthetic Sirt1 Activator, Promotes Cutaneous Wound Healing via the MAPK/Rho Pathway. Int. J. Mol. Med. 2020, 46, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Qiang, L.; Sample, A.; Liu, H.; Wu, X.; He, Y.Y. Epidermal SIRT1 Regulates Inflammation, Cell Migration, and Wound Healing. Sci. Rep. 2017, 7, 14110. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.H.; Lee, H.J.; Kim, J.S.; Lee, S.J.; Han, H.J. EphB2 Signaling-Mediated Sirt3 Expression Reduces MSC Senescence by Maintaining Mitochondrial ROS Homeostasis. Free Radic. Biol. Med. 2017, 110, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Koo, J.H.; Jang, H.Y.; Lee, Y.; Moon, Y.J.; Bae, E.J.; Yun, S.K.; Park, B.H. Myeloid Cell-Specific Sirtuin 6 Deficiency Delays Wound Healing in Mice by Modulating Inflammation and Macrophage Phenotypes. Exp. Mol. Med. 2019, 51, 1–10. [Google Scholar] [CrossRef]
- Wang, Z.H.; Bao, X.G.; Hu, J.J.; Shen, S.B.; Xu, G.H.; Wu, Y.L. Nicotinamide Riboside Enhances Endothelial Precursor Cell Function to Promote Refractory Wound Healing Through Mediating the Sirt1/AMPK Pathway. Front. Pharmacol. 2021, 12, 671563. [Google Scholar] [CrossRef]
- Bai, X.Z.; Liu, J.Q.; Yang, L.L.; Fan, L.; He, T.; Su, L.L.; Shi, J.H.; Tang, C.W.; Zheng, Z.; Hu, D.H. Identification of Sirtuin 1 as a Promising Therapeutic Target for Hypertrophic Scars. Br. J. Pharmacol. 2016, 173, 1589–1601. [Google Scholar] [CrossRef]
- Zhang, Y.; Bai, X.; Shen, K.; Luo, L.; Zhao, M.; Xu, C.; Jia, Y.; Xiao, D.; Li, Y.; Gao, X.; et al. Exosomes Derived from Adipose Mesenchymal Stem Cells Promote Diabetic Chronic Wound Healing through SIRT3/SOD2. Cells 2022, 11, 2568. [Google Scholar] [CrossRef]
- Yang, S.; Xu, M.; Meng, G.; Lu, Y. SIRT3 Deficiency Delays Diabetic Skin Wound Healing via Oxidative Stress and Necroptosis Enhancement. J. Cell. Mol. Med. 2020, 24, 4415–4427. [Google Scholar] [CrossRef]
- Boniakowski, A.M.; denDekker, A.D.; Davis, F.M.; Joshi, A.; Kimball, A.S.; Schaller, M.; Allen, R.; Bermick, J.; Nycz, D.; Skinner, M.E.; et al. SIRT3 Regulates Macrophage-Mediated Inflammation in Diabetic Wound Repair. J. Invest. Dermatol. 2019, 139, 2528–2537.e2. [Google Scholar] [CrossRef]
- Chaurasia, S.; Lim, R.; Lakshminarayanan, R.; Mohan, R. Nanomedicine Approaches for Corneal Diseases. J. Funct. Biomater. 2015, 6, 277–298. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Richardson, A.; Pandzic, E.; Lobo, E.P.; Whan, R.; Watson, S.L.; Lyons, J.G.; Wakefield, D.; Di Girolamo, N. Visualizing the Contribution of Keratin-14+ Limbal Epithelial Precursors in Corneal Wound Healing. Stem Cell Reports 2019, 12, 14–28. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, X.; Shi, D.; Chen, P.; Yu, Y.; Yang, L.; Xie, L. Overexpression of SIRT1 Promotes High Glucose-Attenuated Corneal Epithelial Wound Healing via P53 Regulation of the IGFBP3/IGF-1R/AKT Pathway. Invest. Ophthalmol. Vis. Sci. 2013, 54, 3806–3814. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, Y.; Zhao, X.; Chen, P.; Xie, L. MicroRNA-204-5p–mediated Regulation of SIRT1 Contributes to the Delay of Epithelial Cell Cycle Traversal in Diabetic Corneas. Investig. Ophthalmol. Vis. Sci. 2015, 56, 1493–1504. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Li, Z.; Wang, X.; Zou, D.; Duan, H.; Zhao, C.; Zhou, Q.; Shi, W. SRT1720 Attenuates UVA-Induced Corneal Endothelial Damage via Inhibition of Oxidative Stress and Cellular Apoptosis. Exp. Eye Res. 2023, 231, 109464. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Liu, Q.; Li, L.; Yang, R.; Ye, J.; Yang, S.; Luo, G.; Reinach, P.S.; Yan, D. Sirt1 Regulates Corneal Epithelial Migration by Deacetylating Cortactin. Investig. Ophthalmol. Vis. Sci. 2022, 63, 14. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Kan, T.; Hu, X. Sirt3 Regulates Mitophagy Level to Promote Diabetic Corneal Epithelial Wound Healing. Exp. Eye Res. 2019, 181, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Zhu, S.; Liu, R.; Miller, J.D.; Merkley, K.; Tilton, R.G.; Liu, H. Sirt6 Deficiency Impairs Corneal Epithelial Wound Healing. Aging 2018, 10, 1932–1946. [Google Scholar] [CrossRef]
- Li, X.; Kang, B.; Eom, Y.; Zhong, J.; Lee, H.K.; Kim, H.M.; Song, J.S. SIRT1 Protects against Particulate Matter-Induced Oxidative Stress in Human Corneal and Conjunctival Epithelial Cells. Investig. Ophthalmol. Vis. Sci. 2022, 63, 19. [Google Scholar] [CrossRef]
- Jalil, H.A.; Al-Sudani, B.T.; Jasim, G.A. SIRT1720 Promotes Survival of Corneal Epithelial Cells via the P53 Pathway. J. Popul. Ther. Clin. Pharmacol. 2022, 29, e17–e33. [Google Scholar] [CrossRef]
- Dong, Y.; Ding, Y.Y.; Gao, W.P. Puerarin Alleviates Hyperosmotic Stress-Induced Oxidative Stress, Inflammation, Apoptosis and Barrier Damage of Human Corneal Epithelial Cells by Targeting SIRT1/NLRP3 Signaling. Toxicol. Vitr. 2024, 94, 105722. [Google Scholar] [CrossRef] [PubMed]
- Michalopoulos, G.K.; Bhushan, B. Liver Regeneration: Biological and Pathological Mechanisms and Implications. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Cong, M.; Wu, H.; Wang, M.; Bai, H.; Wang, J.; Que, K.; Zheng, K.; Zhang, W.; Yang, X.; et al. P53/MiR-34a/SIRT1 Positive Feedback Loop Regulates the Termination of Liver Regeneration. Aging 2023, 15, 1859–1877. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.F.; Li, J.X.; Liao, H.T.; Liao, M.H.; Luo, L.; Xu, L.; Yuan, K.F.; Zeng, Y. Nicotinamide Induces Liver Regeneration and Improves Liver Function by Activating SIRT1. Mol. Med. Rep. 2019, 19, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Fan, X.; Jiao, T.; Li, W.; Chen, P.; Jiang, Y.; Sun, J.; Chen, Y.; Chen, P.; Guan, L.; et al. SIRT6 as a Key Event Linking P53 and NRF2 Counteracts APAP-Induced Hepatotoxicity through Inhibiting Oxidative Stress and Promoting Hepatocyte Proliferation. Acta Pharm. Sin. B 2021, 11, 89–99. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, Y.; Fan, X.; Tan, H.; Zeng, H.; Wang, Y.; Chen, P.; Huang, M.; Bi, H. Hepato-Protective Effect of Resveratrol against Acetaminophen-Induced Liver Injury Is Associated with Inhibition of CYP-Mediated Bioactivation and Regulation of SIRT1-P53 Signaling Pathways. Toxicol. Lett. 2015, 236, 82–89. [Google Scholar] [CrossRef]
- Liu, Q.; Pu, S.; Chen, L.; Shen, J.; Cheng, S.; Kuang, J.; Li, H.; Wu, T.; Li, R.; Jiang, W.; et al. Liver-Specific Sirtuin6 Ablation Impairs Liver Regeneration after 2/3 Partial Hepatectomy. Wound Repair Regen. 2019, 27, 366–374. [Google Scholar] [CrossRef]
- Bellet, M.M.; Masri, S.; Astarita, G.; Sassone-Corsi, P.; Della Fazia, M.A.; Servillo, G. Histone Deacetylase SIRT1 Controls Proliferation, Circadian Rhythm, and Lipid Metabolism during Liver Regeneration in Mice. J. Biol. Chem. 2016, 291, 23318–23329. [Google Scholar] [CrossRef]
- Ramirez, T.; Li, Y.M.; Yin, S.; Xu, M.J.; Feng, D.; Zhou, Z.; Zang, M.; Mukhopadhyay, P.; Varga, Z.V.; Pacher, P.; et al. Aging Aggravates Alcoholic Liver Injury and Fibrosis in Mice by Downregulating Sirtuin 1 Expression. J. Hepatol. 2017, 66, 601–609. [Google Scholar] [CrossRef]
- Tian, X.F.; Ji, F.J.; Zang, H.L.; Cao, H. Activation of the MiR-34a/SIRT1/P53 Signaling Pathway Contributes to the Progress of Liver Fibrosis via Inducing Apoptosis in Hepatocytes but Not in HSCs. PLoS ONE 2016, 11, e0158657. [Google Scholar] [CrossRef]
- Jin, J.; Iakova, P.; Jiang, Y.; Medrano, E.E.; Timchenko, N.A. The Reduction of SIRT1 in Livers of Old Mice Leads to Impaired Body Homeostasis and to Inhibition of Liver Proliferation. Hepatology 2011, 54, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Bertero, A.; Murry, C.E. Hallmarks of Cardiac Regeneration. Nat. Rev. Cardiol. 2018, 15, 579–580. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, C.E.; Bakovic, M.; Karra, R. Endothelial Contributions to Zebrafish Heart Regeneration. J. Cardiovasc. Dev. Dis. 2018, 5, 56. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Lei, J.; Han, H.; Li, W.; Qu, Y.; Fu, E.; Fu, F.; Wang, X. SIRT1 Protects against Myocardial Ischemia-Reperfusion Injury via Activating ENOS in Diabetic Rats. Cardiovasc. Diabetol. 2015, 14, 143. [Google Scholar] [CrossRef]
- Shalwala, M.; Zhu, S.G.; Das, A.; Salloum, F.N.; Xi, L.; Kukreja, R.C. Sirtuin 1 (SIRT1) Activation Mediates Sildenafil Induced Delayed Cardioprotection against Ischemia-Reperfusion Injury in Mice. PLoS ONE 2014, 9, e86977. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yang, Q.; Sun, Y.Y.; Xing, Y.F.; Wang, Y.B.; Lu, X.T.; Bai, W.W.; Liu, X.Q.; Zhao, Y.X. Resveratrol-Enhanced Autophagic Flux Ameliorates Myocardial Oxidative Stress Injury in Diabetic Mice. J. Cell. Mol. Med. 2014, 18, 1599–1611. [Google Scholar] [CrossRef]
- Yu, L.; Sun, Y.; Cheng, L.; Jin, Z.; Yang, Y.; Zhai, M.; Pei, H.; Wang, X.; Zhang, H.; Meng, Q.; et al. Melatonin Receptor-Mediated Protection against Myocardial Ischemia/Reperfusion Injury: Role of SIRT1. J. Pineal Res. 2014, 57, 228–238. [Google Scholar] [CrossRef]
- Yang, Y.; Duan, W.; Lin, Y.; Yi, W.; Liang, Z.; Yan, J.; Wang, N.; Deng, C.; Zhang, S.; Li, Y.; et al. SIRT1 Activation by Curcumin Pretreatment Attenuates Mitochondrial Oxidative Damage Induced by Myocardial Ischemia Reperfusion Injury. Free Radic. Biol. Med. 2013, 65, 667–679. [Google Scholar] [CrossRef]
- Luo, Y.; Lu, J.; Wang, Z.; Wang, L.; Wu, G.; Guo, Y.; Dong, Z. Small Ubiquitin-Related Modifier (SUMO)Ylation of SIRT1 Mediates (-)-Epicatechin Inhibited- Differentiation of Cardiac Fibroblasts into Myofibroblasts. Pharm. Biol. 2022, 60, 1762–1770. [Google Scholar] [CrossRef]
- Ozawa, H.; Miyagawa, S.; Fukushima, S.; Itoh, E.; Harada, A.; Saito, A.; Ueno, T.; Toda, K.; Kuratani, T.; Sawa, Y. Sirtuin1 Regulates the Stem Cell Therapeutic Effects on Regenerative Capability for Treating Severe Heart Failure in a Juvenile Animal Model. Ann. Thorac. Surg. 2016, 102, 803–812. [Google Scholar] [CrossRef]
- Klishadi, M.S.; Zarei, F.; Hejazian, S.H.; Moradi, A.; Hemati, M.; Safari, F. Losartan Protects the Heart against Ischemia Reperfusion Injury: Sirtuin3 Involvement. J. Pharm. Pharm. Sci. 2015, 18, 112–123. [Google Scholar] [CrossRef]
- Porter, G.A.; Urciuoli, W.R.; Brookes, P.S.; Nadtochiy, S.M. SIRT3 Deficiency Exacerbates Ischemia-Reperfusion Injury: Implication for Aged Hearts. Am. J. Physiol. Hear. Circ. Physiol. 2014, 306, H1602–H1609. [Google Scholar] [CrossRef] [PubMed]
- Araki, S.; Izumiya, Y.; Rokutanda, T.; Ianni, A.; Hanatani, S.; Kimura, Y.; Onoue, Y.; Senokuchi, T.; Yoshizawa, T.; Yasuda, O.; et al. Sirt7 Contributes to Myocardial Tissue Repair by Maintaining Transforming Growth Factor-β Signaling Pathway. Circulation 2015, 132, 1081–1093. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Li, L.; Chen, J.X. Loss of Sirt3 Limits Bone Marrow Cell-Mediated Angiogenesis and Cardiac Repair in Post-Myocardial Infarction. PLoS ONE 2014, 9, e107011. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, S.; Izumiya, Y.; Araki, S.; Nakamura, T.; Kimura, Y.; Hanatani, S.; Yamada, T.; Ishida, T.; Yamamoto, M.; Onoue, Y.; et al. Cardiomyocyte Sirt (Sirtuin) 7 Ameliorates Stress-Induced Cardiac Hypertrophy by Interacting With and Deacetylating GATA4. Hypertension 2020, 75, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Squire, L.R.; Berg, D.; Bloom, F.E.; Du Lac, S.; Ghosh, A.; Spitzer, N.C. Fundamental Neuroscience, 4th ed.; Academic Press: Cambridge, MA, USA, 2012; ISBN 9780123858719. [Google Scholar]
- Zukor, K.A.; Kent, D.T.; Odelberg, S.J. Meningeal Cells and Glia Establish a Permissive Environment for Axon Regeneration after Spinal Cord Injury in Newts. Neural Dev. 2011, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Anguita-Salinas, C.; Sánchez, M.; Morales, R.A.; Ceci, M.L.; Rojas-Benítez, D.; Allende, M.L. Cellular Dynamics during Spinal Cord Regeneration in Larval Zebrafish. Dev. Neurosci. 2019, 41, 112–122. [Google Scholar] [CrossRef]
- Tsata, V.; Wehner, D. Know How to Regrow—Axon Regeneration in the Zebrafish Spinal Cord. Cells 2021, 10, 1404. [Google Scholar] [CrossRef]
- Lochhead, R.; Sonntag, V.K.H. Laminectomy. In Encyclopedia of the Neurological Sciences; Academic Press: Cambridge, MA, USA, 2014; pp. 829–830. ISBN 9780123851574. [Google Scholar]
- Chen, J.; Qin, R. MicroRNA-138-5p Regulates the Development of Spinal Cord Injury by Targeting SIRT1. Mol. Med. Rep. 2020, 22, 328–336. [Google Scholar] [CrossRef]
- Jiang, T.; Qin, T.; Gao, P.; Tao, Z.; Wang, X.; Wu, M.; Gu, J.; Chu, B.; Zheng, Z.; Yi, J.; et al. SIRT1 Attenuates Blood-Spinal Cord Barrier Disruption after Spinal Cord Injury by Deacetylating P66Shc. Redox Biol. 2023, 60, 102615. [Google Scholar] [CrossRef]
- Zhong, G.; Yang, Y.; Huang, X.; Chen, J.; Feng, D.; Wei, K.; Chen, J.; Chen, H. The Serum SIRT1 Protein Is Associated with the Severity of Injury and Neurological Recovery in Mice with Traumatic Spinal Cord Injury. Neuroscience 2021, 469, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Han, D.; Zhu, K.; Jin, M.; Mei, X.; Lu, H. Effects of Sirtuin 1 on Microglia in Spinal Cord Injury: Involvement of Wnt/β-Catenin Signaling Pathway. Neuroreport 2019, 30, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Niu, J.; Dang, X. Neuroprotection of Melatonin on Spinal Cord Injury by Activating Autophagy and Inhibiting Apoptosis via SIRT1/AMPK Signaling Pathway. Biotechnol. Lett. 2020, 42, 2059–2069. [Google Scholar] [CrossRef]
- Yan, P.; Bai, L.; Lu, W.; Gao, Y.; Bi, Y.; Lv, G. Regulation of Autophagy by AMP-Activated Protein Kinase/Sirtuin 1 Pathway Reduces Spinal Cord Neurons Damage. Iran. J. Basic Med. Sci. 2017, 20, 1029–1036. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Chen, X.; Zaeem, M.; Zhang, W.; Song, L.; Chen, L.; Mubwandarikwa, J.; Chen, X.; Xiao, J.; Xie, L.; et al. Sesamol Attenuates Neuroinflammation by Regulating the AMPK/SIRT1/NF- κ B Signaling Pathway after Spinal Cord Injury in Mice. Oxid. Med. Cell. Longev. 2022, 2022, 8010670. [Google Scholar] [CrossRef]
- Chen, H.; Ji, H.; Zhang, M.; Liu, Z.; Lao, L.; Deng, C.; Chen, J.; Zhong, G. An Agonist of the Protective Factor SIRT1 Improves Functional Recovery and Promotes Neuronal Survival by Attenuating Inflammation after Spinal Cord Injury. J. Neurosci. 2017, 37, 2916–2930. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Mei, X.; Yang, D.; Tu, G. Resveratrol Inhibits Inflammation after Spinal Cord Injury via SIRT-1/NF-ΚB Signaling Pathway. Neurosci. Lett. 2021, 762, 136151. [Google Scholar] [CrossRef]
- Yu, S.; Xie, L.; Liu, Z.; Li, C.; Liang, Y. MLN4924 Exerts a Neuroprotective Effect against Oxidative Stress via Sirt1 in Spinal Cord Ischemia-Reperfusion Injury. Oxid. Med. Cell. Longev. 2019, 2019, 7283639. [Google Scholar] [CrossRef]
- Zhong, G.; Yang, Y.; Feng, D.; Wei, K.; Chen, J.; Chen, J.; Deng, C. Melatonin Protects Injured Spinal Cord Neurons From Apoptosis by Inhibiting Mitochondrial Damage via the SIRT1/Drp1 Signaling Pathway. Neuroscience 2023, 534, 54–65. [Google Scholar] [CrossRef]
- Jablonska, B.; Gierdalski, M.; Chew, L.J.; Hawley, T.; Catron, M.; Lichauco, A.; Cabrera-Luque, J.; Yuen, T.; Rowitch, D.; Gallo, V. Sirt1 Regulates Glial Progenitor Proliferation and Regeneration in White Matter after Neonatal Brain Injury. Nat. Commun. 2016, 7, 13866. [Google Scholar] [CrossRef]
- Gargiolo, C.; Slack, J.M.W. Cell Lineage Tracing during Xenopus Tail Regeneration. Development 2004, 131, 2669–2679. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, T.; Taniguchi, Y.; Tazaki, A.; Ueno, N.; Watanabe, K.; Mochii, M. Differential Gene Expression between the Embryonic Tail Bud and Regenerating Larval Tail in Xenopus Laevis. Dev. Growth Differ. 2004, 46, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.M.; Ayers, J.L.; Koran, L.; Carlson, J.; Anderson, M.C.; Simpson, S.B. Time Course of Salamander Spinal Cord Regeneration and Recovery of Swimming: HRP Retrograde Pathway Tracing and Kinematic Analysis. Exp. Neurol. 1990, 108, 198–213. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.M.; Duffy, M.T.; Simpson, S.B. Bulbospinal and Intraspinal Connections in Normal and Regenerated Salamander Spinal Cord. Exp. Neurol. 1989, 103, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Wang, Z.; Shi, Y.; Dong, L.; Wang, C. Modulating Macrophage Activities to Promote Endogenous Bone Regeneration: Biological Mechanisms and Engineering Approaches. Bioact. Mater. 2021, 6, 244–261. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yeung, K.W.K. Bone Grafts and Biomaterials Substitutes for Bone Defect Repair: A Review. Bioact. Mater. 2017, 2, 224–247. [Google Scholar] [CrossRef]
- Zhou, M.; Graves, D.T. Impact of the Host Response and Osteoblast Lineage Cells on Periodontal Disease. Front. Immunol. 2022, 13, 998244. [Google Scholar] [CrossRef]
- Pountos, I.; Giannoudis, P.V. Fracture Healing: Back to Basics and Latest Advances. In Fracture Reduction and Fixation Techniques: Upper Extremities; Springer: Cham, Switzerland, 2018; pp. 3–17. ISBN 9783319686288. [Google Scholar]
- Bahney, C.S.; Hu, D.P.; Miclau, T.; Marcucio, R.S. The Multifaceted Role of the Vasculature in Endochondral Fracture Repair. Front. Endocrinol. 2015, 6, 4. [Google Scholar] [CrossRef]
- Sheen, J.R.; Garla, V.V. Fracture Healing Overview. StatPearls: St. Petersburg, FL, USA, 2019. [Google Scholar]
- Song, D.; Xu, P.; Liu, S.; Wu, S. Dental Pulp Stem Cells Expressing Sirt1 Improve New Bone Formation during Distraction Osteogenesis. Am. J. Transl. Res. 2019, 11, 832–843. [Google Scholar]
- Huang, X.; Shu, H.; Ren, C.; Zhu, J. SIRT3 Improves Bone Regeneration and Rescues Diabetic Fracture Healing by Regulating Oxidative Stress. Biochem. Biophys. Res. Commun. 2022, 604, 109–115. [Google Scholar] [CrossRef]
- Dave, H.D.; Shook, M.; Varacallo, M. Anatomy, Skeletal Muscle. StatPearls: St. Petersburg, FL, USA, 2023. [Google Scholar]
- Chalkiadaki, A.; Igarashi, M.; Nasamu, A.S.; Knezevic, J.; Guarente, L. Muscle-Specific SIRT1 Gain-of-Function Increases Slow-Twitch Fibers and Ameliorates Pathophysiology in a Mouse Model of Duchenne Muscular Dystrophy. PLoS Genet. 2014, 10, e1004490. [Google Scholar] [CrossRef] [PubMed]
- Mañas-García, L.; Guitart, M.; Duran, X.; Barreiro, E. Satellite Cells and Markers of Muscle Regeneration during Unloading and Reloading: Effects of Treatment with Resveratrol and Curcumin. Nutrients 2020, 12, 1870. [Google Scholar] [CrossRef] [PubMed]
- Myers, M.J.; Shepherd, D.L.; Durr, A.J.; Stanton, D.S.; Mohamed, J.S.; Hollander, J.M.; Alway, S.E. The Role of SIRT1 in Skeletal Muscle Function and Repair of Older Mice. J. Cachexia. Sarcopenia Muscle 2019, 10, 929–949. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Goldberg, A.L. SIRT1 Protein, by Blocking the Activities of Transcription Factors FoxO1 and FoxO3, Inhibits Muscle Atrophy and Promotes Muscle Growth. J. Biol. Chem. 2013, 288, 30515–30526. [Google Scholar] [CrossRef]
- Lee, E.J.; Lee, M.M.; Park, S.Y.; Jeong, K.S. Sirt2 Positively Regulates Muscle Regeneration after Notexin-Induced Muscle Injury. Exp. Mol. Pathol. 2022, 127, 104798. [Google Scholar] [CrossRef]
- Hay, E.D. The Fine Structure of Blastema Cells and Differentiating Cartilage Cells in Regenerating Limbs of Amblystoma Larvae. J. Biophys. Biochem. Cytol. 1958, 4, 583–591. [Google Scholar] [CrossRef]
- Gerber, T.; Murawala, P.; Knapp, D.; Masselink, W.; Schuez, M.; Hermann, S.; Gac-Santel, M.; Nowoshilow, S.; Kageyama, J.; Khattak, S.; et al. Single-Cell Analysis Uncovers Convergence of Cell Identities during Axolotl Limb Regeneration. Science 2018, 362, eaaq0681. [Google Scholar] [CrossRef]
- Bryant, S.V.; Endo, T.; Gardiner, D.M. Vertebrate Limb Regeneration and the Origin of Limb Stem Cells. Int. J. Dev. Biol. 2002, 46, 887–896. [Google Scholar]
- Kragl, M.; Knapp, D.; Nacu, E.; Khattak, S.; Maden, M.; Epperlein, H.H.; Tanaka, E.M. Cells Keep a Memory of Their Tissue Origin during Axolotl Limb Regeneration. Nature 2009, 460, 60–65. [Google Scholar] [CrossRef]
- Lin, Y.F.; Sam, J.; Evans, T. Sirt1 Promotes Tissue Regeneration in Zebrafish through Regulating the Mitochondrial Unfolded Protein Response. iScience 2021, 24, 103118. [Google Scholar] [CrossRef]
- Busse, E.; Simkin, J.; Marrero, L.; Stewart, K.; Brunauer, R.; Muneoka, K.; Guntur, A.; Lacey, M.; Sammarco, M. Sirtuin 3 Deficiency Does Not Impede Digit Regeneration in Mice. Sci. Rep. 2019, 9, 16491. [Google Scholar] [CrossRef] [PubMed]
- Porcu, M.; Chiarugi, A. The Emerging Therapeutic Potential of Sirtuin-Interacting Drugs: From Cell Death to Lifespan Extension. Trends Pharmacol. Sci. 2005, 26, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Gan, L. Therapeutic Potential of Sirtuin-Activating Compounds in Alzheimer’s Disease. Drug News Perspect. 2007, 20, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Balcerczyk, A.; Pirola, L. Therapeutic Potential of Activators and Inhibitors of Sirtuins. BioFactors 2010, 36, 383–393. [Google Scholar] [CrossRef]
- Lavu, S.; Boss, O.; Elliott, P.J.; Lambert, P.D. Sirtuins-Novel Therapeutic Targets to Treat Age-Associated Diseases. Nat. Rev. Drug Discov. 2008, 7, 841–853. [Google Scholar] [CrossRef]

| Activators | Cellular Effects | Mechanisms | References |
|---|---|---|---|
| SRT1720 | It improves the motor restoration and decreases inflammation in lumbar neurons | It reduces the levels of the proinflammatory cytokines TNF-α, IL-12, and IL-10 and it reduces the recruitment of inflammatory macrophages | [166,170] |
| Melatonin | It increases autophagy and decreases the apoptosis of vertebral cells | It increases the autophagic factors beclin 1 and LC-3B | [167] |
| Resveratrol | It increased autophagy and reduces apoptosis | It decreases the levels of BAX and caspase-3 | [168,171] |
| Pevonedistat | It reduces apoptosis | It decreases the levels of BAX and caspase-3 | [172] |
| Sesamol | It accelerates the restoration of motor ability | [169] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Núñez, A.K.; Arenas-Gómez, C.M.; Carbonell Medina, B.A. Sirtuins: Emergent Players in Tissue and Organ Regeneration. Int. J. Transl. Med. 2024, 4, 687-709. https://doi.org/10.3390/ijtm4040048
Núñez AK, Arenas-Gómez CM, Carbonell Medina BA. Sirtuins: Emergent Players in Tissue and Organ Regeneration. International Journal of Translational Medicine. 2024; 4(4):687-709. https://doi.org/10.3390/ijtm4040048
Chicago/Turabian StyleNúñez, Ayla Kyler, Claudia Marcela Arenas-Gómez, and Belfran Alcides Carbonell Medina. 2024. "Sirtuins: Emergent Players in Tissue and Organ Regeneration" International Journal of Translational Medicine 4, no. 4: 687-709. https://doi.org/10.3390/ijtm4040048
APA StyleNúñez, A. K., Arenas-Gómez, C. M., & Carbonell Medina, B. A. (2024). Sirtuins: Emergent Players in Tissue and Organ Regeneration. International Journal of Translational Medicine, 4(4), 687-709. https://doi.org/10.3390/ijtm4040048






