How to Avoid False-Negative and False-Positive COVID-19 PCR Testing
Abstract
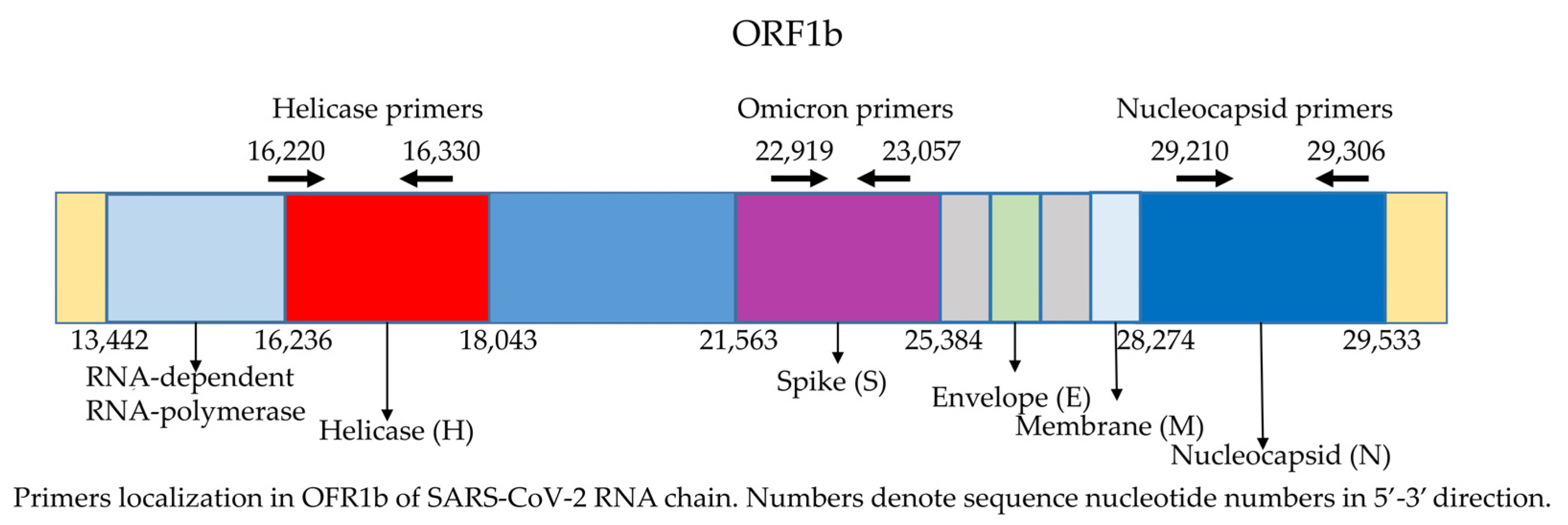
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jing, R.; Kudinha, T.; Zhou, M.L.; Xiao, M.; Wang, H.; Yang, W.H.; Xu, Y.C.; Hsueh, P.R. Laboratory diagnosis of COVID-19 in China: A review of challenging cases and analysis. J. Microbiol. Immunol. Infect. 2021, 54, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Gdoura, M.; Abouda, I.; Mrad, M.; Dhifallah, B.I.; Belaiba, Z.; Fares, W.; Chouikha, A.; Khedhiri, M.; Layouni, K.; Touzi, H.; et al. SARS-CoV2 RT-PCR assays: In vitro comparison of 4 WHO approved protocols on clinical specimens and its implications for real laboratory practice through variant emergence. Virol. J. 2022, 19, 54. [Google Scholar] [CrossRef] [PubMed]
- Onwuamah, C.K.; Okwuraiwe, A.P.; Salu, O.B.; Shaibu, J.O.; Ndodo, N.; Amoo, S.O.; Okoli, L.C.; Ige, F.A.; Ahmed, R.A.; Bankole, M.A.; et al. Comparative performance of SARS-CoV-2 real-time PCR diagnostic assays on samples from Lagos, Nigeria. PLoS ONE 2021, 16, e0246637. [Google Scholar] [CrossRef]
- Tombuloglu, H.; Sabit, H.; Al-Khallaf, H.; Kabanja, J.H.; Alsaeed, M.; Al-Saleh, N.; Al-Suhaimi, E. Multiplex real-time RT-PCR method for the diagnosis of SARS-CoV-2 by targeting viral N, RdRP and human RP genes. Sci. Rep. 2022, 12, 2853. [Google Scholar] [CrossRef]
- Yan, Y.; Chang, L.; Wang, L. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): Current status, challenges, and countermeasures. Rev. Med. Virol. 2020, 30, e2106. [Google Scholar] [CrossRef] [PubMed]
- Philippe, C.; Antonio, M.; Mauro, P.; Gerhard, B.; Maddalena, Q.; Merien, B.; van den Eede, G. Specific Detection of the SARS-CoV-2 Omicron Variant by RT-PCR. Eur. Comm. Jt. Res. Cent. (JRC) 2022. preprint. [Google Scholar] [CrossRef]
- Chan, J.F.; Yip, C.C.; To, K.K.; Tang, T.H.; Wong, S.C.; Leung, K.H.; Fung, A.Y.; Ng, A.C.; Zou, Z.; Tsoi, H.W.; et al. Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and with Clinical Specimens. J. Clin. Microbiol. 2020, 58, e00310–e00320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabert, J.; Beillard, E.; van der Velden, V.H.; Bi, W.; Grimwade, D.; Pallisgaard, N.; Barbany, G.; Cazzaniga, G.; Cayuela, J.M.; Cavé, H.; et al. Standardization and quality control studies of ’real-time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia—A Europe Against Cancer program. Leukemia 2003, 17, 2318–2357. [Google Scholar] [CrossRef] [PubMed]
- Ishige, T.; Murata, S.; Taniguchi, T.; Miyabe, A.; Kitamura, K.; Kawasaki, K.; Nishimura, M.; Igari, H.; Matsushita, K. Highly sensitive detection of SARS-CoV-2 RNA by multiplex rRT-PCR for molecular diagnosis of COVID-19 by clinical laboratories. Clin. Chim. Acta 2020, 507, 139–142. [Google Scholar] [CrossRef] [PubMed]
- Fevraleva, I.; Glinshchikova, O.; Sidorova, J.; Yakutik, I.; Sudarikov, A. On the issue of housekeeping gene primer design for adequate assessment of the expression of the chimeric transcripts. Russ. J. Hematol. Transfusiology 2022, 67, 313. [Google Scholar]

| Gene Target | Primer/ Probe | Sequence 5′-3′ | Ref. |
|---|---|---|---|
| Helicase | Forward Reverse Probe | cgcatacagtcttrcaggct gtgtgatgttgawatgacatggtc FAM-taagatgtggtgcttgcatacgtagac-RTQ2 | [7] |
| Nucleocapsid | Forward Reverse Probe | gcgttcttcggaatgtcg ttggatctttgtcatccaatttg Cy5-aacgtggttgacctacacagst-RTQ2 | [7] |
| Spike (Omicron strain) | Forward Reverse Probe | aacaaaccttgtaatggtgttgc tgctggtgcatgtagaagttc R6G-gatcatatagtttccgacccacttatggtgttggtc-RTQ2 | [6] |
| ABL1—internal control | Forward Reverse Probe | gtccacactgcaatgtttttgtg gagttccaacgagcggcttcactc ROX-ccagtagcatctgactttgagcctcag-RTQ2 | This study |
| Sample | Helicase Tc | Nucleocapsid Tc | ABL1 Tc | Syntol Kit Tc | Synlol Kit Control Tc | Our Result | Syntol Kit Result |
|---|---|---|---|---|---|---|---|
| N | 21 | 20 | 32 | 23 | 25 | Positive | Positive |
| B | 29 | 28 | 31 | 30 | 24 | Positive | Positive |
| K | >45 | >45 | 32 | >45 | 26 | Negative | Negative |
| V | >45 | >45 | >45 | >45 | 25 | Failure | False-negative |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fevraleva, I.; Glinshchikova, O.; Makarik, T.; Sudarikov, A. How to Avoid False-Negative and False-Positive COVID-19 PCR Testing. Int. J. Transl. Med. 2022, 2, 204-209. https://doi.org/10.3390/ijtm2020018
Fevraleva I, Glinshchikova O, Makarik T, Sudarikov A. How to Avoid False-Negative and False-Positive COVID-19 PCR Testing. International Journal of Translational Medicine. 2022; 2(2):204-209. https://doi.org/10.3390/ijtm2020018
Chicago/Turabian StyleFevraleva, Irina, Olga Glinshchikova, Tatiana Makarik, and Andrey Sudarikov. 2022. "How to Avoid False-Negative and False-Positive COVID-19 PCR Testing" International Journal of Translational Medicine 2, no. 2: 204-209. https://doi.org/10.3390/ijtm2020018
APA StyleFevraleva, I., Glinshchikova, O., Makarik, T., & Sudarikov, A. (2022). How to Avoid False-Negative and False-Positive COVID-19 PCR Testing. International Journal of Translational Medicine, 2(2), 204-209. https://doi.org/10.3390/ijtm2020018






