Artificial Intelligence in Translational Medicine
Abstract
:1. Introduction
2. Artificial Intelligence (AI) and Machine Learning (ML) in Translational Medicine
2.1. Drug Discovery and Development, and Drug Target Prediction
2.1.1. Drug Discovery and Development
| AI Technique | Target | Dataset | Statistical Parameters | Outcomes | Ref |
|---|---|---|---|---|---|
| Bayesian ML models | GSK-3β AD | 2368 compounds | Cross-validation, ROC curve = 0.905 | Virtual screening found ruboxistaurin (CHEMBL91829) as GSK-3β (IC50 = 97.3 nM) and GSK-3α (IC50 = 695.9 nM) inhibitor | [63] |
| Bayesian ML and RP algorithms for developing a multi-QSAR approach | 25 crucial cellular targets in AD | 18,741 active compounds against the selected targets | Internal and external validation (area under the ROC curve for the test set 0.741–1.0, average 0.965) | Identification of various MTDLs against AD (seven AChE inhibitors (IC50 = 0.442–72.26 μM); four H3R antagonists (IC50 = 0.308–58.6 μM). The best performing MTDL (DL0410) showed a dual cholinesterase inhibitor behavior (IC50 AChE = 0.442 μM; IC50 BuChE = 3.57 μM), and behaved as a H3R antagonist (IC50 = 0.308 μM) | [82] |
| ML-based approach | DRIAD for drug repurposing in AD | DRIAD was applied to find relationships between the pathology of AD severity (the Braak stage) and molecular mechanisms as determined in records of gene names by using 80 FDA-approved and investigational drugs | Model performance was evaluated through leave-pair-out cross-validation, area under the ROC curve ranging from 0.6 to 0.8 | 33 FDA-approved drugs can be used for repurposing immediately | [69] |
| SVM models coupled with Tanimoto similarity-based clustering analysis | A2A and D2 receptor subtypes as targets for PD | 135 compounds (96 from A2A and 39 from D2) | Experimental validation | Virtual screening of over 13.5 million compounds from PubChem and MDDR databases. Two compounds behaved as multifunctional ligands against human A2A (Ki = 8.7 and 11.2 μM) and D2 receptors (EC50 = 22.5 and 40.2 μM) | [83] |
| SVM and SVR | PD drug discovery A2A vs. A3 receptor subtype selectivity profiles and related binding affinities | For SVM, 104 selective N7- and N8-substituted pyrazolo–triazolo–pyrimidine analogs. For SVR, 104 N8-substituted pyrazolo–triazolo–pyrimidine derivatives. A test set of 51 N8-substituted pyrazolo–triazolo–pyrimidine analogs to validate both SVM and SVR models | LOO-cv Correct prediction 93.3, sensitivity 92.0, specificity 94.4 | 51 novel pyrazolo–triazolo–pyrimidine containing compounds that confirmed the predicted receptor subtype selectivity and the related binding affinity profiles | [84] |
| SVM and RF | Anticancer drug discovery—target FEN1 | The training set contained 1163 FEN1 inhibitors and 281,583 non-inhibitors; the test set 388 inhibitors and 93,861 non-inhibitors | For the test set: sensitivity 0.54, specificity 0.99, MCC 0.67 | The computational tool was used in a virtual screening employing the Maybridge database (53,000 molecules). Five top-ranked compounds were experimentally validated. The molecule JFD00950 behaved as a FEN1 inhibitor in the micromolar range, inhibiting Flap cleavage activity, showing cytotoxic activity against colon cancer cells (DLD-1, IC50 = 16.7 µM) | [85] |
| ML models using naïve Bayesian and RP techniques | Indoleamine 2,3-dioxygenase (IDO), a promising target for cancer immunotherapy | The model was trained using a library of established IDO inhibitors (504 compounds, 242 active and 262 inactive) | The Q values for the test set of the top 10 models are greater than 0.76, the MCC values >0.53, the area under ROC curve >0.89 | Virtual screening campaign using a proprietary chemical library. This step provided 50 potential IDO inhibitors that were experimentally validated. In vitro tests confirmed the prediction of the ML model, since three new IDO inhibitors, belonging to the tanshinone family, were identified (IC50s = 1.30, 4.10, and 4.68 μM) | [86] |
| ML model using naïve Bayesian technique coupled with a molecular docking calculation | VEGFR-2, a drug target for developing anticancer compounds with anti-angiogenic activity | The model was trained using 3464 VEGFR-2 inhibitors | MCC of 0.966 and 0.951 considering the test set and external validation set | Virtual screening protocol for identifying VEGFR-2 inhibitors using a chemical library containing 1841 FDA-approved drugs. Papaverine, rilpivirine, and flubendazole were able to inhibit VEGFR-2 (IC50 = 0.47–6.29 μM) | [87] |
| Four distinct ML algorithms to train the model (LR, naïve Bayesian, SVM, and RF) | Anticancer drug discovery—target BCRP | The dataset contained 433 inhibitors and 545 noninhibitors, collected from 47 publications | Cross-validation (area under ROC curve = 0.9) and predictivity in prospective validation (area under ROC curve = 0.7) | Virtual screening approach using a drug library (1702 compounds). 10 drugs as potential BCRP inhibitors were identified (inhibition of mitoxantrone efflux in BCRP-expressing PLB985 cells). Among the drugs tested two of them behaved as BCRP inhibitors (cisapride and roflumilast, IC50 = 0.4 µM and 0.9 µM, respectively) | [88] |
| ML model, based on Laplacien-modified naïve Bayesian classifiers. The ML model for EGFR was coupled with a structure-based technique regarding the bromodomain | Anticancer drug discovery—target EGFR/BRD4 | Two ML models for EGFR were developed considering ECFP4 based on a total of 591,744 unique kinase compounds (one with 3058 active molecules, pIC50/pKi ≥ 7, and another with 4785 active compounds, pIC50/pKi ≥ 6). | Area under ROC curve values of 0.98 to 0.99 based on 50/50 training/test set and assessed employing LOO-cv | Virtual screening campaign employing a large database (eMolecules > 6 million compounds). Among them, a first-in-class dual EGFR–BRD4 inhibitor (compound 2870) was found (EGFR IC50 = 44 nM; ERBB2, ERBB4, and BRD4 IC50 = 8.73, 24.2, and 9.02 μM, respectively) | [89] |
| ML model based on a GCNN algorithm | DeepMalaria antimalarial drug discovery | 13,446 potential antimalarials contained in GSK database | Accuracy from 44.13% in the whole library to 87.75%. Accuracy of 100% for all nanomolar active compounds | The developed model was validated by predicting hit molecules from an additional chemical collection and a FDA-approved drug database. DeepMalaria identified all molecules showing nanomolar activity and 87.5% of chemicals with greater percentage of inhibition | [92] |
| DL method DNN model | Discovery of novel antibiotic agents, possessing a broad-spectrum antibacterial profile | Dataset of 2335 molecules | Area under ROC curve of 0.896 considering the test data | Virtual screening of various chemical libraries. From this screening step, they identify an existing drug, namely, halicin (SU-3327), showing interesting bactericidal activity in vitro as well as in vivo. It was found to be effective against M. tuberculosis. Virtual screening of ZINC15 (>100 million compounds) provided eight further antibacterial agents, chemically unrelated to known antibiotics. ZINC000100032716 and ZINC000225434673 showed strong broad-spectrum activity, overcoming a range of frequent resistance factors | [94] |
| ML models, employing naïve Bayesian and RP techniques | DNA gyrase to find broad-spectrum antibacterial agents | 137 DNA gyrase inhibitors spanning several orders of magnitude | The overall predictive accuracy, considering the training and test sets, was greater than 80% | ML models used for virtual screening of a chemical library. The potential hits were experimentally validated against DNA gyrase, E. coli, methicillin-resistant S. aureus and other bacteria. For compounds able to inhibit DNA gyrase, MIC values range between 1 and 32 μg/mL, and the relative inhibition rates of inhibitors, range from 42% to 75% at 1 μM | [95] |
| Bayesian ML model | Antiviral research—Ebola virus | 868 molecules viral pseudotype entry assay and the Ebola virus replication assay data | Cross-validation showed ROC values greater than 0.8 | Virtual screening campaign using the MicroSource library of drugs, for selecting possible antiviral compounds. Among the retrieved potential hit compounds, three promising antiviral candidates were found (quinacrine, pyronaridine, and tilorone EC50 = 350, 420, and 230 nM, respectively, against Ebola virus replication). | [96] |
| GENTRL | For de novo small molecule design acting as inhibitors of DDR1 kinase | The model was generated using six data sets: (i) molecules from the ZINC database; (ii) inhibitors of DDR1 kinase; (iii) common kinase inhibitors (positive set); (iv) actives against non-kinase targets (negative set); (v) patent data of biological actives; (vi) 3D structures for DDR1 inhibitors | Experimental validation—GENTRL allowed indication of several compounds for the synthesis, and the authors synthesized six lead compounds | Two molecules strongly inhibited DDR1 activity (IC50 = 10–21 nM), the other two compounds showed moderate potency (IC50 = 0.278–1 μM) | [100] |
| ML models RF and GCNN | Three drug targets (sEH, a hydrolase, ERα, a nuclear receptor, c-KIT, a kinase) | Models were trained on the DEL selection data for classifying molecules (over 2000) | Experimental validation | Virtual screening of large chemical databases (∼88 million compounds). The outcomes revealed that the technique is efficient, with a global hit rate of ∼30% at 30 μM, discovering powerful compounds (IC50 < 10 nM) for each drug target | [101] |
| DL and reinforcement learning DNNs | De novo design of small molecules with desired profile, and JAK2 as the target protein | The generative network was trained with ~1.5 million structures from the ChEMBL21 database | Experimental validation | ReLeaSE was successfully applied for generating a series of libraries containing chemical entities with a precise profile: (a) satisfactory drug-likeness, regarding physchem properties, for which the authors chose Tm and n-octanol/water partition coefficient (logP); (b) desired biological activity, for which the authors selected Janus protein kinase 2 (JAK2) as the target protein | [97] |
2.1.2. Drug Target Prediction and Biomarker Identification
2.1.3. AI/ML in Quantitative Systems Pharmacology (QSP)
2.2. Imaging, Biomarkers, Diagnosis, and Disease Progression
2.2.1. General Consideration
2.2.2. Basic Research
2.2.3. AI, Imaging and Ophthalmology
2.2.4. AI/ML in Central Nervous System (CNS)-Related Disorders
2.2.5. AI in Cardiology and Cardiovascular Diseases
2.2.6. AI in Gastroenterology
| AI Technique | Target | Dataset | Statistical Parameters | Outcomes | Ref |
|---|---|---|---|---|---|
| ML algorithm | Approach for detecting small (<5 mm) adenomatous or sessile polyps | The ML-based model was trained with data from 325 subjects presenting 466 microscopic polyps | The model showed an accuracy of 94% (negative predictive value 96%), with a pathologic prediction rate of 98.1% (457/466) | The application of ML models can be useful for assisting clinicians in detecting gastric pathological state | [307] |
| ML model based on DL algorithm | Approach for detecting polyps in clinical colonoscopy investigations | The model was trained using data from 1290 patients (5545 colonoscopy images containing polyps and images with no polyps). Validation set of 27,113 colonoscopy images of 1138 patients with one detected polyp | The model showed sensitivity of 94.38%, specificity of 95.92%, area under the ROC curve of 0.984 | The developed DL model has great potential in assisting clinicians while conducting colonoscopy, being able to correctly discriminate polyps and adenomas | [308] |
| ML model based on deep CNN technique | Approach for real-time evaluation of endoscopic video images of colorectal polyps | The model was trained and validated using untouched video data derived from routine clinical investigations. An independent set of 125 videos was used for the validation | The ML model showed a sensitivity of 98%, a specificity of 83%, and an accuracy of 94% | The model was able to discriminate hyperplastic from adenomatous polyps | [309] |
| ML model based on deep CNN technique | Approach for detecting polyps from colonoscopy exams | The ML model was trained used 8641 hand-labeled images, with 4088 unique polyps, from colonoscopy derived from over 2000 subjects. The authors tested the model using 20 colonoscopy videos (5 h of duration) | The model showed an area under the ROC curve of 0.991 and an accuracy of 96.4% | The CNN algorithm identified a number of polyps higher than that observed from expert clinicians | [310] |
| ML model based on CNN algorithm | Development of a model for detecting ESCC and assessing its invasiveness | The model was trained using 1751 images of ESCC (white light imaging and narrow-band imaging endoscopic images) | In the validation step, the model identified 95.5% of ESCC properly, estimating the invasion depth of ESCC (sensitivity of 84.1% and accuracy of 80.9%) | The diagnosis assisted by CNN algorithm was more accurate than diagnosis by expert clinicians, indicating a role of ML as ESCC diagnostic tool | [311] |
| ML tool based on DNN algorithm | Approach for diagnosing the invasion depth of ESCC | The model was built using a training set of 8660 non-magnified endoscopic images and 5678 magnified images from 804 patients with superficial ESCC presenting cancer invasion. Validation set consisted of 405 non-magnified ad 509 magnified images from 155 subjects | The model showed specificity 95.8%, sensitivity 90.1%, accuracy 91%, positive predicted value 99.2% negative predictive value 63.9% | These parameters highlighted the capacity of the model to detect invasion depth in patients with superficial ESCC | [312] |
| ML tool based on CNN algorithm | Approach for assessing the severity of IBD and improving its classification | The model was trained on 26,304 colonoscopy images derived from 841 subjects with ulcerative colitis. The model was assessed using an independent test set (3981 images from 114 patients with ulcerative colitis) | The validation was achieved by calculating the areas under the ROC curve, and the results for the ML-based model were 0.86 and 0.98 in identifying normal mucosa (Mayo 0) and mucosal healing state (Mayo 0–1) | This work indicated that the CNN algorithm can assist clinicians in determining severity-based therapies as well as follow-up endoscopy waits for IBD | [313] |
| ML model | ML model for classifying pediatric IBD | The model was trained using data derived from endoscopic and histological imaging of 287 children affected by IBD | Three supervised ML models showed a classification accuracy of 71.0%, 76.9%, and 82.7%. The most promising ML model properly classified patients (accuracy of 83.3%) | This work indicated that for the development of a proper model, it is necessary to consider both endoscopic and histological data for a more accurate disease classification | [314] |
| ML model | Approach for detecting subjects presenting celiac disease and for classifying the disorder | The model was trained using 612 endoscopic images from pediatric patients considering a two-class issue: mucosa affected by celiac disease and unaffected duodenal tissue | The model discriminated celiac disease with an overall accuracy of 88%. The model showed a reduced accuracy (63.7%) in classifying the severity of disorders | The classification method was able to discriminate celiac disease into two groups (disease vs. no disease) | [316] |
| CNN transfer-learning | Approach for classifying luminal endoscopic images of celiac disease | The training set was composed of 1661 images from luminal endoscopic data | The model showed an accuracy of 90.5% in identifying celiac disease considering endoscopic images alone | ML could offer a new method in diagnostic settings, especially where acquiring biopsies is complicated | [317] |
| ML-based models | Approach for detecting undiagnosed celiac disease | The training set was composed of serum samples derived from 47,557 subjects, whit no previous diagnosis of celiac disease | The models showed an area under the ROC curve ranging from 0.49 to 0.53. Two models (RF and bagged classification trees) showed better performance (likelihood >95%) | Considering the selected variables, the development of predictive models could be impractical, since they did not characterize undiagnosed celiac disease | [318] |
| ML algorithm | Approach for an automated classification of duodenal biopsy images | The model was trained using biopsy images extracting features (734 features from each set of data and so, 26,424 features were extracted from three diverse sets of data) from two classes (normal and celiac) | The model showed: accuracy of 88.89%, sensitivity of 89.67% specificity of 86.67% in the two-class classification | The approach for an automatic classification of biopsy pictures can help with the process of evaluating villous atrophy, suggesting that automation of biopsy images is a feasible task | [319] |
| DL decision support method based on DNN algorithm | Approach for detecting Helicobacter pylori considering gastric biopsies | The model was trained considering Giemsa and H&E slides (191 H&E- and 286 Giemsa-stained slides for a total of 2629 tiles containing Giemsa and 790 H&E; additionally, 4241 (Giemsa) and 1533 (H&E) tiles without H. pylori-like bacterial structures) | Several validation approaches were used showing an area under the ROC curve >0.8 | The model was able to detect H. pylori, indicating that ML tools can assist clinicians in diagnosis regarding the presence of H. pylori in gastric biopsies | [320] |
2.2.7. AI in Dermatology
3. Conclusions and Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Available online: https://www.brookings.edu/techstream/what-investment-trends-reveal-about-the-global-ai-landscape/ (accessed on 14 October 2021).
- Available online: https://outsideinsight.com/insights/global-ai-investment-150-billion-2025/ (accessed on 14 October 2021).
- Hinton, G. Deep Learning-A Technology With the Potential to Transform Health Care. JAMA 2018, 320, 1101–1102. [Google Scholar] [CrossRef]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Cui, C.; Corrado, G.; Thrun, S.; Dean, J. A guide to deep learning in healthcare. Nat. Med. 2019, 25, 24–29. [Google Scholar] [CrossRef]
- He, J.; Baxter, S.L.; Xu, J.; Xu, J.; Zhou, X.; Zhang, K. The practical implementation of artificial intelligence technologies in medicine. Nat. Med. 2019, 25, 30–36. [Google Scholar] [CrossRef]
- Toh, T.S.; Dondelinger, F.; Wang, D. Looking beyond the hype: Applied AI and machine learning in translational medicine. EBioMedicine 2019, 47, 607–615. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device#whatis (accessed on 14 October 2021).
- Murdoch, T.B.; Detsky, A.S. The inevitable application of big data to health care. JAMA 2013, 309, 1351–1352. [Google Scholar] [CrossRef]
- Jiang, F.; Jiang, Y.; Zhi, H.; Dong, Y.; Li, H.; Ma, S.; Wang, Y.; Dong, Q.; Shen, H.; Wang, Y. Artificial intelligence in healthcare: Past, present and future. Stroke Vasc. Neurol. 2017, 2, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine Learning in Medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef] [PubMed]
- Beam, A.L.; Kohane, I.S. Big Data and Machine Learning in Health Care. JAMA 2018, 319, 1317–1318. [Google Scholar] [CrossRef]
- Muhammad, L.J.; Algehyne, E.A.; Usman, S.S.; Ahmad, A.; Chakraborty, C.; Mohammed, I.A. Supervised Machine Learning Models for Prediction of COVID-19 Infection using Epidemiology Dataset. SN Comput. Sci. 2021, 2, 11. [Google Scholar] [CrossRef]
- Mirri, S.; Delnevo, G.; Roccetti, M. Is a COVID-19 Second Wave Possible in Emilia-Romagna (Italy)? Forecasting a Future Outbreak with Particulate Pollution and Machine Learning. Computation 2020, 8, 74. [Google Scholar] [CrossRef]
- Patel, L.; Shukla, T.; Huang, X.; Ussery, D.W.; Wang, S. Machine Learning Methods in Drug Discovery. Molecules 2020, 25, 5277. [Google Scholar] [CrossRef] [PubMed]
- Reda, C.; Kaufmann, E.; Delahaye-Duriez, A. Machine learning applications in drug development. Comput. Struct. Biotechnol. J. 2020, 18, 241–252. [Google Scholar] [CrossRef]
- Gupta, R.; Srivastava, D.; Sahu, M.; Tiwari, S.; Ambasta, R.K.; Kumar, P. Artificial intelligence to deep learning: Machine intelligence approach for drug discovery. Mol. Divers. 2021, 25, 1315–1360. [Google Scholar] [CrossRef]
- Ramesh, N.; Tasdizen, T. Detection and segmentation in microscopy images. In Computer Vision for Microscopy Image Analysis; Academic Press Books; Elsevier: New York, NY, USA, 2021; pp. 43–71. [Google Scholar] [CrossRef]
- Alloghani, M.; Al-Jumeily, D.; Mustafina, J.; Hussain, A.; Aljaaf, A.J. A Systematic Review on Supervised and Unsupervised Machine Learning Algorithms for Data Science. In Supervised and Unsupervised Learning for Data Science; Springer Nature: London, UK, 2020; pp. 3–21. [Google Scholar] [CrossRef]
- Cleret de Langavant, L.; Bayen, E.; Yaffe, K. Unsupervised Machine Learning to Identify High Likelihood of Dementia in Population-Based Surveys: Development and Validation Study. J. Med. Internet Res. 2018, 20, e10493. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Ghoman, S.K.; Cutumisu, M.; Schmolzer, G.M. Unsupervised Machine Learning Algorithms Examine Healthcare Providers’ Perceptions and Longitudinal Performance in a Digital Neonatal Resuscitation Simulator. Front. Pediatr. 2020, 8, 544. [Google Scholar] [CrossRef]
- Roohi, A.; Faust, K.; Djuric, U.; Diamandis, P. Unsupervised Machine Learning in Pathology: The Next Frontier. Surg. Pathol. Clin. 2020, 13, 349–358. [Google Scholar] [CrossRef]
- Lopez, C.; Tucker, S.; Salameh, T.; Tucker, C. An unsupervised machine learning method for discovering patient clusters based on genetic signatures. J. Biomed. Inf. 2018, 85, 30–39. [Google Scholar] [CrossRef]
- Ceriotti, M. Unsupervised machine learning in atomistic simulations, between predictions and understanding. J. Chem. Phys. 2019, 150, 150901. [Google Scholar] [CrossRef] [Green Version]
- Ma, E.Y.; Kim, J.W.; Lee, Y.; Cho, S.W.; Kim, H.; Kim, J.K. Combined unsupervised-supervised machine learning for phenotyping complex diseases with its application to obstructive sleep apnea. Sci. Rep. 2021, 11, 4457. [Google Scholar] [CrossRef]
- Omta, W.A.; van Heesbeen, R.G.; Shen, I.; de Nobel, J.; Robers, D.; van der Velden, L.M.; Medema, R.H.; Siebes, A.; Feelders, A.J.; Brinkkemper, S.; et al. Combining Supervised and Unsupervised Machine Learning Methods for Phenotypic Functional Genomics Screening. SLAS Discov. 2020, 25, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Neftci, E.O.; Averbeck, B.B. Reinforcement learning in artificial and biological systems. Nat. Mach. Intell. 2019, 1, 133–143. [Google Scholar] [CrossRef]
- Yauney, G.; Shah, P. Reinforcement Learning with Action-Derived Rewards for Chemotherapy and Clinical Trial Dosing Regimen Selection. In Proceedings of the 3rd Machine Learning for Healthcare Conference, Palo Alto, CA, USA, 17–18 August 2018; pp. 161–226. [Google Scholar]
- Sirous, H.; Campiani, G.; Brogi, S.; Calderone, V.; Chemi, G. Computer-Driven Development of an in Silico Tool for Finding Selective Histone Deacetylase 1 Inhibitors. Molecules 2020, 25, 1952. [Google Scholar] [CrossRef] [Green Version]
- Brogi, S.; Brindisi, M.; Joshi, B.P.; Sanna Coccone, S.; Parapini, S.; Basilico, N.; Novellino, E.; Campiani, G.; Gemma, S.; Butini, S. Exploring clotrimazole-based pharmacophore: 3D-QSAR studies and synthesis of novel antiplasmodial agents. Bioorg Med. Chem. Lett. 2015, 25, 5412–5418. [Google Scholar] [CrossRef] [PubMed]
- Brogi, S.; Papazafiri, P.; Roussis, V.; Tafi, A. 3D-QSAR using pharmacophore-based alignment and virtual screening for discovery of novel MCF-7 cell line inhibitors. Eur. J. Med. Chem. 2013, 67, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Brogi, S.; Kladi, M.; Vagias, C.; Papazafiri, P.; Roussis, V.; Tafi, A. Pharmacophore modeling for qualitative prediction of antiestrogenic activity. J. Chem. Inf. Model. 2009, 49, 2489–2497. [Google Scholar] [CrossRef]
- Muratov, E.N.; Bajorath, J.; Sheridan, R.P.; Tetko, I.V.; Filimonov, D.; Poroikov, V.; Oprea, T.I.; Baskin, I.I.; Varnek, A.; Roitberg, A.; et al. QSAR without borders. Chem. Soc. Rev. 2020, 49, 3525–3564. [Google Scholar] [CrossRef]
- Lin, X.; Li, X.; Lin, X. A Review on Applications of Computational Methods in Drug Screening and Design. Molecules 2020, 25, 1375. [Google Scholar] [CrossRef] [Green Version]
- Kohlbacher, S.M.; Langer, T.; Seidel, T. QPHAR: Quantitative pharmacophore activity relationship: Method and validation. J. Cheminform. 2021, 13, 57. [Google Scholar] [CrossRef] [PubMed]
- Flori, L.; Petrarolo, G.; Brogi, S.; La Motta, C.; Testai, L.; Calderone, V. Identification of novel SIRT1 activators endowed with cardioprotective profile. Eur. J. Pharm. Sci. 2021, 165, 105930. [Google Scholar] [CrossRef]
- Sirous, H.; Chemi, G.; Campiani, G.; Brogi, S. An integrated in silico screening strategy for identifying promising disruptors of p53-MDM2 interaction. Comput. Biol. Chem. 2019, 83, 107105. [Google Scholar] [CrossRef]
- Battah, B.; Chemi, G.; Butini, S.; Campiani, G.; Brogi, S.; Delogu, G.; Gemma, S. A Repurposing Approach for Uncovering the Anti-Tubercular Activity of FDA-Approved Drugs with Potential Multi-Targeting Profiles. Molecules 2019, 24, 4373. [Google Scholar] [CrossRef] [Green Version]
- Sirous, H.; Chemi, G.; Gemma, S.; Butini, S.; Debyser, Z.; Christ, F.; Saghaie, L.; Brogi, S.; Fassihi, A.; Campiani, G.; et al. Identification of Novel 3-Hydroxy-pyran-4-One Derivatives as Potent HIV-1 Integrase Inhibitors Using in silico Structure-Based Combinatorial Library Design Approach. Front. Chem. 2019, 7, 574. [Google Scholar] [CrossRef] [Green Version]
- Batool, M.; Ahmad, B.; Choi, S. A Structure-Based Drug Discovery Paradigm. Int. J. Mol. Sci. 2019, 20, 2783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maia, E.H.B.; Assis, L.C.; de Oliveira, T.A.; da Silva, A.M.; Taranto, A.G. Structure-Based Virtual Screening: From Classical to Artificial Intelligence. Front. Chem. 2020, 8, 343. [Google Scholar] [CrossRef]
- Fischer, A.; Smiesko, M.; Sellner, M.; Lill, M.A. Decision Making in Structure-Based Drug Discovery: Visual Inspection of Docking Results. J. Med. Chem. 2021, 64, 2489–2500. [Google Scholar] [CrossRef] [PubMed]
- De Vivo, M.; Masetti, M.; Bottegoni, G.; Cavalli, A. Role of Molecular Dynamics and Related Methods in Drug Discovery. J. Med. Chem. 2016, 59, 4035–4061. [Google Scholar] [CrossRef] [PubMed]
- Decherchi, S.; Grisoni, F.; Tiwary, P.; Cavalli, A. Editorial: Molecular Dynamics and Machine Learning in Drug Discovery. Front. Mol. Biosci. 2021, 8, 673773. [Google Scholar] [CrossRef]
- Brogi, S.; Ramalho, T.C.; Kuca, K.; Medina-Franco, J.L.; Valko, M. Editorial: In silico Methods for Drug Design and Discovery. Front. Chem. 2020, 8, 612. [Google Scholar] [CrossRef]
- Zaccagnini, L.; Brogi, S.; Brindisi, M.; Gemma, S.; Chemi, G.; Legname, G.; Campiani, G.; Butini, S. Identification of novel fluorescent probes preventing PrP(Sc) replication in prion diseases. Eur. J. Med. Chem. 2017, 127, 859–873. [Google Scholar] [CrossRef]
- Brogi, S. Computational Approaches for Drug Discovery. Molecules 2019, 24, 3061. [Google Scholar] [CrossRef] [Green Version]
- Sirous, H.; Campiani, G.; Calderone, V.; Brogi, S. Discovery of novel hit compounds as potential HDAC1 inhibitors: The case of ligand and structure-based virtual screening. Comput. Biol. Med. 2021, 137, 104808. [Google Scholar] [CrossRef]
- Vazquez, J.; Lopez, M.; Gibert, E.; Herrero, E.; Luque, F.J. Merging Ligand-Based and Structure-Based Methods in Drug Discovery: An Overview of Combined Virtual Screening Approaches. Molecules 2020, 25, 4723. [Google Scholar] [CrossRef]
- Ferreira, L.L.G.; Andricopulo, A.D. Editorial: Chemoinformatics Approaches to Structure- and Ligand-Based Drug Design. Front. Pharm. 2018, 9, 1416. [Google Scholar] [CrossRef]
- Ivanov, J.; Polshakov, D.; Kato-Weinstein, J.; Zhou, Q.; Li, Y.; Granet, R.; Garner, L.; Deng, Y.; Liu, C.; Albaiu, D.; et al. Quantitative Structure-Activity Relationship Machine Learning Models and their Applications for Identifying Viral 3CLpro- and RdRp-Targeting Compounds as Potential Therapeutics for COVID-19 and Related Viral Infections. ACS Omega 2020, 5, 27344–27358. [Google Scholar] [CrossRef]
- Ancuceanu, R.; Dinu, M.; Neaga, I.; Laszlo, F.G.; Boda, D. Development of QSAR machine learning-based models to forecast the effect of substances on malignant melanoma cells. Oncol. Lett. 2019, 17, 4188–4196. [Google Scholar] [CrossRef]
- Keyvanpour, M.R.; Shirzad, M.B. An Analysis of QSAR Research Based on Machine Learning Concepts. Curr. Drug Discov. Technol. 2021, 18, 17–30. [Google Scholar] [CrossRef]
- Simoes, R.S.; Maltarollo, V.G.; Oliveira, P.R.; Honorio, K.M. Transfer and Multi-task Learning in QSAR Modeling: Advances and Challenges. Front. Pharm. 2018, 9, 74. [Google Scholar] [CrossRef] [Green Version]
- Lill, M.A. Multi-dimensional QSAR in drug discovery. Drug Discov. Today 2007, 12, 1013–1017. [Google Scholar] [CrossRef] [PubMed]
- Sadik, K.; Byadi, S.; Hachim, M.E.; Hamdani, N.E.; Podlipnik, Č.; Aboulmouhajir, A. Multi-QSAR approaches for investigating the relationship between chemical structure descriptors of Thiadiazole derivatives and their corrosion inhibition performance. J. Mol. Struct. 2021, 1240, 130571. [Google Scholar] [CrossRef]
- Speck-Planche, A.; Cordeiro, M.N. Multi-Target QSAR Approaches for Modeling Protein Inhibitors. Simultaneous Prediction of Activities Against Biomacromolecules Present in Gram-Negative Bacteria. Curr. Top. Med. Chem. 2015, 15, 1801–1813. [Google Scholar] [CrossRef] [PubMed]
- Solimeo, R.; Zhang, J.; Kim, M.; Sedykh, A.; Zhu, H. Predicting chemical ocular toxicity using a combinatorial QSAR approach. Chem. Res. Toxicol. 2012, 25, 2763–2769. [Google Scholar] [CrossRef]
- Zhu, H.; Tropsha, A.; Fourches, D.; Varnek, A.; Papa, E.; Gramatica, P.; Oberg, T.; Dao, P.; Cherkasov, A.; Tetko, I.V. Combinatorial QSAR modeling of chemical toxicants tested against Tetrahymena pyriformis. J. Chem. Inf. Model. 2008, 48, 766–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neves, B.J.; Braga, R.C.; Melo-Filho, C.C.; Moreira-Filho, J.T.; Muratov, E.N.; Andrade, C.H. QSAR-Based Virtual Screening: Advances and Applications in Drug Discovery. Front. Pharm. 2018, 9, 1275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melo-Filho, C.C.; Dantas, R.F.; Braga, R.C.; Neves, B.J.; Senger, M.R.; Valente, W.C.; Rezende-Neto, J.M.; Chaves, W.T.; Muratov, E.N.; Paveley, R.A.; et al. QSAR-Driven Discovery of Novel Chemical Scaffolds Active against Schistosoma mansoni. J. Chem. Inf. Model. 2016, 56, 1357–1372. [Google Scholar] [CrossRef] [Green Version]
- Vamathevan, J.; Clark, D.; Czodrowski, P.; Dunham, I.; Ferran, E.; Lee, G.; Li, B.; Madabhushi, A.; Shah, P.; Spitzer, M.; et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 2019, 18, 463–477. [Google Scholar] [CrossRef]
- Dara, S.; Dhamercherla, S.; Jadav, S.S.; Babu, C.M.; Ahsan, M.J. Machine Learning in Drug Discovery: A Review. Artif. Intell. Rev. 2021, 2021, 1–53. [Google Scholar] [CrossRef]
- Vignaux, P.A.; Minerali, E.; Foil, D.H.; Puhl, A.C.; Ekins, S. Machine Learning for Discovery of GSK3beta Inhibitors. ACS Omega 2020, 5, 26551–26561. [Google Scholar] [CrossRef]
- Huang, D.Z.; Kouznetsova, V.L.; Tsigelny, I.F. Deep-learning- and pharmacophore-based prediction of RAGE inhibitors. Phys. Biol. 2020, 17, 036003. [Google Scholar] [CrossRef]
- Shi, C.; Dong, F.; Zhao, G.; Zhu, N.; Lao, X.; Zheng, H. Applications of machine-learning methods for the discovery of NDM-1 inhibitors. Chem. Biol. Drug Des. 2020, 96, 1232–1243. [Google Scholar] [CrossRef] [PubMed]
- Tinivella, A.; Pinzi, L.; Rastelli, G. Prediction of activity and selectivity profiles of human Carbonic Anhydrase inhibitors using machine learning classification models. J. Cheminform. 2021, 13, 18. [Google Scholar] [CrossRef]
- Ballester, P.J. Machine Learning for Molecular Modelling in Drug Design. Biomolecules 2019, 9, 216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medina-Franco, J.L. Grand Challenges of Computer-Aided Drug Design: The Road Ahead. Front. Drug Discov. 2021, 1, 2. [Google Scholar]
- Rodriguez, S.; Hug, C.; Todorov, P.; Moret, N.; Boswell, S.A.; Evans, K.; Zhou, G.; Johnson, N.T.; Hyman, B.T.; Sorger, P.K.; et al. Machine learning identifies candidates for drug repurposing in Alzheimer’s disease. Nat. Commun. 2021, 12, 1033. [Google Scholar] [CrossRef] [PubMed]
- Tanoli, Z.; Vaha-Koskela, M.; Aittokallio, T. Artificial intelligence, machine learning, and drug repurposing in cancer. Expert Opin. Drug Discov. 2021, 16, 977–989. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, F.; Tang, J.; Nussinov, R.; Cheng, F. Artificial intelligence in COVID-19 drug repurposing. Lancet Digit. Health 2020, 2, e667–e676. [Google Scholar] [CrossRef]
- Beck, B.R.; Shin, B.; Choi, Y.; Park, S.; Kang, K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. 2020, 18, 784–790. [Google Scholar] [CrossRef]
- Xu, Y.; Verma, D.; Sheridan, R.P.; Liaw, A.; Ma, J.; Marshall, N.M.; McIntosh, J.; Sherer, E.C.; Svetnik, V.; Johnston, J.M. Deep Dive into Machine Learning Models for Protein Engineering. J. Chem. Inf. Model. 2020, 60, 2773–2790. [Google Scholar] [CrossRef]
- Gao, W.; Mahajan, S.P.; Sulam, J.; Gray, J.J. Deep Learning in Protein Structural Modeling and Design. Patterns 2020, 1, 100142. [Google Scholar] [CrossRef]
- Hameduh, T.; Haddad, Y.; Adam, V.; Heger, Z. Homology modeling in the time of collective and artificial intelligence. Comput. Struct. Biotechnol. J. 2020, 18, 3494–3506. [Google Scholar] [CrossRef]
- Torrisi, M.; Pollastri, G.; Le, Q. Deep learning methods in protein structure prediction. Comput. Struct. Biotechnol. J. 2020, 18, 1301–1310. [Google Scholar] [CrossRef]
- Lavecchia, A. Machine-learning approaches in drug discovery: Methods and applications. Drug Discov. Today 2015, 20, 318–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gawehn, E.; Hiss, J.A.; Schneider, G. Deep Learning in Drug Discovery. Mol. Inf. 2016, 35, 3–14. [Google Scholar] [CrossRef]
- Kimber, T.B.; Chen, Y.; Volkamer, A. Deep Learning in Virtual Screening: Recent Applications and Developments. Int. J. Mol. Sci. 2021, 22, 4435. [Google Scholar] [CrossRef] [PubMed]
- Melville, J.L.; Burke, E.K.; Hirst, J.D. Machine learning in virtual screening. Comb. Chem. High Throughput Screen. 2009, 12, 332–343. [Google Scholar] [CrossRef]
- Mendez, D.; Gaulton, A.; Bento, A.P.; Chambers, J.; De Veij, M.; Felix, E.; Magarinos, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Li, Y.; Liu, R.; Pang, X.; Li, C.; Yang, R.; He, Y.; Lian, W.; Liu, A.L.; Du, G.H. Discovery of multitarget-directed ligands against Alzheimer’s disease through systematic prediction of chemical-protein interactions. J. Chem. Inf. Model. 2015, 55, 149–164. [Google Scholar] [CrossRef]
- Shao, Y.M.; Ma, X.; Paira, P.; Tan, A.; Herr, D.R.; Lim, K.L.; Ng, C.H.; Venkatesan, G.; Klotz, K.N.; Federico, S.; et al. Discovery of indolylpiperazinylpyrimidines with dual-target profiles at adenosine A2A and dopamine D2 receptors for Parkinson’s disease treatment. PLoS ONE 2018, 13, e0188212. [Google Scholar] [CrossRef] [Green Version]
- Michielan, L.; Bolcato, C.; Federico, S.; Cacciari, B.; Bacilieri, M.; Klotz, K.N.; Kachler, S.; Pastorin, G.; Cardin, R.; Sperduti, A.; et al. Combining selectivity and affinity predictions using an integrated Support Vector Machine (SVM) approach: An alternative tool to discriminate between the human adenosine A(2A) and A(3) receptor pyrazolo-triazolo-pyrimidine antagonists binding sites. Bioorg. Med. Chem. 2009, 17, 5259–5274. [Google Scholar] [CrossRef]
- Deshmukh, A.L.; Chandra, S.; Singh, D.K.; Siddiqi, M.I.; Banerjee, D. Identification of human flap endonuclease 1 (FEN1) inhibitors using a machine learning based consensus virtual screening. Mol. Biosyst. 2017, 13, 1630–1639. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, W.; Liu, Z.; Ju, Y.; Xu, M.; Zhang, Y.; Wu, X.; Gu, Q.; Wang, Z.; Xu, J. Discovery of indoleamine 2,3-dioxygenase inhibitors using machine learning based virtual screening. Medchemcomm 2018, 9, 937–945. [Google Scholar] [CrossRef]
- Kang, D.; Pang, X.; Lian, W.; Xu, L.; Wang, J.; Jia, H.; Zhang, B.; Liu, A.-L.; Du, G.-H. Discovery of VEGFR2 inhibitors by integrating naïve Bayesian classification, molecular docking and drug screening approaches. RSC Adv. 2018, 8, 5286–5297. [Google Scholar] [CrossRef] [Green Version]
- Montanari, F.; Cseke, A.; Wlcek, K.; Ecker, G.F. Virtual Screening of DrugBank Reveals Two Drugs as New BCRP Inhibitors. SLAS Discov. 2017, 22, 86–93. [Google Scholar] [CrossRef] [Green Version]
- Allen, B.K.; Mehta, S.; Ember, S.W.; Schonbrunn, E.; Ayad, N.; Schurer, S.C. Large-Scale Computational Screening Identifies First in Class Multitarget Inhibitor of EGFR Kinase and BRD4. Sci. Rep. 2015, 5, 16924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winkler, D.A. Use of Artificial Intelligence and Machine Learning for Discovery of Drugs for Neglected Tropical Diseases. Front. Chem. 2021, 9, 614073. [Google Scholar] [CrossRef] [PubMed]
- Lima Guimaraes, G.; Sanchez-Lengeling, B.; Outeiral, C.; Cunha Farias, P.L.; Aspuru-Guzik, A. Objective-Reinforced Generative Adversarial Networks (ORGAN) for Sequence Generation Models. arXiv 2017, arXiv:1705.10843. [Google Scholar]
- Keshavarzi Arshadi, A.; Salem, M.; Collins, J.; Yuan, J.S.; Chakrabarti, D. DeepMalaria: Artificial Intelligence Driven Discovery of Potent Antiplasmodials. Front. Pharm. 2019, 10, 1526. [Google Scholar] [CrossRef] [Green Version]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; MacNair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A Deep Learning Approach to Antibiotic Discovery. Cell 2020, 180, 688–702. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Le, X.; Wang, L.; Gu, Q.; Zhou, H.; Xu, J. Discovering new DNA gyrase inhibitors using machine learning approaches. RSC Adv. 2015, 5, 105600–105608. [Google Scholar] [CrossRef]
- Ekins, S.; Freundlich, J.S.; Clark, A.M.; Anantpadma, M.; Davey, R.A.; Madrid, P. Machine learning models identify molecules active against the Ebola virus in vitro. F1000Research 2015, 4, 1091. [Google Scholar] [CrossRef]
- Popova, M.; Isayev, O.; Tropsha, A. Deep reinforcement learning for de novo drug design. Sci. Adv. 2018, 4, eaap7885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, A.; Muller, A.T.; Huisman, B.J.H.; Fuchs, J.A.; Schneider, P.; Schneider, G. Generative Recurrent Networks for De Novo Drug Design. Mol. Inf. 2018, 37, 1700111. [Google Scholar] [CrossRef] [Green Version]
- Merk, D.; Friedrich, L.; Grisoni, F.; Schneider, G. De Novo Design of Bioactive Small Molecules by Artificial Intelligence. Mol. Inf. 2018, 37, 1700153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhavoronkov, A.; Ivanenkov, Y.A.; Aliper, A.; Veselov, M.S.; Aladinskiy, V.A.; Aladinskaya, A.V.; Terentiev, V.A.; Polykovskiy, D.A.; Kuznetsov, M.D.; Asadulaev, A.; et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol. 2019, 37, 1038–1040. [Google Scholar] [CrossRef] [PubMed]
- McCloskey, K.; Sigel, E.A.; Kearnes, S.; Xue, L.; Tian, X.; Moccia, D.; Gikunju, D.; Bazzaz, S.; Chan, B.; Clark, M.A.; et al. Machine Learning on DNA-Encoded Libraries: A New Paradigm for Hit Finding. J. Med. Chem. 2020, 63, 8857–8866. [Google Scholar] [CrossRef]
- Zeng, X.; Zhu, S.; Lu, W.; Liu, Z.; Huang, J.; Zhou, Y.; Fang, J.; Huang, Y.; Guo, H.; Li, L.; et al. Target identification among known drugs by deep learning from heterogeneous networks. Chem. Sci. 2020, 11, 1775–1797. [Google Scholar] [CrossRef] [Green Version]
- Dennis, M.J.; Beijnen, J.H.; Grochow, L.B.; van Warmerdam, L.J. An overview of the clinical pharmacology of topotecan. Semin. Oncol. 1997, 24, S5–S12. [Google Scholar]
- Kollmannsberger, C.; Mross, K.; Jakob, A.; Kanz, L.; Bokemeyer, C. Topotecan—A novel topoisomerase I inhibitor: Pharmacology and clinical experience. Oncology 1999, 56, 1–12. [Google Scholar] [CrossRef]
- White, S.C.; Cheeseman, S.; Thatcher, N.; Anderson, H.; Carrington, B.; Hearn, S.; Ross, G.; Ranson, M. Phase II study of oral topotecan in advanced non-small cell lung cancer. Clin. Cancer Res. 2000, 6, 868–873. [Google Scholar]
- Yang, Y.; Winger, R.C.; Lee, P.W.; Nuro-Gyina, P.K.; Minc, A.; Larson, M.; Liu, Y.; Pei, W.; Rieser, E.; Racke, M.K.; et al. Impact of suppressing retinoic acid-related orphan receptor gamma t (ROR)gammat in ameliorating central nervous system autoimmunity. Clin. Exp. Immunol. 2015, 179, 108–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Luo, X.Y.; Wu, D.H.; Xu, Y. ROR nuclear receptors: Structures, related diseases, and drug discovery. Acta Pharm. Sin. 2015, 36, 71–87. [Google Scholar] [CrossRef] [Green Version]
- Madhukar, N.S.; Khade, P.K.; Huang, L.; Gayvert, K.; Galletti, G.; Stogniew, M.; Allen, J.E.; Giannakakou, P.; Elemento, O. A Bayesian machine learning approach for drug target identification using diverse data types. Nat. Commun. 2019, 10, 5221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dezso, Z.; Ceccarelli, M. Machine learning prediction of oncology drug targets based on protein and network properties. BMC Bioinform. 2020, 21, 104. [Google Scholar] [CrossRef] [Green Version]
- Terranova, N.; Venkatakrishnan, K.; Benincosa, L.J. Application of Machine Learning in Translational Medicine: Current Status and Future Opportunities. AAPS J. 2021, 23, 74. [Google Scholar] [CrossRef]
- van der Graaf, P.H.; Benson, N. Systems pharmacology: Bridging systems biology and pharmacokinetics-pharmacodynamics (PKPD) in drug discovery and development. Pharm. Res. 2011, 28, 1460–1464. [Google Scholar] [CrossRef]
- Stern, A.M.; Schurdak, M.E.; Bahar, I.; Berg, J.M.; Taylor, D.L. A Perspective on Implementing a Quantitative Systems Pharmacology Platform for Drug Discovery and the Advancement of Personalized Medicine. J. Biomol. Screen. 2016, 21, 521–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agoram, B.M.; Martin, S.W.; van der Graaf, P.H. The role of mechanism-based pharmacokinetic-pharmacodynamic (PK-PD) modelling in translational research of biologics. Drug Discov. Today 2007, 12, 1018–1024. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, E.L.; Spilker, M.E.; Zang, R.; Bansal, L.; He, H.; Jones, R.D.O.; Le, K.; Penney, M.; Schuck, E.; Topp, B.; et al. Applications of Quantitative Systems Pharmacology in Model-Informed Drug Discovery: Perspective on Impact and Opportunities. CPT Pharmacomet. Syst. Pharm. 2019, 8, 777–791. [Google Scholar] [CrossRef] [Green Version]
- Lazarou, G.; Chelliah, V.; Small, B.G.; Walker, M.; van der Graaf, P.H.; Kierzek, A.M. Integration of Omics Data Sources to Inform Mechanistic Modeling of Immune-Oncology Therapies: A Tutorial for Clinical Pharmacologists. Clin. Pharm. 2020, 107, 858–870. [Google Scholar] [CrossRef] [Green Version]
- Ramm, S.; Todorov, P.; Chandrasekaran, V.; Dohlman, A.; Monteiro, M.B.; Pavkovic, M.; Muhlich, J.; Shankaran, H.; Chen, W.W.; Mettetal, J.T.; et al. A Systems Toxicology Approach for the Prediction of Kidney Toxicity and Its Mechanisms In Vitro. Toxicol. Sci. 2019, 169, 54–69. [Google Scholar] [CrossRef]
- Song, J.; Wang, L.; Ng, N.N.; Zhao, M.; Shi, J.; Wu, N.; Li, W.; Liu, Z.; Yeom, K.W.; Tian, J. Development and Validation of a Machine Learning Model to Explore Tyrosine Kinase Inhibitor Response in Patients With Stage IV EGFR Variant-Positive Non-Small Cell Lung Cancer. JAMA Netw. Open 2020, 3, e2030442. [Google Scholar] [CrossRef]
- Lu, J.; Bender, B.; Jin, J.Y.; Guan, Y. Deep learning prediction of patient response time course from early data via neural-pharmacokinetic/pharmacodynamic modelling. Nat. Mach. Intell. 2021, 3, 696–704. [Google Scholar] [CrossRef]
- Obermeyer, Z.; Emanuel, E.J. Predicting the Future—Big Data, Machine Learning, and Clinical Medicine. N. Engl. J. Med. 2016, 375, 1216–1219. [Google Scholar] [CrossRef] [Green Version]
- Aggarwal, R.; Sounderajah, V.; Martin, G.; Ting, D.S.W.; Karthikesalingam, A.; King, D.; Ashrafian, H.; Darzi, A. Diagnostic accuracy of deep learning in medical imaging: A systematic review and meta-analysis. NPJ Digit. Med. 2021, 4, 65. [Google Scholar] [CrossRef] [PubMed]
- Briganti, G.; Le Moine, O. Artificial Intelligence in Medicine: Today and Tomorrow. Front. Med. 2020, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.A.; Herbert Chan, H.W.; Baker, M.A.B. Machine learning: Applications of artificial intelligence to imaging and diagnosis. Biophys. Rev. 2019, 11, 111–118. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.; van Ginneken, B.; Sanchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Benjamens, S.; Dhunnoo, P.; Mesko, B. The state of artificial intelligence-based FDA-approved medical devices and algorithms: An online database. NPJ Digit. Med. 2020, 3, 118. [Google Scholar] [CrossRef]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Muehlematter, U.J.; Daniore, P.; Vokinger, K.N. Approval of artificial intelligence and machine learning-based medical devices in the USA and Europe (2015–2020): A comparative analysis. Lancet Digit. Health 2021, 3, e195–e203. [Google Scholar] [CrossRef]
- Chartrand, G.; Cheng, P.M.; Vorontsov, E.; Drozdzal, M.; Turcotte, S.; Pal, C.J.; Kadoury, S.; Tang, A. Deep Learning: A Primer for Radiologists. Radiographics 2017, 37, 2113–2131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adadi, A.; Adadi, S.; Berrada, M. Gastroenterology Meets Machine Learning: Status Quo and Quo Vadis. Adv. Bioinform. 2019, 2019, 1870975. [Google Scholar] [CrossRef] [Green Version]
- van der Sommen, F.; de Groof, J.; Struyvenberg, M.; van der Putten, J.; Boers, T.; Fockens, K.; Schoon, E.J.; Curvers, W.; de With, P.; Mori, Y.; et al. Machine learning in GI endoscopy: Practical guidance in how to interpret a novel field. Gut 2020, 69, 2035–2045. [Google Scholar] [CrossRef] [PubMed]
- Auger, S.D.; Jacobs, B.M.; Dobson, R.; Marshall, C.R.; Noyce, A.J. Big data, machine learning and artificial intelligence: A neurologist’s guide. Pract. Neurol. 2020, 21, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Jiang, B.; Tong, L.; Xie, Y.; Zaharchuk, G.; Wintermark, M. Applications of Deep Learning to Neuro-Imaging Techniques. Front. Neurol. 2019, 10, 869. [Google Scholar] [CrossRef]
- Tong, Y.; Lu, W.; Yu, Y.; Shen, Y. Application of machine learning in ophthalmic imaging modalities. Eye Vis. 2020, 7, 22. [Google Scholar] [CrossRef] [Green Version]
- Ting, D.S.W.; Pasquale, L.R.; Peng, L.; Campbell, J.P.; Lee, A.Y.; Raman, R.; Tan, G.S.W.; Schmetterer, L.; Keane, P.A.; Wong, T.Y. Artificial intelligence and deep learning in ophthalmology. Br. J. Ophthalmol. 2019, 103, 167–175. [Google Scholar] [CrossRef] [Green Version]
- Quer, G.; Arnaout, R.; Henne, M.; Arnaout, R. Machine Learning and the Future of Cardiovascular Care: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2021, 77, 300–313. [Google Scholar] [CrossRef] [PubMed]
- Cuocolo, R.; Perillo, T.; De Rosa, E.; Ugga, L.; Petretta, M. Current applications of big data and machine learning in cardiology. J. Geriatr. Cardiol. 2019, 16, 601–607. [Google Scholar] [CrossRef]
- Chan, S.; Reddy, V.; Myers, B.; Thibodeaux, Q.; Brownstone, N.; Liao, W. Machine Learning in Dermatology: Current Applications, Opportunities, and Limitations. Dermatol. Ther. 2020, 10, 365–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serag, A.; Ion-Margineanu, A.; Qureshi, H.; McMillan, R.; Saint Martin, M.J.; Diamond, J.; O’Reilly, P.; Hamilton, P. Translational AI and Deep Learning in Diagnostic Pathology. Front. Med. 2019, 6, 185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamamoto, R.; Suvarna, K.; Yamada, M.; Kobayashi, K.; Shinkai, N.; Miyake, M.; Takahashi, M.; Jinnai, S.; Shimoyama, R.; Sakai, A.; et al. Application of Artificial Intelligence Technology in Oncology: Towards the Establishment of Precision Medicine. Cancers 2020, 12, 3532. [Google Scholar] [CrossRef]
- Davenport, T.; Kalakota, R. The potential for artificial intelligence in healthcare. Future Healthc. J. 2019, 6, 94–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cutillo, C.M.; Sharma, K.R.; Foschini, L.; Kundu, S.; Mackintosh, M.; Mandl, K.D.; MI in Healthcare Workshop Working Group. Machine intelligence in healthcare-perspectives on trustworthiness, explainability, usability, and transparency. NPJ Digit. Med. 2020, 3, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, P.; Kendall, F.; Khozin, S.; Goosen, R.; Hu, J.; Laramie, J.; Ringel, M.; Schork, N. Artificial intelligence and machine learning in clinical development: A translational perspective. NPJ Digit. Med. 2019, 2, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adlung, L.; Cohen, Y.; Mor, U.; Elinav, E. Machine learning in clinical decision making. Med 2021, 2, 642–665. [Google Scholar] [CrossRef]
- Komatsu, M.; Sakai, A.; Dozen, A.; Shozu, K.; Yasutomi, S.; Machino, H.; Asada, K.; Kaneko, S.; Hamamoto, R. Towards Clinical Application of Artificial Intelligence in Ultrasound Imaging. Biomedicines 2021, 9, 720. [Google Scholar] [CrossRef]
- Khemasuwan, D.; Sorensen, J.S.; Colt, H.G. Artificial intelligence in pulmonary medicine: Computer vision, predictive model and COVID-19. Eur. Respir. Rev. 2020, 29, 200181. [Google Scholar] [CrossRef]
- Kaplan, A.; Cao, H.; FitzGerald, J.M.; Iannotti, N.; Yang, E.; Kocks, J.W.H.; Kostikas, K.; Price, D.; Reddel, H.K.; Tsiligianni, I.; et al. Artificial Intelligence/Machine Learning in Respiratory Medicine and Potential Role in Asthma and COPD Diagnosis. J. Allergy Clin. Immunol. Pr. 2021, 9, 2255–2261. [Google Scholar] [CrossRef]
- Available online: https://www.maxq.ai/products (accessed on 14 October 2021).
- Available online: https://global.medical.canon/products/computed-tomography/aice (accessed on 14 October 2021).
- Available online: https://www.siemens-healthineers.com/en-us/digital-health-solutions/digital-solutions-overview/clinical-decision-support/ai-rad-companion (accessed on 14 October 2021).
- Chamberlin, J.; Kocher, M.R.; Waltz, J.; Snoddy, M.; Stringer, N.F.C.; Stephenson, J.; Sahbaee, P.; Sharma, P.; Rapaka, S.; Schoepf, U.J.; et al. Automated detection of lung nodules and coronary artery calcium using artificial intelligence on low-dose CT scans for lung cancer screening: Accuracy and prognostic value. BMC Med. 2021, 19, 55. [Google Scholar] [CrossRef]
- Available online: https://www.varian.com/fi/resources-support/events/virtual-2020/artificial-intelligence-ai (accessed on 14 October 2021).
- Available online: https://www.amcad.com.tw/product/uo (accessed on 14 October 2021).
- Available online: https://www.amcad.com.tw/product/future_detail/3 (accessed on 14 October 2021).
- Available online: https://www.amcad.com.tw/product/ut (accessed on 14 October 2021).
- Li, T.; Jiang, Z.; Lu, M.; Zou, S.; Wu, M.; Wei, T.; Wang, L.; Li, J.; Hu, Z.; Cheng, X.; et al. Computer-aided diagnosis system of thyroid nodules ultrasonography: Diagnostic performance difference between computer-aided diagnosis and 111 radiologists. Medicine 2020, 99, e20634. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.amcad.com.tw/product/future_detail/2 (accessed on 14 October 2021).
- Available online: https://www.arterys.com/clinicalapp/cardioapp (accessed on 14 October 2021).
- Available online: https://www.arterys.com/clinicalapp/lungapp (accessed on 14 October 2021).
- Available online: https://www.arterys.com/ (accessed on 14 October 2021).
- Available online: https://www.verathon.com/bladderscan-prime-plus/ (accessed on 14 October 2021).
- Available online: https://www.gehealthcare.co.uk/products/computed-tomography/advanced-visualization/bone-vcar (accessed on 14 October 2021).
- Available online: https://www.brainomix.com/e-cta/ (accessed on 14 October 2021).
- Zelenak, K.; Krajina, A.; Meyer, L.; Fiehler, J.; Esmint Artificial, I.; Robotics Ad Hoc, C.; Behme, D.; Bulja, D.; Caroff, J.; Chotai, A.A.; et al. How to Improve the Management of Acute Ischemic Stroke by Modern Technologies, Artificial Intelligence, and New Treatment Methods. Life 2021, 11, 488. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.aidoc.com/ (accessed on 14 October 2021).
- Available online: https://www.circlecvi.com/cvi42/cardiac-mri/deep-learning/ (accessed on 14 October 2021).
- Karimi-Bidhendi, S.; Arafati, A.; Cheng, A.L.; Wu, Y.; Kheradvar, A.; Jafarkhani, H. Fully-automated deep-learning segmentation of pediatric cardiovascular magnetic resonance of patients with complex congenital heart diseases. J. Cardiovasc. Magn. Reson. 2020, 22, 80. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.claripi.com/clarict-ai/ (accessed on 14 October 2021).
- Available online: https://www.riveraintech.com/clearread-ai-solutions/clearread-ct/ (accessed on 14 October 2021).
- Wagner, A.K.; Hapich, A.; Psychogios, M.N.; Teichgraber, U.; Malich, A.; Papageorgiou, I. Computer-Aided Detection of Pulmonary Nodules in Computed Tomography Using ClearReadCT. J. Med. Syst. 2019, 43, 58. [Google Scholar] [CrossRef]
- Available online: https://curemetrix.com/cm-triage-2/ (accessed on 14 October 2021).
- Available online: https://www.viz.ai/ (accessed on 14 October 2021).
- Available online: https://www.gehealthcare.co.uk/products/radiography-systems/mobile-xray-systems/critical-care-suite-on-optima-xr240amx (accessed on 14 October 2021).
- Available online: https://www.keyamedical.com/curarad-ich/ (accessed on 14 October 2021).
- Available online: https://www.gehealthcare.co.uk/ (accessed on 14 October 2021).
- Available online: https://www.deep-voxel.com/#/home (accessed on 14 October 2021).
- Available online: https://captionhealth.com/ (accessed on 14 October 2021).
- Available online: https://ferriscan.com/ferrismart/ (accessed on 14 October 2021).
- Available online: https://www.resonancehealth.com/products/ferriscan-mri-measurement-of-liver-iron-concentration.html (accessed on 14 October 2021).
- Padeniya, P.; Siriwardana, S.; Ediriweera, D.; Samarasinghe, N.; Silva, S.; Silva, I.; Ahamed, N.; Niriella, M.; Premawardhena, A. Comparison of liver MRI R2(FerriScan(R)) VS liver MRI T2* as a measure of body iron load in a cohort of beta thalassaemia major patients. Orphanet J. Rare Dis. 2020, 15, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Available online: https://www.zebra-med.com/solutions/triage/healthcxr (accessed on 14 October 2021).
- Available online: https://www.zebra-med.com/ (accessed on 14 October 2021).
- Available online: https://icometrix.com/services/icobrain-ms (accessed on 14 October 2021).
- Struyfs, H.; Sima, D.M.; Wittens, M.; Ribbens, A.; Pedrosa de Barros, N.; Phan, T.V.; Ferraz Meyer, M.I.; Claes, L.; Niemantsverdriet, E.; Engelborghs, S.; et al. Automated MRI volumetry as a diagnostic tool for Alzheimer’s disease: Validation of icobrain dm. Neuroimage Clin. 2020, 26, 102243. [Google Scholar] [CrossRef]
- Available online: https://www.usa.philips.com/healthcare/solutions/diagnostic-informatics (accessed on 14 October 2021).
- Available online: https://global.infervision.com/product/5/ (accessed on 14 October 2021).
- Available online: https://clinicaltrials.gov/ct2/show/NCT04119960 (accessed on 14 October 2021).
- Available online: https://www.infinittna.com/solutions/radiology/infinitt-pacs/ (accessed on 14 October 2021).
- Available online: https://www.imagebiopsy.com/product/koala-ce (accessed on 14 October 2021).
- Available online: https://koiosmedical.com/ (accessed on 14 October 2021).
- Available online: https://perspectum.com/products/livermultiscan (accessed on 14 October 2021).
- Available online: https://www.dia-analysis.com/lvivo-ef-app (accessed on 14 October 2021).
- Available online: https://thirona.eu/solutions/chestct/ (accessed on 14 October 2021).
- Available online: https://perspectum.com/products/mrcp (accessed on 14 October 2021).
- Available online: https://www.philips.com.au/healthcare/product/HCNMRF320/mrcat-brain-mr-rt-clinical-application (accessed on 14 October 2021).
- Available online: https://imagen.ai/ (accessed on 14 October 2021).
- Available online: https://www.algomedica.com/low-radation-ct-scans-algomedica (accessed on 14 October 2021).
- Available online: https://www.icadmed.com/powerlook-density-assessment.html (accessed on 14 October 2021).
- Available online: https://www.icadmed.com/profoundai.html (accessed on 14 October 2021).
- Available online: https://www.qlarityimaging.com/ (accessed on 14 October 2021).
- Available online: https://quibim.com/ (accessed on 14 October 2021).
- Available online: https://www.rapidai.com/rapid-aspects (accessed on 14 October 2021).
- Available online: https://www.rapidai.com/rapid-ich (accessed on 14 October 2021).
- Available online: https://www.raysearchlabs.com/data-handling-and-machine-learning/ (accessed on 14 October 2021).
- Available online: https://www.raysearchlabs.com/machine-learning-in-raystation/ (accessed on 14 October 2021).
- Available online: https://www.radiobotics.com/products (accessed on 14 October 2021).
- Available online: https://behold.ai/ (accessed on 14 October 2021).
- Available online: https://www.imagingbiometrics.com/what-we-offer/product-services/ib-stonechecker/ (accessed on 14 October 2021).
- Available online: https://strokeviewer.nico-lab.com/ (accessed on 14 October 2021).
- Available online: https://subtlemedical.com/subtlemr/ (accessed on 14 October 2021).
- Available online: https://subtlemedical.com/subtlepet/ (accessed on 14 October 2021).
- Available online: https://www.siemens-healthineers.com/computed-tomography/options-upgrades/clinical-applications/syngo-ct-cardiac-function (accessed on 14 October 2021).
- Available online: https://screenpoint-medical.com/fusion-ai/ (accessed on 14 October 2021).
- Sasaki, M.; Tozaki, M.; Rodriguez-Ruiz, A.; Yotsumoto, D.; Ichiki, Y.; Terawaki, A.; Oosako, S.; Sagara, Y.; Sagara, Y. Artificial intelligence for breast cancer detection in mammography: Experience of use of the ScreenPoint Medical Transpara system in 310 Japanese women. Breast Cancer 2020, 27, 642–651. [Google Scholar] [CrossRef]
- Available online: https://www.veolity.com/ (accessed on 14 October 2021).
- Available online: https://mirada-medical.com/ (accessed on 14 October 2021).
- Available online: https://www.carewell.com.cn/en/AI-ECG.html (accessed on 14 October 2021).
- Available online: https://www.carewellhealth.com/products_aiecg.html (accessed on 14 October 2021).
- Available online: https://www.biotricity.com/bioflux-2/ (accessed on 14 October 2021).
- Available online: https://www.ultromics.com/echogo (accessed on 14 October 2021).
- Available online: https://www.ekohealth.com/ (accessed on 14 October 2021).
- Available online: https://emurmur.com/ai/ (accessed on 14 October 2021).
- Available online: https://www.kardia.com/ (accessed on 14 October 2021).
- Available online: https://kosmosplatform.com/ (accessed on 14 October 2021).
- Available online: https://www.ventripoint.com/explore-vms-3-0 (accessed on 14 October 2021).
- Available online: https://altoida.com/ (accessed on 14 October 2021).
- Meier, I.B.; Buegler, M.; Harms, R.; Seixas, A.; Coltekin, A.; Tarnanas, I. Using a Digital Neuro Signature to measure longitudinal individual-level change in Alzheimer’s disease: The Altoida large cohort study. NPJ Digit. Med. 2021, 4, 101. [Google Scholar] [CrossRef]
- Available online: https://www.brainscope.com/ (accessed on 14 October 2021).
- Available online: https://cognoa.com/providers/ (accessed on 14 October 2021).
- Available online: https://coaptengineering.com/user-manual/complete-control-room (accessed on 14 October 2021).
- Available online: https://www.ensodata.com/ensosleep/ (accessed on 14 October 2021).
- Available online: https://www.qbtech.com/adhd-tests# (accessed on 14 October 2021).
- Hult, N.; Kadesjo, J.; Kadesjo, B.; Gillberg, C.; Billstedt, E. ADHD and the QbTest: Diagnostic Validity of QbTest. J. Atten. Disord. 2018, 22, 1074–1080. [Google Scholar] [CrossRef]
- Available online: https://www.zeiss.com/meditec/int/product-portfolio/retinal-cameras/zeiss-clarus-700.html (accessed on 14 October 2021).
- Available online: https://www.eyenuk.com/en/products/eyeart/ (accessed on 14 October 2021).
- Ipp, E.L.I.; Shah, V.N.; Bode, B.W.; Sadda, S.R. 599-P: Diabetic Retinopathy (DR) Screening Performance of General Ophthalmologists, Retina Specialists, and Artificial Intelligence (AI): Analysis from a Pivotal Multicenter Prospective Clinical Trial. Diabetes 2019, 68, 599. [Google Scholar] [CrossRef]
- Available online: https://www.digitaldiagnostics.com/ (accessed on 14 October 2021).
- Abramoff, M.D.; Lou, Y.; Erginay, A.; Clarida, W.; Amelon, R.; Folk, J.C.; Niemeijer, M. Improved Automated Detection of Diabetic Retinopathy on a Publicly Available Dataset Through Integration of Deep Learning. Invest. Ophthalmol. Vis. Sci. 2016, 57, 5200–5206. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://dreamed-diabetes.com/advisor/ (accessed on 14 October 2021).
- Available online: https://www.medtronicdiabetes.com/products/guardian-connect-continuous-glucose-monitoring-system (accessed on 14 October 2021).
- Available online: https://cleverculturesystems.com/apas-independence/ (accessed on 14 October 2021).
- Brenton, L.; Waters, M.J.; Stanford, T.; Giglio, S. Clinical evaluation of the APAS(R) Independence: Automated imaging and interpretation of urine cultures using artificial intelligence with composite reference standard discrepant resolution. J. Microbiol. Methods 2020, 177, 106047. [Google Scholar] [CrossRef]
- Available online: https://nightowl.care/ (accessed on 14 October 2021).
- Available online: https://www.nuvasive.com/surgical-solutions/pulse/ (accessed on 14 October 2021).
- Available online: https://www.sightdx.com/product (accessed on 14 October 2021).
- Bachar, N.; Benbassat, D.; Brailovsky, D.; Eshel, Y.; Gluck, D.; Levner, D.; Levy, S.; Pecker, S.; Yurkovsky, E.; Zait, A.; et al. An artificial intelligence-assisted diagnostic platform for rapid near-patient hematology. Am. J. Hematol. 2021, 96, 1264–1274. [Google Scholar] [CrossRef]
- Available online: https://www.impedimed.com/sozo-launched-first-product-for-health-and-wellness-markets/ (accessed on 14 October 2021).
- Available online: https://respiri.co/products/ (accessed on 14 October 2021).
- Yu, K.H.; Zhang, C.; Berry, G.J.; Altman, R.B.; Re, C.; Rubin, D.L.; Snyder, M. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun. 2016, 7, 12474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozaki, Y.; Yamada, H.; Kikuchi, H.; Hirotsu, A.; Murakami, T.; Matsumoto, T.; Kawabata, T.; Hiramatsu, Y.; Kamiya, K.; Yamauchi, T.; et al. Label-free classification of cells based on supervised machine learning of subcellular structures. PLoS ONE 2019, 14, e0211347. [Google Scholar] [CrossRef] [PubMed]
- Simm, J.; Klambauer, G.; Arany, A.; Steijaert, M.; Wegner, J.K.; Gustin, E.; Chupakhin, V.; Chong, Y.T.; Vialard, J.; Buijnsters, P.; et al. Repurposing High-Throughput Image Assays Enables Biological Activity Prediction for Drug Discovery. Cell Chem. Biol. 2018, 25, 611–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nassar, M.; Doan, M.; Filby, A.; Wolkenhauer, O.; Fogg, D.K.; Piasecka, J.; Thornton, C.A.; Carpenter, A.E.; Summers, H.D.; Rees, P.; et al. Label-Free Identification of White Blood Cells Using Machine Learning. Cytometry Part A 2019, 95, 836–842. [Google Scholar] [CrossRef] [Green Version]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyo, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef]
- Chang, C.H.; Lin, C.H.; Lane, H.Y. Machine Learning and Novel Biomarkers for the Diagnosis of Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 2761. [Google Scholar] [CrossRef]
- Kang, C.; Wang, D.; Zhang, X.; Wang, L.; Wang, F.; Chen, J. Construction and Validation of a Lung Cancer Diagnostic Model Based on 6-Gene Methylation Frequency in Blood, Clinical Features, and Serum Tumor Markers. Comput. Math. Methods Med. 2021, 2021, 9987067. [Google Scholar] [CrossRef]
- Brown, J.M.; Campbell, J.P.; Beers, A.; Chang, K.; Ostmo, S.; Chan, R.V.P.; Dy, J.; Erdogmus, D.; Ioannidis, S.; Kalpathy-Cramer, J.; et al. Automated Diagnosis of Plus Disease in Retinopathy of Prematurity Using Deep Convolutional Neural Networks. JAMA Ophthalmol. 2018, 136, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Wu, L.; Li, H.; Cai, C.; Wu, Q.; Kong, H.; Liu, R.; Wang, X.; Hou, X.; Liu, Y.; et al. A deep learning system for detecting diabetic retinopathy across the disease spectrum. Nat. Commun. 2021, 12, 3242. [Google Scholar] [CrossRef]
- Gargeya, R.; Leng, T. Automated Identification of Diabetic Retinopathy Using Deep Learning. Ophthalmology 2017, 124, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Ariya, A.; Asha, S. Automated detection of macular Edema using machine learning algorithm. AIP Conf. Proc. 2020, 2222, 030015. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, Z.; Li, J.; Liu, G. Identifying Diabetic Macular Edema and Other Retinal Diseases by Optical Coherence Tomography Image and Multiscale Deep Learning. Diabetes Metab. Syndr. Obes. 2020, 13, 4787–4800. [Google Scholar] [CrossRef] [PubMed]
- Kuwayama, S.; Ayatsuka, Y.; Yanagisono, D.; Uta, T.; Usui, H.; Kato, A.; Takase, N.; Ogura, Y.; Yasukawa, T. Automated Detection of Macular Diseases by Optical Coherence Tomography and Artificial Intelligence Machine Learning of Optical Coherence Tomography Images. J. Ophthalmol. 2019, 2019, 6319581. [Google Scholar] [CrossRef] [Green Version]
- Burlina, P.M.; Joshi, N.; Pekala, M.; Pacheco, K.D.; Freund, D.E.; Bressler, N.M. Automated Grading of Age-Related Macular Degeneration From Color Fundus Images Using Deep Convolutional Neural Networks. JAMA Ophthalmol. 2017, 135, 1170–1176. [Google Scholar] [CrossRef]
- Li, Z.; He, Y.; Keel, S.; Meng, W.; Chang, R.T.; He, M. Efficacy of a Deep Learning System for Detecting Glaucomatous Optic Neuropathy Based on Color Fundus Photographs. Ophthalmology 2018, 125, 1199–1206. [Google Scholar] [CrossRef] [Green Version]
- Ran, A.R.; Tham, C.C.; Chan, P.P.; Cheng, C.Y.; Tham, Y.C.; Rim, T.H.; Cheung, C.Y. Deep learning in glaucoma with optical coherence tomography: A review. Eye 2021, 35, 188–201. [Google Scholar] [CrossRef]
- Asaoka, R.; Murata, H.; Hirasawa, K.; Fujino, Y.; Matsuura, M.; Miki, A.; Kanamoto, T.; Ikeda, Y.; Mori, K.; Iwase, A.; et al. Using Deep Learning and Transfer Learning to Accurately Diagnose Early-Onset Glaucoma From Macular Optical Coherence Tomography Images. Am. J. Ophthalmol. 2019, 198, 136–145. [Google Scholar] [CrossRef]
- Kamal, H.; Lopez, V.; Sheth, S.A. Machine Learning in Acute Ischemic Stroke Neuroimaging. Front. Neurol. 2018, 9, 945. [Google Scholar] [CrossRef]
- Plis, S.M.; Hjelm, D.R.; Salakhutdinov, R.; Allen, E.A.; Bockholt, H.J.; Long, J.D.; Johnson, H.J.; Paulsen, J.S.; Turner, J.A.; Calhoun, V.D. Deep learning for neuroimaging: A validation study. Front. Neurosci. 2014, 8, 229. [Google Scholar] [CrossRef] [Green Version]
- Myszczynska, M.A.; Ojamies, P.N.; Lacoste, A.M.B.; Neil, D.; Saffari, A.; Mead, R.; Hautbergue, G.M.; Holbrook, J.D.; Ferraiuolo, L. Applications of machine learning to diagnosis and treatment of neurodegenerative diseases. Nat. Rev. Neurol. 2020, 16, 440–456. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, M.K.; Morales-Menendez, R.; Huang, X.; Hussain, N. A review of epileptic seizure detection using machine learning classifiers. Brain Inform. 2020, 7, 1–18. [Google Scholar] [CrossRef]
- Abdelhameed, A.; Bayoumi, M. A Deep Learning Approach for Automatic Seizure Detection in Children With Epilepsy. Front. Comput. Neurosci. 2021, 15, 650050. [Google Scholar] [CrossRef]
- Abbasi, B.; Goldenholz, D.M. Machine learning applications in epilepsy. Epilepsia 2019, 60, 2037–2047. [Google Scholar] [CrossRef]
- Tenev, A.; Markovska-Simoska, S.; Kocarev, L.; Pop-Jordanov, J.; Muller, A.; Candrian, G. Machine learning approach for classification of ADHD adults. Int. J. Psychophysiol. 2014, 93, 162–166. [Google Scholar] [CrossRef]
- Slobodin, O.; Yahav, I.; Berger, I. A Machine-Based Prediction Model of ADHD Using CPT Data. Front. Hum. Neurosci. 2020, 14, 560021. [Google Scholar] [CrossRef] [PubMed]
- Kautzky, A.; Vanicek, T.; Philippe, C.; Kranz, G.S.; Wadsak, W.; Mitterhauser, M.; Hartmann, A.; Hahn, A.; Hacker, M.; Rujescu, D.; et al. Machine learning classification of ADHD and HC by multimodal serotonergic data. Transl. Psychiatry 2020, 10, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubreuil-Vall, L.; Ruffini, G.; Camprodon, J.A. Deep Learning Convolutional Neural Networks Discriminate Adult ADHD From Healthy Individuals on the Basis of Event-Related Spectral EEG. Front. Neurosci. 2020, 14, 251. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Popuri, K.; Ding, G.W.; Balachandar, R.; Beg, M.F.; Alzheimer’s Disease Neuroimaging Initiative. Multimodal and Multiscale Deep Neural Networks for the Early Diagnosis of Alzheimer’s Disease using structural MR and FDG-PET images. Sci. Rep. 2018, 8, 5697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Zheng, X.; Li, Y.; Zhang, Q.; Ying, S. Multimodal Neuroimaging Feature Learning With Multimodal Stacked Deep Polynomial Networks for Diagnosis of Alzheimer’s Disease. IEEE J. Biomed. Health Inf. 2018, 22, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.W.; Hui, R. A deep learning based approach to classification of CT brain images. In Proceedings of the 2016 SAI Computing Conference (SAI), London, UK, 13–15 July 2016; pp. 28–31. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Zhao, S.; Chen, H.; Wang, A. A new machine learning method for identifying Alzheimer’s disease. Simul. Model. Pract. Theory 2020, 99, 102023. [Google Scholar] [CrossRef]
- Grassi, M.; Perna, G.; Caldirola, D.; Schruers, K.; Duara, R.; Loewenstein, D.A. A Clinically-Translatable Machine Learning Algorithm for the Prediction of Alzheimer’s Disease Conversion in Individuals with Mild and Premild Cognitive Impairment. J. Alzheimers Dis. 2018, 61, 1555–1573. [Google Scholar] [CrossRef] [PubMed]
- Landolfi, A.; Ricciardi, C.; Donisi, L.; Cesarelli, G.; Troisi, J.; Vitale, C.; Barone, P.; Amboni, M. Machine Learning Approaches in Parkinson’s Disease. Curr. Med. Chem. 2021, 28, 6548–6568. [Google Scholar] [CrossRef]
- Ahmadi Rastegar, D.; Ho, N.; Halliday, G.M.; Dzamko, N. Parkinson’s progression prediction using machine learning and serum cytokines. NPJ Parkinsons Dis. 2019, 5, 14. [Google Scholar] [CrossRef]
- Shu, Z.Y.; Cui, S.J.; Wu, X.; Xu, Y.; Huang, P.; Pang, P.P.; Zhang, M. Predicting the progression of Parkinson’s disease using conventional MRI and machine learning: An application of radiomic biomarkers in whole-brain white matter. Magn. Reson. Med. 2021, 85, 1611–1624. [Google Scholar] [CrossRef] [PubMed]
- Juutinen, M.; Wang, C.; Zhu, J.; Haladjian, J.; Ruokolainen, J.; Puustinen, J.; Vehkaoja, A. Parkinson’s disease detection from 20-step walking tests using inertial sensors of a smartphone: Machine learning approach based on an observational case-control study. PLoS ONE 2020, 15, e0236258. [Google Scholar] [CrossRef]
- Severson, K.A.; Chahine, L.M.; Smolensky, L.A.; Dhuliawala, M.; Frasier, M.; Ng, K.; Ghosh, S.; Hu, J. Discovery of Parkinson’s disease states and disease progression modelling: A longitudinal data study using machine learning. Lancet Digit. Health 2021, 3, e555–e564. [Google Scholar] [CrossRef]
- Park, K.W.; Lee, E.J.; Lee, J.S.; Jeong, J.; Choi, N.; Jo, S.; Jung, M.; Do, J.Y.; Kang, D.W.; Lee, J.G.; et al. Machine Learning-Based Automatic Rating for Cardinal Symptoms of Parkinson Disease. Neurology 2021, 96, e1761–e1769. [Google Scholar] [CrossRef]
- Mei, J.; Desrosiers, C.; Frasnelli, J. Machine Learning for the Diagnosis of Parkinson’s Disease: A Review of Literature. Front. Aging Neurosci. 2021, 13, 633752. [Google Scholar] [CrossRef]
- Maitín, A.M.; García-Tejedor, A.J.; Muñoz, J.P.R. Machine Learning Approaches for Detecting Parkinson’s Disease from EEG Analysis: A Systematic Review. Appl. Sci. 2020, 10, 8662. [Google Scholar] [CrossRef]
- Sriram, T.V.S.; Rao, M.V.; Narayana, G.V.S.; Kaladhar, D.S.V.G.K. Diagnosis of Parkinson Disease Using Machine Learning and Data Mining Systems from Voice Dataset. Adv. Intell. Syst. Comput. 2015, 327, 151–157. [Google Scholar] [CrossRef]
- Karapinar Senturk, Z. Early diagnosis of Parkinson’s disease using machine learning algorithms. Med. Hypotheses 2020, 138, 109603. [Google Scholar] [CrossRef]
- Seetharam, K.; Shrestha, S.; Sengupta, P.P. Artificial Intelligence in Cardiac Imaging. US Cardiol. Rev. 2020, 13, 110–116. [Google Scholar] [CrossRef]
- Constantinides, P.; Fitzmaurice, D.A. Artificial intelligence in cardiology: Applications, benefits and challenges. Br. J. Cardiol. 2018, 25, 86–87. [Google Scholar] [CrossRef]
- Seetharam, K.; Shrestha, S.; Sengupta, P.P. Artificial Intelligence in Cardiovascular Medicine. Curr. Treat. Options Cardiovasc. Med. 2019, 21, 25. [Google Scholar] [CrossRef]
- Sevakula, R.K.; Au-Yeung, W.M.; Singh, J.P.; Heist, E.K.; Isselbacher, E.M.; Armoundas, A.A. State-of-the-Art Machine Learning Techniques Aiming to Improve Patient Outcomes Pertaining to the Cardiovascular System. J. Am. Heart Assoc. 2020, 9, e013924. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.; Sengupta, P.P. The Mechanics of Machine Learning: From a Concept to Value. J. Am. Soc. Echocardiogr. 2018, 31, 1285–1287. [Google Scholar] [CrossRef] [PubMed]
- Madani, A.; Arnaout, R.; Mofrad, M.; Arnaout, R. Fast and accurate view classification of echocardiograms using deep learning. NPJ Digit. Med. 2018, 1, 1–8. [Google Scholar] [CrossRef]
- Madani, A.; Ong, J.R.; Tibrewal, A.; Mofrad, M.R.K. Deep echocardiography: Data-efficient supervised and semi-supervised deep learning towards automated diagnosis of cardiac disease. NPJ Digit. Med. 2018, 1, 59. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gajjala, S.; Agrawal, P.; Tison, G.H.; Hallock, L.A.; Beussink-Nelson, L.; Lassen, M.H.; Fan, E.; Aras, M.A.; Jordan, C.; et al. Fully Automated Echocardiogram Interpretation in Clinical Practice. Circulation 2018, 138, 1623–1635. [Google Scholar] [CrossRef] [PubMed]
- Samad, M.D.; Ulloa, A.; Wehner, G.J.; Jing, L.; Hartzel, D.; Good, C.W.; Williams, B.A.; Haggerty, C.M.; Fornwalt, B.K. Predicting Survival From Large Echocardiography and Electronic Health Record Datasets: Optimization With Machine Learning. JACC Cardiovasc. Imaging 2019, 12, 681–689. [Google Scholar] [CrossRef]
- Strodthoff, N.; Strodthoff, C. Detecting and interpreting myocardial infarction using fully convolutional neural networks. Physiol. Meas. 2019, 40, 015001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hannun, A.Y.; Rajpurkar, P.; Haghpanahi, M.; Tison, G.H.; Bourn, C.; Turakhia, M.P.; Ng, A.Y. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med. 2019, 25, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Elul, Y.; Rosenberg, A.A.; Schuster, A.; Bronstein, A.M.; Yaniv, Y. Meeting the unmet needs of clinicians from AI systems showcased for cardiology with deep-learning-based ECG analysis. Proc. Natl. Acad. Sci. USA 2021, 118, 118. [Google Scholar] [CrossRef]
- Attia, Z.I.; Kapa, S.; Lopez-Jimenez, F.; McKie, P.M.; Ladewig, D.J.; Satam, G.; Pellikka, P.A.; Enriquez-Sarano, M.; Noseworthy, P.A.; Munger, T.M.; et al. Screening for cardiac contractile dysfunction using an artificial intelligence-enabled electrocardiogram. Nat. Med. 2019, 25, 70–74. [Google Scholar] [CrossRef]
- Pandey, A.; Kagiyama, N.; Yanamala, N.; Segar, M.W.; Cho, J.S.; Tokodi, M.; Sengupta, P.P. Deep-Learning Models for the Echocardiographic Assessment of Diastolic Dysfunction. JACC Cardiovasc. Imaging 2021, 14, 1887–1900. [Google Scholar] [CrossRef]
- Ma, W.; Cheng, X.; Xu, X.; Wang, F.; Zhou, R.; Fenster, A.; Ding, M. Multilevel Strip Pooling-Based Convolutional Neural Network for the Classification of Carotid Plaque Echogenicity. Comput. Math. Methods Med. 2021, 2021, 3425893. [Google Scholar] [CrossRef]
- Yang, Y.J.; Bang, C.S. Application of artificial intelligence in gastroenterology. World J. Gastroenterol. 2019, 25, 1666–1683. [Google Scholar] [CrossRef]
- Namikawa, K.; Hirasawa, T.; Yoshio, T.; Fujisaki, J.; Ozawa, T.; Ishihara, S.; Aoki, T.; Yamada, A.; Koike, K.; Suzuki, H.; et al. Utilizing artificial intelligence in endoscopy: A clinician’s guide. Expert Rev. Gastroenterol. Hepatol. 2020, 14, 689–706. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.J.; Poon, N.C. Artificial intelligence in gastroenterology: Where are we heading? Front. Med. 2020, 14, 511–517. [Google Scholar] [CrossRef]
- Mori, Y.; Kudo, S.E.; Misawa, M.; Saito, Y.; Ikematsu, H.; Hotta, K.; Ohtsuka, K.; Urushibara, F.; Kataoka, S.; Ogawa, Y.; et al. Real-Time Use of Artificial Intelligence in Identification of Diminutive Polyps During Colonoscopy: A Prospective Study. Ann. Intern. Med. 2018, 169, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xiao, X.; Glissen Brown, J.R.; Berzin, T.M.; Tu, M.; Xiong, F.; Hu, X.; Liu, P.; Song, Y.; Zhang, D.; et al. Development and validation of a deep-learning algorithm for the detection of polyps during colonoscopy. Nat. Biomed. Eng. 2018, 2, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Byrne, M.F.; Chapados, N.; Soudan, F.; Oertel, C.; Linares Perez, M.; Kelly, R.; Iqbal, N.; Chandelier, F.; Rex, D.K. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut 2019, 68, 94–100. [Google Scholar] [CrossRef] [Green Version]
- Urban, G.; Tripathi, P.; Alkayali, T.; Mittal, M.; Jalali, F.; Karnes, W.; Baldi, P. Deep Learning Localizes and Identifies Polyps in Real Time With 96% Accuracy in Screening Colonoscopy. Gastroenterology 2018, 155, 1069–1078. [Google Scholar] [CrossRef]
- Tokai, Y.; Yoshio, T.; Aoyama, K.; Horie, Y.; Yoshimizu, S.; Horiuchi, Y.; Ishiyama, A.; Tsuchida, T.; Hirasawa, T.; Sakakibara, Y.; et al. Application of artificial intelligence using convolutional neural networks in determining the invasion depth of esophageal squamous cell carcinoma. Esophagus 2020, 17, 250–256. [Google Scholar] [CrossRef]
- Nakagawa, K.; Ishihara, R.; Aoyama, K.; Ohmori, M.; Nakahira, H.; Matsuura, N.; Shichijo, S.; Nishida, T.; Yamada, T.; Yamaguchi, S.; et al. Classification for invasion depth of esophageal squamous cell carcinoma using a deep neural network compared with experienced endoscopists. Gastrointest. Endosc. 2019, 90, 407–414. [Google Scholar] [CrossRef]
- Ozawa, T.; Ishihara, S.; Fujishiro, M.; Saito, H.; Kumagai, Y.; Shichijo, S.; Aoyama, K.; Tada, T. Novel computer-assisted diagnosis system for endoscopic disease activity in patients with ulcerative colitis. Gastrointest. Endosc. 2019, 89, 416–421.e1. [Google Scholar] [CrossRef]
- Mossotto, E.; Ashton, J.J.; Coelho, T.; Beattie, R.M.; MacArthur, B.D.; Ennis, S. Classification of Paediatric Inflammatory Bowel Disease using Machine Learning. Sci. Rep. 2017, 7, 2427. [Google Scholar] [CrossRef] [Green Version]
- Molder, A.; Balaban, D.V.; Jinga, M.; Molder, C.C. Current Evidence on Computer-Aided Diagnosis of Celiac Disease: Systematic Review. Front. Pharm. 2020, 11, 341. [Google Scholar] [CrossRef]
- Vecsei, A.; Amann, G.; Hegenbart, S.; Liedlgruber, M.; Uhl, A. Automated Marsh-like classification of celiac disease in children using local texture operators. Comput. Biol. Med. 2011, 41, 313–325. [Google Scholar] [CrossRef] [Green Version]
- Wimmer, G.; Vecsei, A.; Uhl, A. CNN transfer learning for the automated diagnosis of celiac disease. In Proceedings of the 2016 Sixth International Conference on Image Processing Theory, Tools and Applications (IPTA) (IEEE 2016), Oulu, Finland, 12–15 December 2016; pp. 1–6. [Google Scholar] [CrossRef]
- Hujoel, I.A.; Murphree, D.H., Jr.; Van Dyke, C.T.; Choung, R.S.; Sharma, A.; Murray, J.A.; Rubio-Tapia, A. Machine Learning in Detection of Undiagnosed Celiac Disease. Clin. Gastroenterol. Hepatol. 2018, 16, 1354–1355. [Google Scholar] [CrossRef] [PubMed]
- Koh, J.E.W.; De Michele, S.; Sudarshan, V.K.; Jahmunah, V.; Ciaccio, E.J.; Ooi, C.P.; Gururajan, R.; Gururajan, R.; Oh, S.L.; Lewis, S.K.; et al. Automated interpretation of biopsy images for the detection of celiac disease using a machine learning approach. Comput. Methods Programs Biomed. 2021, 203, 106010. [Google Scholar] [CrossRef]
- Klein, S.; Gildenblat, J.; Ihle, M.A.; Merkelbach-Bruse, S.; Noh, K.W.; Peifer, M.; Quaas, A.; Buttner, R. Deep learning for sensitive detection of Helicobacter Pylori in gastric biopsies. BMC Gastroenterol. 2020, 20, 417. [Google Scholar] [CrossRef] [PubMed]
- Gomolin, A.; Netchiporouk, E.; Gniadecki, R.; Litvinov, I.V. Artificial Intelligence Applications in Dermatology: Where Do We Stand? Front. Med. 2020, 7, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spyridonos, P.; Gaitanis, G.; Likas, A.; Bassukas, I.D. Automatic discrimination of actinic keratoses from clinical photographs. Comput. Biol. Med. 2017, 88, 50–59. [Google Scholar] [CrossRef]
- Hirota, M.; Kouzuki, H.; Ashikaga, T.; Sono, S.; Tsujita, K.; Sasa, H.; Aiba, S. Artificial neural network analysis of data from multiple in vitro assays for prediction of skin sensitization potency of chemicals. Toxicol. Vitr. 2013, 27, 1233–1246. [Google Scholar] [CrossRef]
- Tsujita-Inoue, K.; Hirota, M.; Ashikaga, T.; Atobe, T.; Kouzuki, H.; Aiba, S. Skin sensitization risk assessment model using artificial neural network analysis of data from multiple in vitro assays. Toxicol. Vitr. 2014, 28, 626–639. [Google Scholar] [CrossRef] [PubMed]
- Zang, Q.; Paris, M.; Lehmann, D.M.; Bell, S.; Kleinstreuer, N.; Allen, D.; Matheson, J.; Jacobs, A.; Casey, W.; Strickland, J. Prediction of skin sensitization potency using machine learning approaches. J. Appl. Toxicol. 2017, 37, 792–805. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Haenssle, H.A.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Kalloo, A.; Hassen, A.B.H.; Thomas, L.; Enk, A.; et al. Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 2018, 29, 1836–1842. [Google Scholar] [CrossRef]
- Haenssle, H.A.; Fink, C.; Toberer, F.; Winkler, J.; Stolz, W.; Deinlein, T.; Hofmann-Wellenhof, R.; Lallas, A.; Emmert, S.; Buhl, T.; et al. Man against machine reloaded: Performance of a market-approved convolutional neural network in classifying a broad spectrum of skin lesions in comparison with 96 dermatologists working under less artificial conditions. Ann. Oncol. 2020, 31, 137–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, S.S.; Kim, M.S.; Lim, W.; Park, G.H.; Park, I.; Chang, S.E. Classification of the Clinical Images for Benign and Malignant Cutaneous Tumors Using a Deep Learning Algorithm. J. Investig. Derm. 2018, 138, 1529–1538. [Google Scholar] [CrossRef] [Green Version]
- Yap, J.; Yolland, W.; Tschandl, P. Multimodal skin lesion classification using deep learning. Exp. Derm. 2018, 27, 1261–1267. [Google Scholar] [CrossRef] [Green Version]
- Phillips, M.; Marsden, H.; Jaffe, W.; Matin, R.N.; Wali, G.N.; Greenhalgh, J.; McGrath, E.; James, R.; Ladoyanni, E.; Bewley, A.; et al. Assessment of Accuracy of an Artificial Intelligence Algorithm to Detect Melanoma in Images of Skin Lesions. JAMA Netw. Open 2019, 2, e1913436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kharazmi, P.; Kalia, S.; Lui, H.; Wang, Z.J.; Lee, T.K. A feature fusion system for basal cell carcinoma detection through data-driven feature learning and patient profile. Ski. Res. Technol. 2018, 24, 256–264. [Google Scholar] [CrossRef]
- Hekler, A.; Utikal, J.S.; Enk, A.H.; Hauschild, A.; Weichenthal, M.; Maron, R.C.; Berking, C.; Haferkamp, S.; Klode, J.; Schadendorf, D.; et al. Superior skin cancer classification by the combination of human and artificial intelligence. Eur. J. Cancer 2019, 120, 114–121. [Google Scholar] [CrossRef] [Green Version]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Berking, C.; Haferkamp, S.; Hauschild, A.; Weichenthal, M.; Klode, J.; Schadendorf, D.; Holland-Letz, T.; et al. Deep neural networks are superior to dermatologists in melanoma image classification. Eur. J. Cancer 2019, 119, 11–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Klode, J.; Hauschild, A.; Berking, C.; Schilling, B.; Haferkamp, S.; Schadendorf, D.; Holland-Letz, T.; et al. Deep learning outperformed 136 of 157 dermatologists in a head-to-head dermoscopic melanoma image classification task. Eur. J. Cancer 2019, 113, 47–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tschandl, P.; Rosendahl, C.; Akay, B.N.; Argenziano, G.; Blum, A.; Braun, R.P.; Cabo, H.; Gourhant, J.Y.; Kreusch, J.; Lallas, A.; et al. Expert-Level Diagnosis of Nonpigmented Skin Cancer by Combined Convolutional Neural Networks. JAMA Derm. 2019, 155, 58–65. [Google Scholar] [CrossRef]
- Arevalo, J.; Cruz-Roa, A.; Arias, V.; Romero, E.; Gonzalez, F.A. An unsupervised feature learning framework for basal cell carcinoma image analysis. Artif. Intell. Med. 2015, 64, 131–145. [Google Scholar] [CrossRef]
- De Guzman, L.C.; Maglaque, R.P.C.; Torres, V.M.B.; Zapido, S.P.A.; Cordel, M.O. Design and Evaluation of a Multi-model, Multi-level Artificial Neural Network for Eczema Skin Lesion Detection. In Proceedings of the 2015 3rd International Conference on Artificial Intelligence, Modelling and Simulation (AIMS), Kota Kinabalu, Malaysia, 2–4 December 2015; pp. 42–47. [Google Scholar] [CrossRef]
- Han, S.S.; Park, G.H.; Lim, W.; Kim, M.S.; Na, J.I.; Park, I.; Chang, S.E. Deep neural networks show an equivalent and often superior performance to dermatologists in onychomycosis diagnosis: Automatic construction of onychomycosis datasets by region-based convolutional deep neural network. PLoS ONE 2018, 13, e0191493. [Google Scholar] [CrossRef] [Green Version]
- Binol, H.; Plotner, A.; Sopkovich, J.; Kaffenberger, B.; Niazi, M.K.K.; Gurcan, M.N. Ros-NET: A deep convolutional neural network for automatic identification of rosacea lesions. Ski. Res. Technol. 2020, 26, 413–421. [Google Scholar] [CrossRef]
- Meienberger, N.; Anzengruber, F.; Amruthalingam, L.; Christen, R.; Koller, T.; Maul, J.T.; Pouly, M.; Djamei, V.; Navarini, A.A. Observer-independent assessment of psoriasis-affected area using machine learning. J. Eur. Acad. Derm. Venereol. 2020, 34, 1362–1368. [Google Scholar] [CrossRef] [PubMed]
- Fadzil, M.H.; Ihtatho, D.; Affandi, A.M.; Hussein, S.H. Area assessment of psoriasis lesions for PASI scoring. J. Med. Eng. Technol. 2009, 33, 426–436. [Google Scholar] [CrossRef]
- George, Y.; Aldeen, M.; Garnavi, R. Automatic Scale Severity Assessment Method in Psoriasis Skin Images Using Local Descriptors. IEEE J. Biomed. Health Inf. 2020, 24, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Garain, U.; Chandra, A.; Chatterjee, R.; Senapati, S. Psoriasis skin biopsy image segmentation using Deep Convolutional Neural Network. Comput. Methods Programs Biomed. 2018, 159, 59–69. [Google Scholar] [CrossRef] [PubMed]
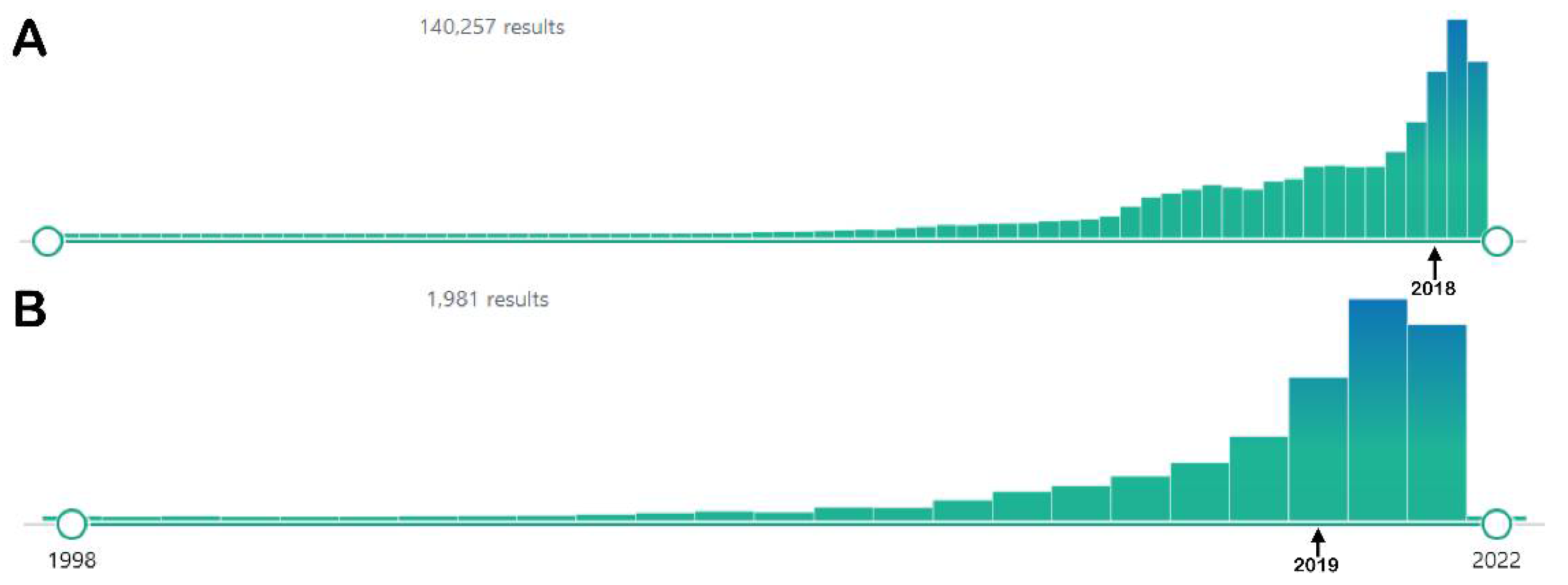
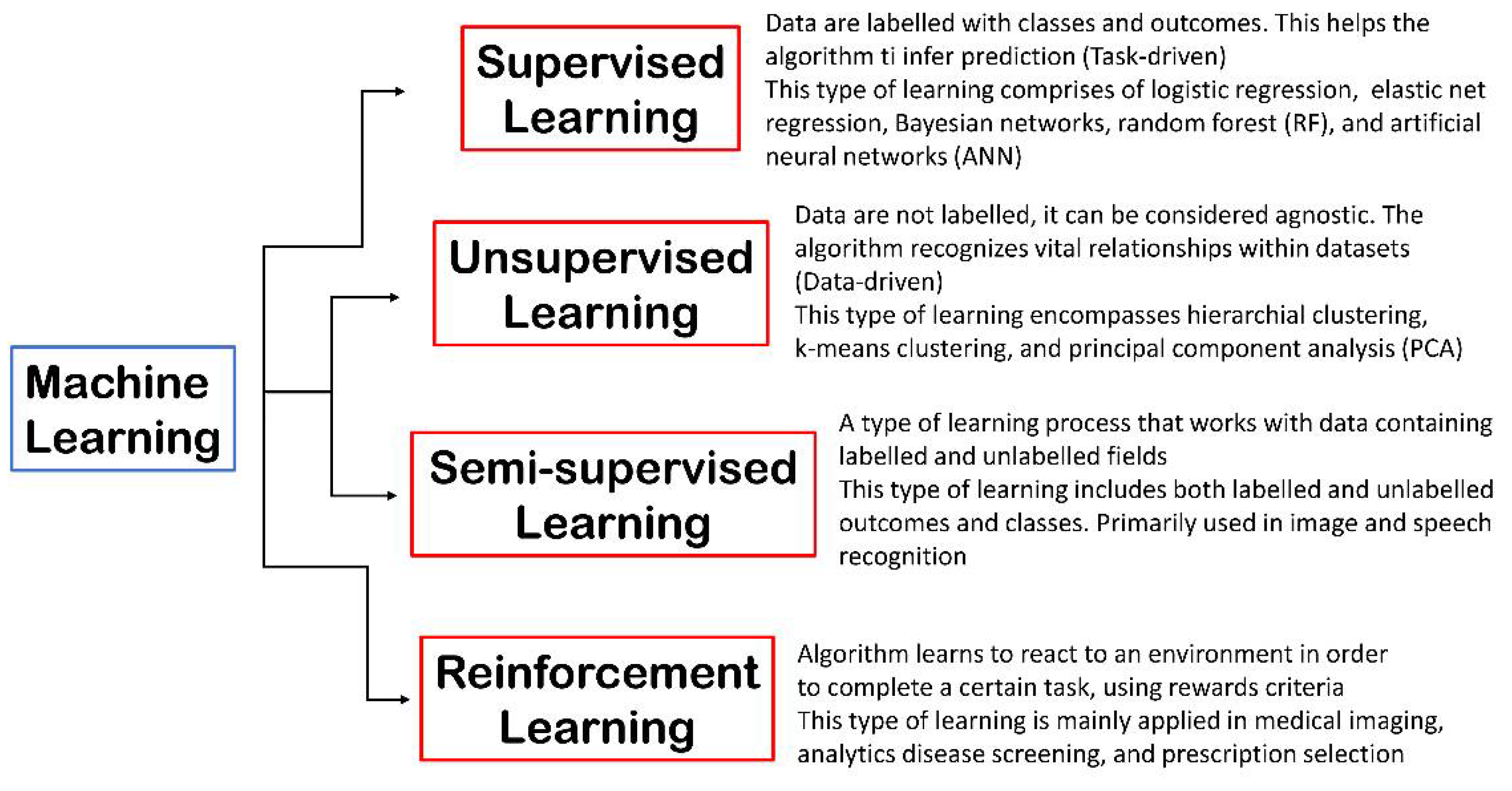
| AI Technique | Target | Dataset | Statistical Parameters | Outcomes | Ref |
|---|---|---|---|---|---|
| DL methodology deepDTnet | Multiple sclerosis | DeepDTnet was generated using 732 FDA-approved for training | Area under the ROC curve = 0.963 | Topotecan was predicted as an inhibitor of ROR-γt, (IC50 = 0.43 μM), showing potential therapeutic effects in multiple sclerosis, being effective in reverting the pathological phenotype in vivo in an EAE mouse model at 10 mg/kg | [102] |
| Bayesian ML algorithm BANDIT | Prediction of drug targets combining various kinds of data | A total of 20 million data points derived from six diverse types of data such as drug efficacy, post-treatment transcriptional response, drug structure, described undesirable effects, bioassay results, and well-established targets | Using over 2000 compounds, BANDIT showed an accuracy of ~90% in identifying correct targets | BANDIT was validated using 14,000 molecules with no target, producing ~4000 molecule target predictions. Fourteen molecules were predicted as microtubule binders and validated in vitro, supporting the predictions. BANDIT was applied to ONC201 (anticancer in clinical with no target). ONC201 was predicted and validated as a D2 receptor antagonist and will be evaluated in pheochromocytomas, a rare cancer overexpressing D2 receptor NCT03034200 | [108] |
| ML-based approach RF algorithm | Druggability score of novel unidentified drug targets | The ML model included 70 features obtained from drug targets, generating 10,000 ML models using a training set enclosing 102 complexes drug targets/drugs, and a “negative” set enclosing 102 non-drug targets | The ML models discriminated drug targets. The approach was validated using an external test set of 277 clinically relevant drug targets (area under the ROC curve of 0.89) | The output reported in this work provided new potential drug targets for developing innovative anticancer drugs | [109] |
| Device/Algorithm (Company) | Type of Algorithm | Description | FDA Approval Number | Medical Field(s) | Date and Reference |
|---|---|---|---|---|---|
| Accipio Ix (MaxQ-Al Ltd.), Tel Aviv, Israel | AI | The tool is used for an automatic, rapid, highly accurate identification and prioritization of suspected intracranial hemorrhage | K182177 | Radiology Neurology | October 2018 [146] |
| Advanced Intelligent Clear-IQ Engine (AiCE) (Canon Medical Systems Corporation, Ōtawara, Japan) | Deep CNN | AiCE system is used for reducing noise-boosting signals to quickly deliver sharp, clear, and distinct images | K183046 | Radiology | June 2019 [147] |
| AI-Rad Companion (Cardiovascular) (Siemens Medical Solutions USA, Inc., Malvern, PA, USA) | DL | The software is used for detecting cardiovascular risks from CT images | K183268 | Radiology | October 2019 [148,149] |
| AI-Rad Companion (Pulmonary) (Siemens Medical Solutions USA, Inc., Malvern, PA, USA) | DL | The software is used for detecting lung nodules from CT images | K183271 | Radiology | July 2019 [148,149] |
| AI Segmentation (Varian Medical Systems, Inc., Crawley, UK) | AI | The software is used for providing fast, accurate, and intelligent contouring for improving the reproducibility of structure delineation in radiation oncology | K203469 | Radiology Oncology | April 2021 [150] |
| AmCAD-UO (AmCad BioMed Corporation, Taipei City, Taiwan) | AI | The tool is used for detecting OSA in awake patients; it can precisely scan upper airway and analyze the gap between normal breathing and Müller Maneuver models | K180867 | Radiology | December 2018 [151] |
| AmCAD-US (AmCad BioMed Corporation, Taipei City, Taiwan) | AI | The tool is used to view and quantify ultrasound image data of backscattered signals acquired from ultrasound data | K162574 | Radiology | May 2017 [152] |
| AmCAD-UT Detection 2.2 (AmCad BioMed Corporation, Taipei City, Taiwan) | AI | The software is used for facilitating the detection, visualization, and characterization of thyroid nodule features on sonographic images | K180006 | Radiology | August 2018 [153,154] |
| AmCAD-UV (AmCad BioMed Corporation, Taipei City, Taiwan) | AI | The tool is used for classifying the ultrasonic color intensity data from signals of flow Doppler ultrasound images | K170069 | Radiology | April 2017 [155] |
| Arterys Cardio DL (Arterys Inc., San Francisco, CA, USA) | DL | The software is used for the analysis of cardiac MRI images | K163253 | Radiology Cardiology | January 2017 [156] |
| Arterys Oncology DL (Arterys Inc., San Francisco, CA, USA) | DL | The software is used for measuring and tracking lesions and nodules from MRI and CT images | K173542 | Radiology Oncology | January 2018 [157] |
| Arterys MICA (Arterys Inc., San Francisco, CA, USA) | AI | AI platform used for liver and lung cancer diagnosis from MRI and CT images | K182034 | Radiology Oncology | October 2018 [158] |
| BladderScan Prime PLUS System (Verathon Inc., Bothell, WA, USA) | DL | The tool provides improved bladder volume measurement accuracy | K172356 | Radiology | Sepember 2017 [159] |
| Bone VCAR (BVCAR) (GE Medical Systems SCS, Buc, France) | DL | The tool is used for automated spine labeling (segments or whole spine) from CT images | K183204 | Radiology | April 2019 [160] |
| Brainomix 360° e-CTA (Brainomix Limited, Oxford, UK) | AI | The tool is used for automatically detecting LVO on CT angiography | K192692 | Radiology | May 2020 [161,162] |
| BriefCase (Aidoc Medical, Ltd., Tel Aviv, Israel) | DL | The tool is used for detecting acute abnormalities across the body, helping radiologists to prioritize life-threatening cases, expediting patient care | K180647 | Radiology Emergency Medicine | August 2018 [163] |
| cvi42 for cardiac CT/MRI (Circle Cardiovascular Imaging Inc., Calgary, AB, Canada) | ML/DL | The software is used for assessing heart function, flow, and tissue attributes from CT/MRI images | K141480 | Radiology Cardiology | August 2014 [164,165] |
| ClariCT.AI (ClariPI Inc., Seoul, South-Korea) | DL | The tool is used for processing and enhancing CT images reducing noise | K183460 | Radiology | Jun2019 [166] |
| ClearRead CT (Riverain Technologies, LLC, Miamisburg, OH, USA) | DL | The software is used to detect pulmonary nodules and abnormalities in CT | K161201 | Radiology Oncology | Sepember 2016 [167,168] |
| cmTriage (CureMetrix, Inc., La Jolla, CA, USA) | AI | cmTriage is a tool enabling radiologists to triage, sort, and prioritize mammography | K183285 | Radiology Oncology | March 2019 [169] |
| ContaCT (Viz.AI, San Francisco, CA, USA) | AI | The software is used for detecting stroke from CT angiogram images of the brain | DEN170073 | Radiology Neurology | February 2018 [170] |
| Critical Care Suite (GE Medical Systems, LLC, Waukesha, WI, USA) | AI | The platform is used for automatically detecting PNX from X-rays, triaging critical cases | K183182 | Radiology Emergency Medicine | August 2019 [171] |
| CuraRad-ICH (CuraCloud Corp., Seattle, WA, USA) | DL | The tool is used for triaging suspected intracranial hemorrhage | K192167 | Radiology | April 2020 [172] |
| Deep Learning Image Reconstruction (GE Medical Systems, LLC, Waukesha, WI, USA) | DL | The application is used for CT image reconstruction Follow-up—K201745 DL Image Reconstruction for Gemstone Spectral Imaging (December 2020) | K183202 | Radiology | April 2019 [173] |
| DV.Target (Deepvoxel Inc., Irvine, CA, USA) | DL | The algorithm is used to automatically delineate OARs. Contours generated by DV.Target may be used as an input to clinical workflows in radiation therapy. | K202928 | Radiology | April 2021 [174] |
| EchoMD Automated Ejection Fraction Software (Bay Labs, Inc., San Francisco, CA, USA) | ML | This software is used for automated ECG analysis | K173780 | Radiology Cardiology | June 2018 [175] |
| FerriSmart Analysis System (Resonance Health Analysis Service Pty Ltd., Burswood, Australia) | ML/CNN | The software is used for measuring liver iron concentration from R2-MRI images. The system is based on the previously approved (K043271, Jan2005) R2-MRI Analysis System | K182218 | Radiology Internal Medicine | November 2018 [176,177,178] |
| HealthCXR (Zebra Medical Vision Ltd., HaMerkaz, Israel) | AI | The software is used for identifying and triaging pleural effusion in chest X-rays | K192320 | Radiology Emergency Medicine | November 2019 [179] |
| HealthMammo (Zebra Medical Vision Ltd., HaMerkaz, Israel) | DL | The tool is used for supporting identifying and prioritizing suspicious mammograms | K200905 | Radiology Oncology | June 2020 [180] |
| HealthPNX (Zebra Medical Vision Ltd., HaMerkaz, Israel) | AI | The tool increases the radiologist’s confidence in making PNX diagnosis from chest X-rays imaging output | K190362 | Radiology Emergency Medicine | May 2019 [180] |
| icobrain (icometrix NV, Leuven, Belgium) | ML and DL | The software is used for interpreting MRI images from the brain for detecting neurological disorders | K181939 | Radiology Neurology | November 2018 [181,182] |
| Illumeo System (Philips Medical Systems Technologies, Ltd., Haifa, Israel) | AI | The tool is used for acquiring, storing, distributing, processing, and displaying images | K173588 | Radiology | January 2018 [183] |
| lnferRead Lung CT (Beijing Infervision Technology Co. Ltd., Beijing, China) | AI | The tool is used for assisting radiologists fin detecting pulmonary nodules from CT (NCT04119960) | K192880 | Radiology Oncology | June 2020 [184,185] |
| Infinitt PACS 7.0 (Infinitt Healthcare Co. Ltd., Seoul, South-Korea) | AI | The software is used to analyze incoming tasks, identifying high-priority cases | K172803 | Radiology | Sepember 2017 [186] |
| KOALA (IB Lab GmbH, Wien, Austria) | DL | The algorithm is used to detect radiographic signs of knee osteoarthritis | K192109 | Radiology | November 2019 [187] |
| Koios DS for Breast (Koios Medical, Inc., Chicago, IL, USA) | AI | The software is used for analyzing ultrasound images for providing improved accuracy and efficiency in cancer diagnosis | K190442 | Radiology Oncology | July 2019 [188] |
| LiverMultiScan (Perspectum Diagnostics Ltd., Oxford, UK) | ML | This platform is used to assess liver tissue to enable diagnostic and patient management decisions. | K190017 | Radiology | June 2019 [189] |
| LVivo Software Application (DiA Imaging Analysis Ltd., Beer-Sheva, Israel) | AI | The software provides an automated AI-based ejection fraction analysis, allowing a fast assessment of cardiac functions | K210053 | Radiology | January 2021 [190] |
| LungQ (Thirona Corp., Nijmegen, Netherlands) | AI | The software is used for automatically identifying lung abnormalities from CT images | K173821 | Radiology | June 2018 [191] |
| MRCP+ V1.0 (Perspectum Diagnostics Ltd., Oxford, UK) | AI | The software is used for quantitatively analyzing the biliary tree and pancreatic duct from MRCP images | K183133 | Radiology | January 2019 [192] |
| MRCAT brain (Philips Medical Systems MR, Vantaa, Finland) | AI | The tool is used for radiotherapy planning of primary and metastatic tumors using MRI | K193109 | Radiology | January 2020 [193] |
| OsteoDetect (Imagen Technologies, Inc., New York, NY, USA) | DL | The software is used for detecting signs of distal radius fracture from X-ray | DEN180005 | Radiology Emergency Medicine | May 2018 [194] |
| PixelShine (ALGOMEDICA, Palo Alto, CA, USA) | DL | The software is used for improving the quality of scans obtained from any CT images, reducing noise | K161625 | Radiology | Sepember 2016 [195] |
| PowerLook Density Assessment Software (iCAD, Inc., Nashua, NH, USA) | ML | The algorithm is used for assessing breast density in 2D and 3D mammography | K180125 | Radiology | April 2018 [196] |
| ProFound™ AI Software (iCAD, Inc., Nashua, NH, USA) | DL | The software is used for detecting both malignant soft tissue densities and calcifications from DBT images | K191994 | Radiology Oncology | April 2019 [197] |
| QuantX (Qlarity Imaging, Chicago, IL, USA) | AI | The software is used for assessing and characterizing breast abnormalities from MRIdata | DEN170022 | Radiology Oncology | July 2017 [198] |
| qp-Prostate (Quibim S.L., Valencia, Spain) | AI | The tool is used for analyzing prostate MRI images | K203582 | Radiology Oncology | December 2020 [199] |
| Rapid ASPECTS (iSchemaView, Inc., San Mateo, CA, USA) | AI | The tool is used as assisted diagnostic software for lesions suspicious of cancer | K200760 | Radiology | May 2020 [200] |
| RAPID-ICH (iSchemaView, Inc., San Mateo, CA, USA) | AI | The tool is used to triage non-contrast CT (NCCT) cases for rapidly detecting suspicious intracranial hemorrhage | K193087 | Radiology | March 2020 [201] |
| RayCare 3.1 (RaySearch Laboratories AB, Stockholm, Sweden) | ML/DL | The software is used for improving workflow efficiency across different treatments in medical, radiation, and surgical oncology to support decisions in the clinic | K200487 | Radiology Oncology | June 2020 [202] |
| RayStation 10.1 (RaySearch Laboratories AB, Stockholm, Sweden) | ML | The platform is used to automatically generate treatment plans | K210645 | Radiology Oncology | June 2021 [203] |
| RBknee (Radiobotics ApS, Copenaghen, Denmark) | ML | The software is used for automatically identifying osteoarthritis in the knees based on X-ray images | K203696 | Radiology | August 2021 [204] |
| Red DotTM (Behold.AI Technologies Ltd., London, UK) | AI | The software is used for assessing PNX from chest X-ray images | K191556 | Radiology | January 2020 [205] |
| StoneChecker (Imaging Biometrics, LLC, Elm Grove, WI, USA) | AI | The software is used with standard CT scans in people with kidney stones for measuring stone parameters and to inform clinical decisions | K191530 | Radiology | June 2019 [206] |
| StrokeViewer (NiCo-Lab B.V., Amsterdam, Netherlands) | AI | This tool is used for the localization and quantification of stroke biomarkers from CT scans | K200873 | Radiology | October 2020 [207] |
| SubtleMR (Subtle Medical, Inc., Menlo Park, CA, USA) | CNN | The application is used for improving the quality of MRI images increasing resolution and reducing noise | K191688 | Radiology | Sepember 2019 [208] |
| SubtlePET (Subtle Medical, Inc., Menlo Park, CA, USA) | DNN | The application is used for processing PET images | K182336 | Radiology | November 2018 [209] |
| syngo.CT Cardiac Planning (Siemens Medical Solutions USA, Inc., Malvern, PA, USA) | AI | The software is used forenhancing CT images; analysis of morphology and pathology of vascular and cardiac structures | K200515 | Radiology | March 2020 [210] |
| TransparaTM (Screenpoint Medical B.V., Nijmegen, Netherlands) | ML | The software provides a support solution for mammography, identifying suspicious areas in 2D and 3D mammograms | K192287 | Radiology Oncology | December 2019 [211,212] |
| Veolity (MeVis Medical Solutions AG, Bremen, Germany) | ML | The software is used to recognize even the subtlest potential signs of lung cancer | K201501 | Radiology | February 2021 [213] |
| Workflow Box including DCLExpertTM (Mirada Medical Ltd., Oxford, UK) | AI | The software is used for autocontouring organs for cancer treatment planning | K181572 | Radiology | July 2018 [214] |
| AI-ECG Platform (Shenzhen Carewell Electronics, Ltd., Shenzhen, China) | AI | AI platform for assisting physicians in measuring and interpreting ECG | K180432 | Cardiology | November 2018 [215] |
| AI-ECG Tracker (Shenzhen Carewell Electronics, Ltd., Shenzhen, China) | AI | The tool is used for improving the detection efficiency of non-persistent arrhythmias (irregular heartbeats) | K200036 | Cardiology | March 2020 [216] |
| BioFlux Device (Biotricity Inc., Redwood City, CA, USA) | AI | The tool is used for detecting arrhythmias | K172311 | Cardiology | December 2017 [217] |
| EchoGo Core (Ultromics Ltd., Oxford, UK) | ML | The application is used to automatically evaluate cardiac functions from echocardiography, enabling physicians to diagnose heart failure and coronary artery disease | K191171 | Cardiology | November 2019 [218] |
| Eko Analysis Software (Eko Devices Inc., Oakland, CA, USA) | ANN | The software is used for detecting suspected murmurs in the heart sounds and atrial fibrillation from ECG data | K192004 | Cardiology | January 2020 [219] |
| eMurmur ID (CSD Labs GmbH, Graz, Austria) | ML | The software is used to understand, identify, and detect heart murmurs | K181988 | Cardiology | April 2019 [220] |
| KardiaAI (AliveCor, Inc., Mountain View, CA, USA) | AI | The tool is used for enhancing cardiac MRI to improve diagnosis of heart disorders | K181823 | Cardiology | November 2019 [221] |
| KOSMOS (EchoNous Inc., Redmond, WA, USA) | DL | This tool combining ultrasound with DL is used for clinical assessment of the heart, lungs, and abdomen | K193518 | Cardiology | March 2020 [222] |
| Ventripoint Medical System Plus (VMS+) 3.0 (Ventripoint Diagnostics Ltd., Toronto, ON, Canada) | AI | The tool is used for measuring whole heart function using conventional ultrasound (NCT01557582) | K191493 | Cardiology | October 2019 [223] |
| Altoida (Altoida, Inc., Washington, DC, USA) | ML | The software is used for detecting AD up to 10 years prior to the onset. ML is used for classifying patients’ risk of MCI due to AD (NCT02843529) | FDA-ClassII | Neurology | August 2021 [224,225] |
| BrainScope Ahead 100 (Brainscope Company, Inc., Bethesda, MD, USA) | AI | The software is used for interpreting the structural condition of the patient’s brain after head injury from EEG data | DEN140025 | Neurology | November 2014 [226] |
| Cognoa ASD Diagnosis Aid (Cognoa, Inc., Palo Alto, CA, USA) | ML | The software is used for evaluating patients at risk of ASD | DEN200069 | Neurology | June 2021 [227] |
| complete control system gen2 (Coapt, LLC, Chicago, IL, USA) | AI/ML | The platform provides a human–bionic interface that learns and adapts to users, giving them unrivaled control of their prosthetic arms | K191083 | Neurology | April 2019 [228] |
| EnsoSleep (EnsoData, Inc., Madison, WI, USA) | AI | The application assists clinicians in the diagnosis of sleep disorders | K162627 | Neurology | March 2017 [229] |
| QbTest/QbCheck (QbTech AB, Goteborg, Sweden) | AI/ML | The tools are used for braingazing using eye-tracking technology to capture eye vergence and AI algorithms for classifying ADHD patients vs. non-ADHD | K040894 K143468 | Neurology Psychiatry | June 2004 March 2016 [230,231] |
| Clarus 700 (Carl Zeiss Meditec Inc., Dublin, CA, USA) | DL | The algorithm is applied to diagnosing and monitoring retina disorders | K191194 | Ophthalmology | May 2019 [232] |
| EyeArt (EyeNuk, Inc., Woodland Hills, CA, USA) | AI | The software is used as a screening tool for detecting diabetic retinopathy | K200667 | Ophthalmology | March 2020 [233,234] |
| IDx (Digital Diagnostics Inc. -IDx LLC., Coralville, IA, USA) | AI | The software is used for detecting diabetic retinopathy | DEN180001 | Ophthalmology | January 2018 [235,236] |
| DreaMed Advisor Pro (DreaMed Diabetes, Ltd., Petah Tikva, Israel) | AI | The application is used for automatically determining the optimal therapy to maintain balanced glucose levels | DEN170043 | Endocrinology | June 2018 [237] |
| Guardian Connect System (Medtronic Minimed, Northridge, CA, USA) | AI | The application is used with diabetic patients for monitoring blood glucose content, predicting changes | P160007 | Endocrinology | March 2018 [238] |
| APAS Independence (Clever Culture Systems AG, Bäch, Switzerland) | AI/ML | The tool is used to automate culture plate imaging, analysis, and interpretation | K183648 | Microbiology | Sepember 2019 [239,240] |
| NightOwl (Ectosense nv, Leuven, Belgium) | AI | The algorithm is used for analyzing biophysical parameters for evaluating sleep-related breathing disorders of patients suspected of sleep apnea (NCT03774199; NCT04194073) | K191031 | Anesthesiology | March 2020 [241] |
| NuVasive Pulse System (NuVasive, Inc., San Diego, CA. USA) | AI | The tool is used during spinal surgery, neck dissection, and thoracic surgeries, improving surgical procedures | K180038 | Surgery | June 2018 [242] |
| Sight OLO (Sight Diagnostics Ltd., Tel Aviv, Israel) | AI | The algorithm is used for inspecting blood samples (NCT03595501) | K190898 | Hematology | November 2019 [243,244] |
| SOZO (ImpediMed Ltd., Carlsbad, CA, USA) | AI | The tool is use for the clinical assessment of unilateral lymphedema, combining BIS with AI to create a rapid, non-invasive scan of a person’s body | K190529 | Gastroenterology Urology | November 2019 [245] |
| wheezo WheezeRate Detector (Respiri Ltd., Melbourne, Australia) | ML | The tool is used for asthma management and remote monitoring | K202062 | Pneumology | March 2021 [246] |
| AI Technique | Target | Dataset | Statistical Parameters | Outcomes | Ref |
|---|---|---|---|---|---|
| ML-based method DNN algorithm | Predicting the activity of a given compound from images | Image-based fingerprints of morphological descriptions for each molecule considering GCR as a target for HTI assays used. A total of 842-dimensional feature vectors per cell related to activity data for selected orthogonal assays. Supervised ML for training models | Area under the ROC curve >0.9 as threshold for selecting the best performing models | The resulting ML model was successfully used for selecting novel chemical entities for biological evaluation | [249] |
| ML-based method (evaluating six ML algorithms: AdaBoost, GB, k-NN, RF, and SVM) | Classifying WBC for assessing the immune system status of a person | By using the proposed label-free approach only employing an imaging flow cytometer combined with ML methods, unstained WBCs were classified | The developed model discriminated B and T lymphocytes. Validation was achieved performing WBC analyses from unstained samples from 85 donors. The approach allows a precise classification of WBC avoiding cell disruption, leaving marker channels open to address further biological issues | The proposed method enables the use of ML for liquid biopsy, applying the powerful information in cell morphology for several diagnostics (e.g., detection of tumor products or circulating tumor cells in the blood | [250] |
| ML algorithms CNN | Classifying and predicting mutations from histopathological images from non-small cell lung cancer into LUAD, LUSC or normal lung tissue | Whole-slide images acquired from The Cancer Genome Atlas. The network was also trained for predicting most frequently mutated genes in LUAD (STK11, EGFR, FAT1, SETBP1, KRAS, and TP53) | The ML model performance was equivalent to that of pathologists (area under the ROC curve = 0.97). Validation using independent datasets of frozen tissues, formalin-fixed paraffin-embedded tissues and biopsies. Mutated genes in LUAD correctly predicted from pathology images (area under the ROC curve 0.733–0.856) | Aid pathologists in detecting gene mutations related to cancer subtypes | [251] |
| ML-based model, SVM algorithm | Implementing a diagnostic tool for identifying lung cancer risk of suspected cases | Tissues from samples of patients with lung cancer and tissue from healthy persons (70 pairs). Evaluation of the methylation rates of six genes (FHIT, p16, MGMT, RASSF1A, APC, DAPK) in lung cancer patients, the critical clinical data, tumor marker concentrations | Area under the ROC curve of 0.963, sensitivity of 0.900, specificity of 0.971, and accuracy of 0.936 | ML models as diagnostic tools for the early diagnosis of cancers that can contribute to increasing the survival rate of patients | [253] |
| AI Technique | Target | Dataset | Statistical Parameters | Outcomes | Ref |
|---|---|---|---|---|---|
| ML approach based on DL algorithms | Development of DeepDR, an intriguing screening platform for detecting diabetic retinopathy | DeepDR was generated considering 666,383 fundus images (173,346 patients), and it was trained for real-time image quality valuation, lesion detection and grading by 466,247 fundus images from 121,342 patients with diabetes. The evaluation was conducted considering 52,004 patients. Validation set: 200,136 fundus images, three external datasets, 209,322 images | Area under ROC curves of 0.901, 0.941, 0.954, and 0.967 regarding the detection of microaneurysms, cotton-wool spots, hard exudates, and hemorrhages, respectively. Area under the ROC curves of 0.943, 0.955, 0.960, and 0.972, regarding the grading of diabetic retinopathy (mild, moderate, severe, and proliferative). External validation, area under the ROC curves ranging from 0.916 to 0.970 | DeepDR showed significant accuracy and high sensitivity in detecting diabetic retinopathy from early-to-late stages | [255] |
| ML approach based on deep and transfer learning | Diagnosis regarding early-onset glaucoma using OCT images | The DL model was built from 4316 OCT images (1565 eyes from patients suffering from glaucoma and 193 normal eyes) used as a pre-training set. A set of OCT images trained the model (94 eyes from patient with early glaucoma, 84 healthy eyes). Test set comprised 114 eyes from 114 patients at early stages of glaucoma and 82 eyes from 82 healthy people | The DL model displayed an area under the ROC curve of 93.7%, considerably decreasing (to 76.6 and 78.8%) with no pre-training procedure, suggesting a relevant sensitivity and specificity of the DL model to diagnose glaucoma | The use of ML approaches can offer a significant improvement in diagnostic performances, assisting clinicians in making a decision | [263] |
| ML-based model CNN algorithm | Diagnostic model for DME | The model was generated from 38,057 OCT images (drusen, CNV, DME, healthy) by CNN technique. Training set 37,457 samples (9891 CNV, 9633 DME, 7975 drusen, and 9958 healthy). Validation set 600 samples (150 CNV, 150 DME, 150 drusen and 150 healthy) | The developed computational tool showed 94.5% accuracy, 97.2% precision, 97.7% sensitivity, and 97% specificity in the independent testing dataset | OCT images can be used for assessing the health of patients, automatically and accurately diagnosing several eye health conditions | [258] |
| AI Technique | Target | Dataset | Statistical Parameters | Outcomes | Ref |
|---|---|---|---|---|---|
| ML model based on CNN technique | Automatic detection of seizure for identifying epileptic events | Extraction and selection of specific characteristics of the EEG signal dataset was recorded at Boston Children’s Hospital from 23 pediatric patients with intractable seizures | The model showed accuracy 98.79%, sensitivity 98.72%, specificity 98.86%, precision 98.86%, F1-score of 98.79% | ML-based models can be useful in detecting seizure in epilepsy | [268] |
| ML algorithm based on SVM technique | Classification of adult ADHD using EEG data | The model was trained using 117 adults (67 ADHD, 50 healthy) from four conditions: two resting conditions (eyes open and eyes closed) and two neuropsychological tasks (visual and emotional continuous performance tests). Four datasets (one for each condition) independently trained diverse SVM classifiers | Model performances: normal vs. ADHD >70% ADHD II vs. ADHD III >90% ADHD III vs. ADHD IV >87% | ML-based model discriminated patients with ADHD from healthy subjects, differentiating ADHD subtypes | [270] |
| ML-based model | Prediction of ADHD by employing CPT indices | CPT indices from 458 children were used for training, cross-validating, and testing ML models (age 6–12 years, 213 ADHD patients and 245 healthy) | The tool was capable of discriminating patients with ADHD, showing an accuracy of 87%, sensitivity of 89%, and specificity of 84% | ML models can accurately classify ADHD using CPT data | [271] |
| ML model based on RF technique | Approach for discriminating ADHD patients from healthy subjects using multivariate, genetic, and PET imaging data | The model was built considering 16 ADHD patients and 22 healthy subjects. These groups were scanned via PET for measuring the SERT binding potential. The subjects were analyzed on the basis of 30 possible SNPs | The results regarding the model performances revealed an accuracy of 0.82, sensitivity of 0.75, and specificity of 0.86 | The outcomes highlighted the relevance of SERT along with SNPs in ADHD, indicating that a diagnostic tool based on these features supports clinical decisions | [272] |
| ML model based on the CNN technique | Discrimination of ADHD patients from healthy subjects using data extracted from EEG analysis | EEG data obtained from 20 ADHD patients and 20 healthy controls were used to train the model | The computational tool can correctly categorize ADHD patients with an accuracy of 88% | CNN algorithm built using EEG data is suitable for developing diagnostic tools for ADHD | [273] |
| ML-based approach based on DL technique | Approach for an early diagnosis of AD from MRI and FDG-PET images | Data from 1242 subjects with both a T1-weighted MRI scan and FDG-PET images from ADNI database were used for developing and validating the model. Subjects were clustered into 5 classes: (1) sNC 360 subjects; (2) sMCI 409 subjects; (3) pNC 18 subjects; (4) pMCI 217 subjects; (5) sAD 238 subjects | The classifier trained using pNC, pMCI, and sAD samples showed the highest classification accuracy of 82.4% (identification of individuals with MCI who will convert to AD), a 94.23% sensitivity in classifying persons with probable AD, a 86.3% specificity in classifying non-dementia controls | The results indicate that DNN classifiers may be useful as a potential tool for providing evidence in support of the clinical diagnosis of probable AD | [274] |
| DL algorithm based on DPN | Approaches for AD diagnosis and progression | Data from ADNI dataset (MRI and PET images from 51 AD patients, 99 MCI patients (43 MCI-C, who progressed to AD, and 56 MCI-NC, who did not progress to AD in 18 months), and 52 NC | Validation results using ROC curve showed an area under the curve of 0.897 | ML-based approaches for correct AD diagnosis, classifying all stages of AD progression | [275] |
| ML approach based on CNN | Classification of CT brain images for AD patients | Three main groups containing subjects with AD (1000 images), lesions (e.g., cancer) (947 images), or normal aging (2129 images). These data were used for training the model | Accuracy of 88.8%, 76.7%, and 95% for groups of AD, lesion and normal, respectively (average of 86.8%) | ML approach based on CNN is suitable for classifying CT brain images for AD | [276] |
| ML-based models, LRCV technique | Extraction of extracting spectrogram features from speech data for identifying early AD | Info from speech dataset, based on the spectrogram features (extracted based on audio data using an algorithm ad hoc), that enclosed AD patients and healthy subjects as controls. A total of 36 subjects were included in the collected speech dataset (23 AD 13 healthy) | LRCV accuracy 0.833, precision 0.869, recall 0.869, F1-score 0.869 | Identification of AD at early stages for providing therapies for delaying the disorder progression | [277] |
| ML approach EN, SVM, GP, k-NN | Prediction of possible progression of patients with MCI and preMCI to AD in 3 years | ML models were trained employing information from 90 patients with MCI and 94 subjects with PreMCI | The best performing ML model based on SVM technique showed an area under the ROC curve of 0.962 and an accuracy of 0.913 | Possible use of ML applications in medical practice and clinical trials | [278] |
| AI Technique | Target | Dataset | Statistical Parameters | Outcomes | Ref |
|---|---|---|---|---|---|
| DL approach CNN algorithm | AI tool to interpret echocardiograms | The model was trained using images and video (267 transthoracic echocardiograms) consisting of 200,000 images (240 studies) for arranging a training and validation set and a test set of over 20,000 images | The developed computer-based model showed an overall accuracy of 97.8% on videos (F-score 0.964) and of 100% on seven of the 12 video view | The use of CNN algorithms is suitable for a correct interpretation of echocardiograms | [294] |
| ML-based approach using CNN technique | Development of DL classifiers for automatically interpreting echocardiography data | The model was built using a dataset of 347,726 echocardiogram images (325,980 images were in the training set) | The model showed accuracy of 94.4% considering 15 echocardiographic view classifications of still images and 91.2% accuracy for binary left ventricular hypertrophy view classification | Efficient DL solutions for medical imaging assessment in cardiology | [295] |
| ML models based on CNN technique | Approach for an automatic classification of echocardiograms to detect three cardiovascular diseases: hypertrophic cardiomyopathy, cardiac amyloid, and pulmonary arterial hypertension | For training and validating the models 14,035 echocardiograms spanning a 10-year period were used. Results were assessed by comparing data from manual segmentation and measurements considering 8666 echocardiograms from clinical assessment | CNN models appropriately detect hypertrophic cardiomyopathy, cardiac amyloidosis, and pulmonary arterial hypertension showing C statistical parameters of 0.93, 0.87, and 0.85, respectively | ML models are useful for classifying echocardiograms and for detecting cardiovascular disorders | [296] |
| Supervised ML model based on RF algorithm | Approach for predicting future adverse cardiac events—RF algorithm for predicting survival from echocardiography data | The model was trained using echocardiograms from 171,510 patients | The ML model showed an area under the ROC curve >0.82 greater than conventional clinical risk scores (area under the ROC curve ranging from 0.61 to 0.79) | ML can successfully be used for predicting survival considering echocardiography data | [297] |
| ML model based on CNN technique | ML model for detecting myocardial infarction directly from ECG with no preprocessing | Dataset of 549 ECG outcomes from 290 subjects (PTB database) was used | The ML model showed sensitivity of 93.3% and specificity of 89.7% as assessed employing 10-fold cross-validation with sampling established on patients | The model detected myocardial infarction with performances comparable with those obtained from human cardiologists | [298] |
| ML model based on DNN technique | Approach for detecting arrhythmias employing ECG data | The model was trained using 91,232 single-lead ECG records from 53,549 patients for classifying 12 rhythm classes (10 arrhythmias, sinus rhythm and noise). Validation test set 328 ECGs collected from 328 patients | The ML model showed an area under the ROC curve of 0.97. The F1 score of 0.837 surpassed that of average cardiologists (0.780) for all rhythm classes | The results clearly indicate that the ML approach based on DNN can be used for correctly classifying different types of arrhythmias from ECG outcomes | [299] |
| CNN algorithm | ALVD can be predicted employing ECG data | Dataset composed of ECG/echocardiogram data from 44,959 patients for training a CNN algorithm. The developed model was tested against an independent set of 52,870 subjects | The model showed an area under the ROC curve, accuracy, specificity, and sensitivity of 0.93, 85.7%, 85.7%, and 86.3%, respectively | AI/ML to ECG data is versatile for predicting possible outputs for finding potential subjects who will develop ALVD | [301] |
| Unsupervised ML approach based on DNN technique | Approach for assessing diastolic dysfunction integrating multidimensional echocardiographic data with the aim to detect distinct patient subgroups with HFpEF | The established DNN model predicted high- and low-risk phenogroups in a derivation group (n = 1242). Two external groups for validating the model to identify high left ventricular filling pressure (n = 84) and assessing its prognostic capacity in patients (n = 219) showing different degrees of systolic and diastolic dysfunction | The relevance of the ML model was evaluated in three HFpEF clinical trials by assessing the relationships of the groups with adverse clinical outcomes. The developed model showed an area under ROC curve higher than that reported by the American Society of Echocardiography guidelines for predicting high left ventricular filling pressure (0.88 vs. 0.67; p = 0.01) | The DNN classifier can depict the severity of diastolic dysfunction and identify a specific subgroup of patients with HFpEF | [302] |
| ML model employing CNN technique | Approach for accurately classifying carotid plaques to estimate their stability to predict cardiovascular events | The model was trained using 1463 carotid plaque images (335 echo-rich plaques, 405 intermediate plaques, and 723 echolucent plaques) | The model showed sensitivity of 92.1%, specificity of 95.6%, accuracy of 92.1%, F1-score of 92.1% | The findings of this work proved that this strategy is relevant for enhancing the applicability of CNN using any input size of samples | [303] |
| AI Technique | Target | Dataset | Statistical Parameters | Outcomes | Ref |
|---|---|---|---|---|---|
| ML model based on SVM technique | Computational approach for discriminating actinic keratoses from healthy skin based on color texture examination of typical clinical photographs | Dataset composed of 6010 (actinic keratoses) and 13,915 (healthy) ROI from 22 patients. The model was tested using 157 actinic keratoses and 216 healthy skin rectangular regions of arbitrary size | The SVM model achieved a sensitivity of 80.1% and a specificity of 81.1% at ROI level, while a sensitivity of 89.8% and a specificity of 91.7% at region level | This work indicated that combining clinical photos with ML algorithms for a detailed image analysis is a useful, non-invasive, cost-effective method to monitor actinic keratoses for early therapeutic strategies against such skin lesions | [322] |
| ML approach based on ANN algorithm | Approach for assessing the skin sensitization risk derived from several chemicals | Dataset obtained for 62 compounds in murine LLNA (53 composed the training set, while the others were used for validating the computational tool) | The model was assessed using a 10-fold cross-validation method. The accuracy, specificity, and sensitivity of the model were 84.9%, 92.3%, and 82.5%, respectively | ML approaches for evaluating the risk estimation of compounds regarding skin sensitization can represent a valuable resource for replacing animal testing | [324] |
| ML model based on SVM technique | Approach for assessing the skin sensitization risk derived from several chemicals | Dataset composed of 120 chemicals with data on human skin sensitization, including LLNA. The molecules were distributed into the training set (75%) and test set (25%) | The validation step was performed applying LOO-cv. SVM was found to be the best method in modeling LLNA output with an accuracy of 89% and a sensitivity of 86%, and specificity of 92% on the test set | SVM model showed interesting results regarding the prediction of human outcomes | [325] |
| Deep CNN-based model | Approach for classifying skin lesions | The model was trained using a set of 129,450 clinical images | Results show an area under the ROC curve of 0.96 for carcinoma, and of 0.94 for melanoma | Computational tools based on CNN algorithms correctly classified skin lesions | [326] |
| ML-based model generated using CNN technique | Approach for evaluating the accuracy of melanoma skin cancer diagnosis considering the performance of 58 experts in comparison with the ML-based model | ML model was developed, validated, and tested for classifying dermoscopic images of lesions. The dataset enclosed a test set composed of 300 images containing 20% melanomas and 80% benign melanocytic nevi | The average of the calculated area under the ROC curves was 0.79, considering the results from the 58 dermatologists, and 0.86, considering the ML model, respectively | ML models appropriately trained have the ability to perform accurate diagnostic classification of dermoscopic images of melanocytic origin | [327,328] |
| ML model using CNN algorithm | Approach for classifying clinical images from 12 skin diseases | ML model was trained, tested, and validated employing the Asan dataset, MED-NODE dataset, and atlas site images, for a total of 19,398 images, opportunely divided in training set and test set | Considering the Asan dataset, the area under the ROC curve concerning the diagnosis of basal cell carcinoma, squamous cell carcinoma, intraepithelial carcinoma, and melanoma was 0.96, 0.83, 0.82, and 0.96, respectively. Considering the Edinburgh dataset, the area under the ROC curve for the same disorders was 0.90, 0.91, 0.83, and 0.88, respectively. | The ML-based model showed comparable performances to those obtained from 16 dermatologists | [329] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brogi, S.; Calderone, V. Artificial Intelligence in Translational Medicine. Int. J. Transl. Med. 2021, 1, 223-285. https://doi.org/10.3390/ijtm1030016
Brogi S, Calderone V. Artificial Intelligence in Translational Medicine. International Journal of Translational Medicine. 2021; 1(3):223-285. https://doi.org/10.3390/ijtm1030016
Chicago/Turabian StyleBrogi, Simone, and Vincenzo Calderone. 2021. "Artificial Intelligence in Translational Medicine" International Journal of Translational Medicine 1, no. 3: 223-285. https://doi.org/10.3390/ijtm1030016
APA StyleBrogi, S., & Calderone, V. (2021). Artificial Intelligence in Translational Medicine. International Journal of Translational Medicine, 1(3), 223-285. https://doi.org/10.3390/ijtm1030016







