Regional Differences in Gene Expression of Proliferating Human Choroidal Endothelial Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of CEC
2.2. Confirmation of EC Purity
2.3. RNA Extraction
2.4. Microarray Analysis
2.5. Data Analysis
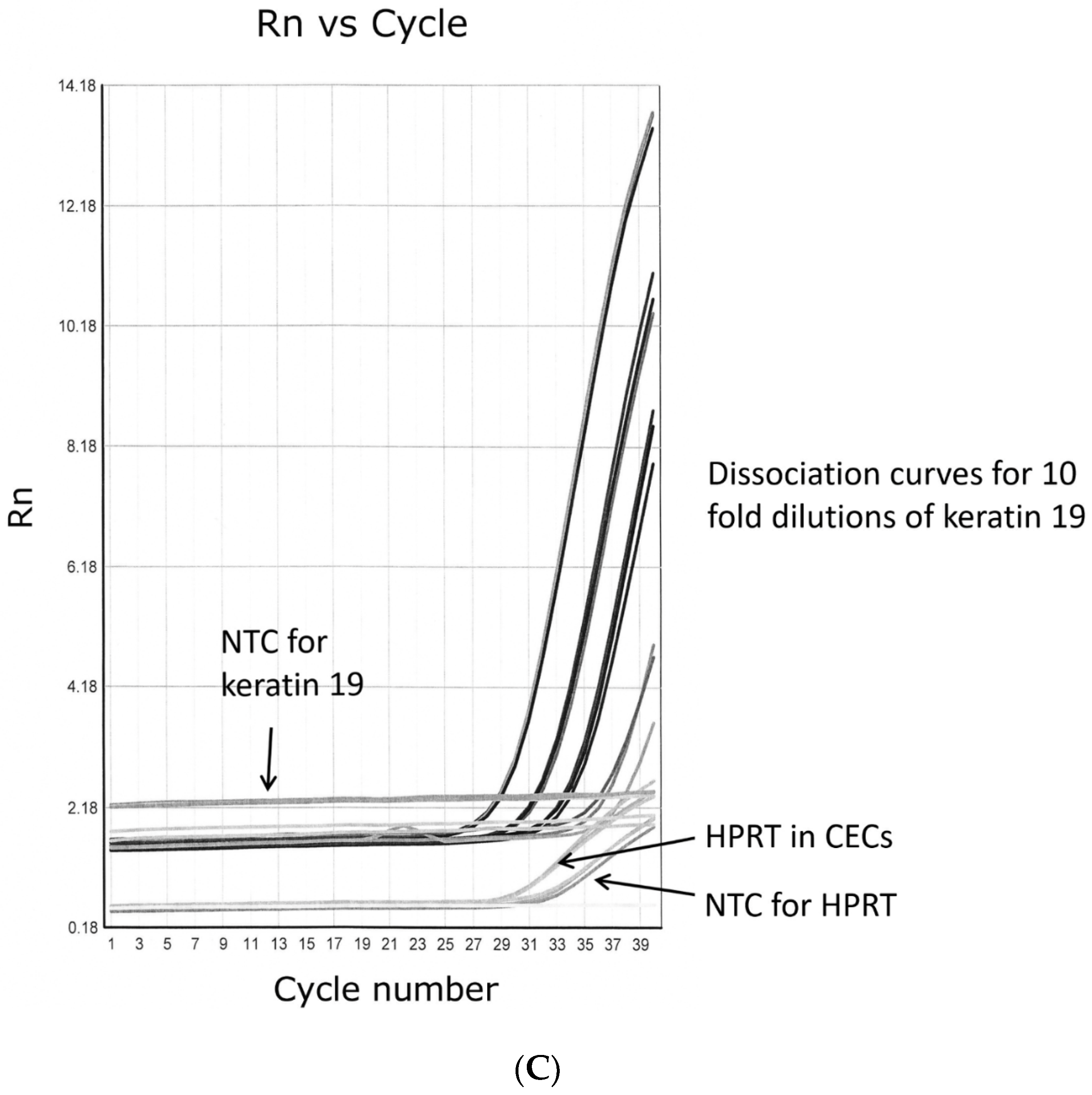
2.6. QPCR
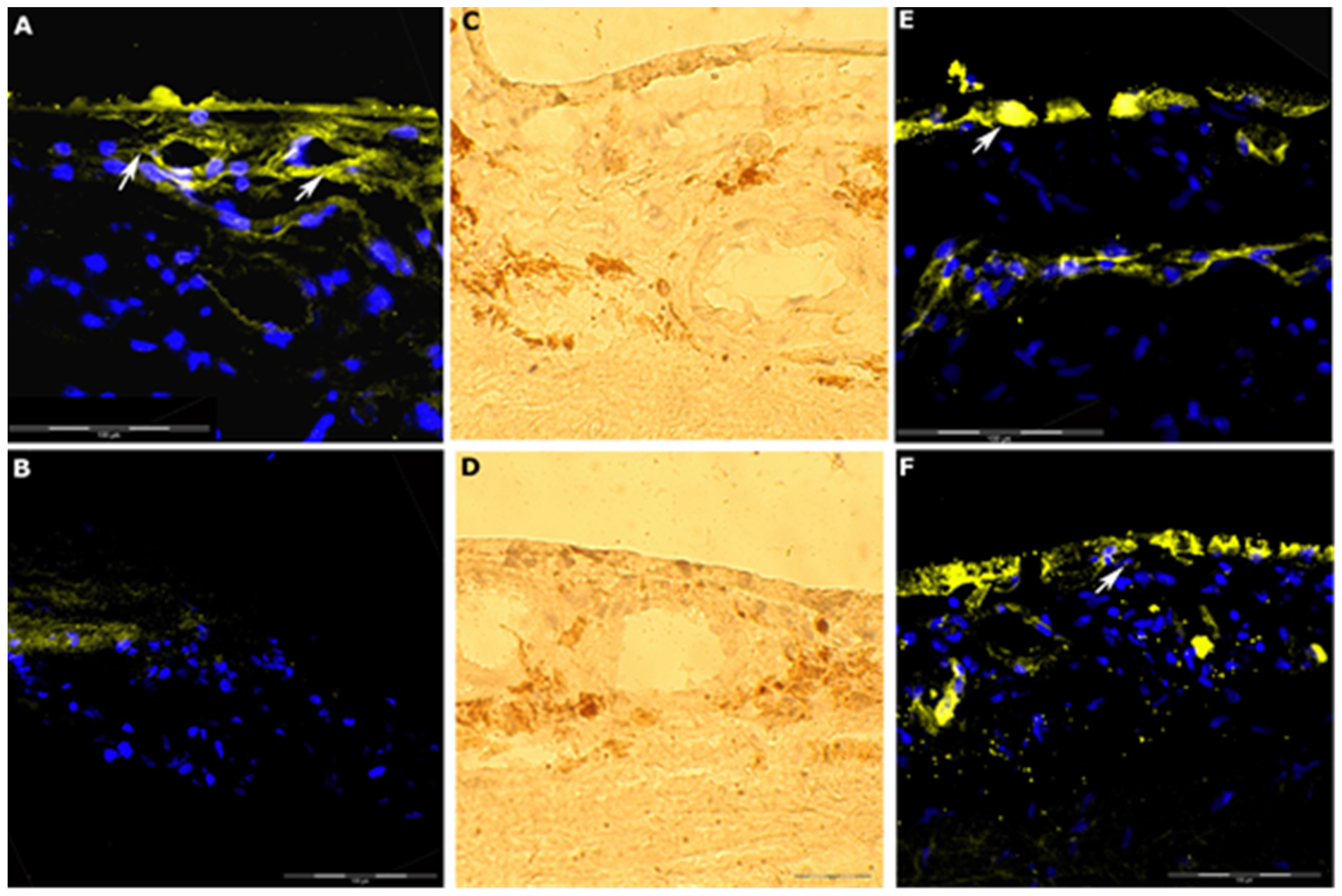
2.7. Immunohistochemistry
3. Results
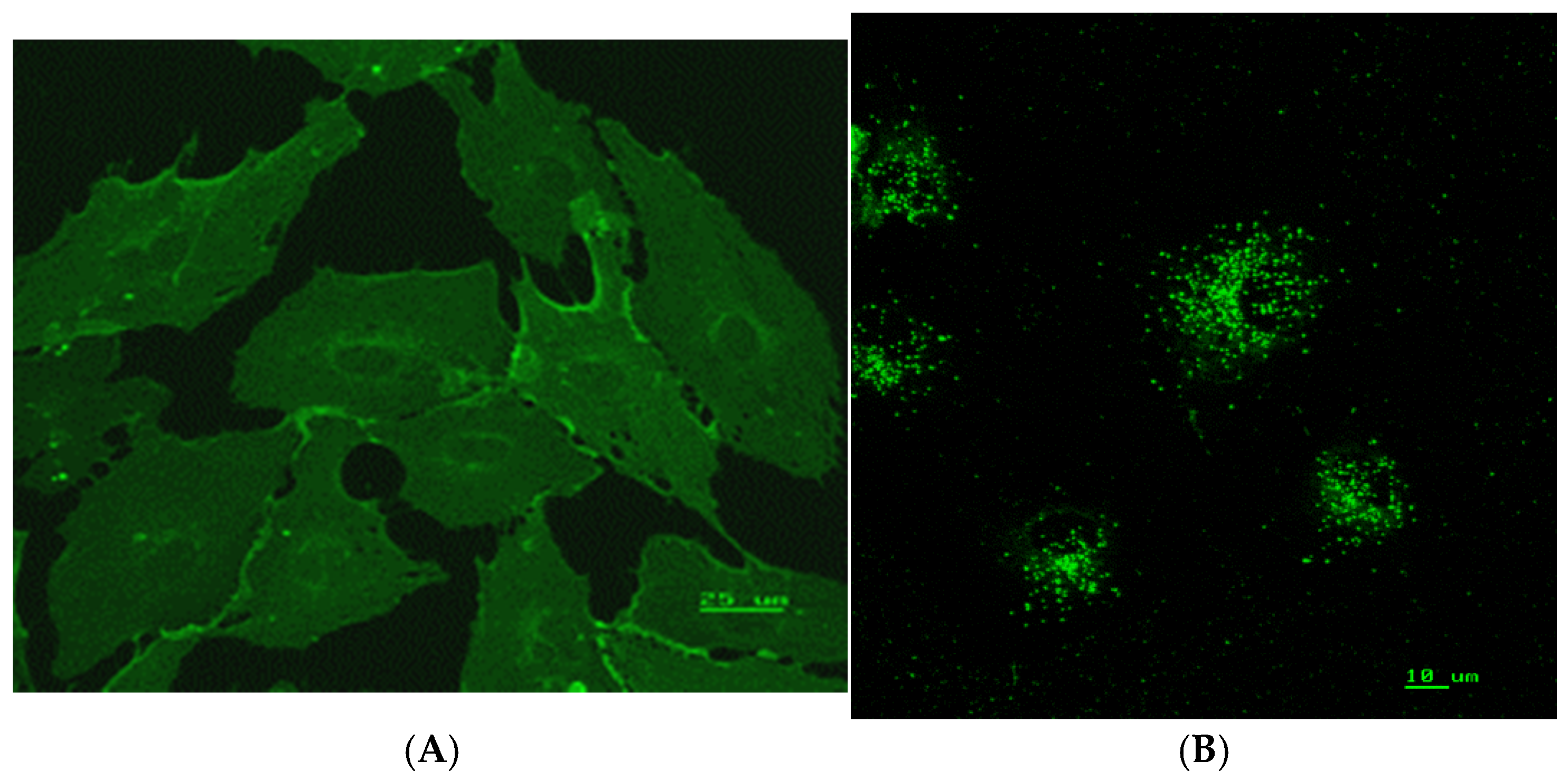
3.1. Confirmation of Human Endothelial Cell Identity
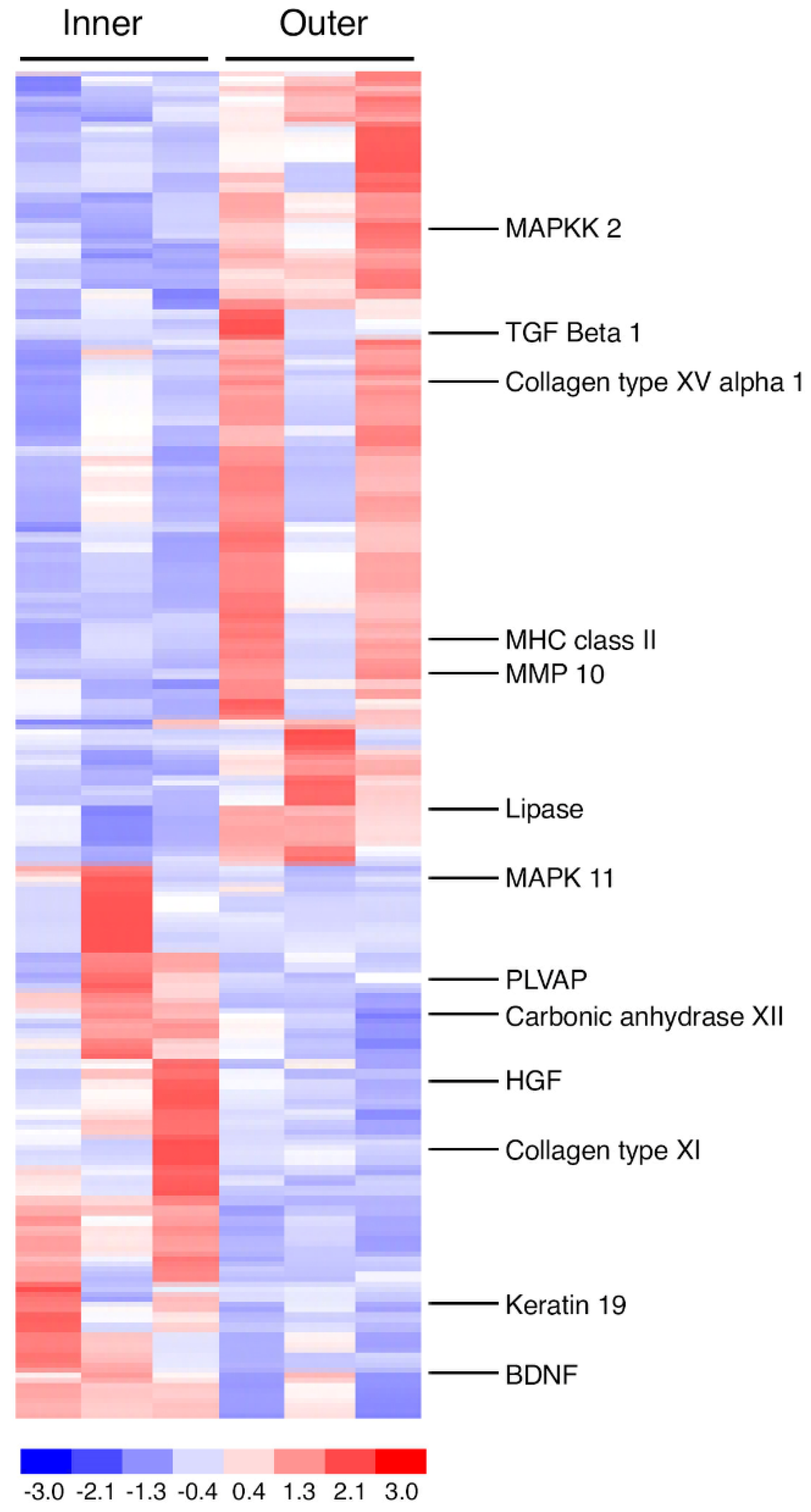
3.2. Overview of Gene Expression Patterns
3.3. Proliferating Human Macular Inner CEC Versus Peripheral Inner CEC
3.4. Proliferating Human Macular Inner CEC Versus Macular Outer CEC
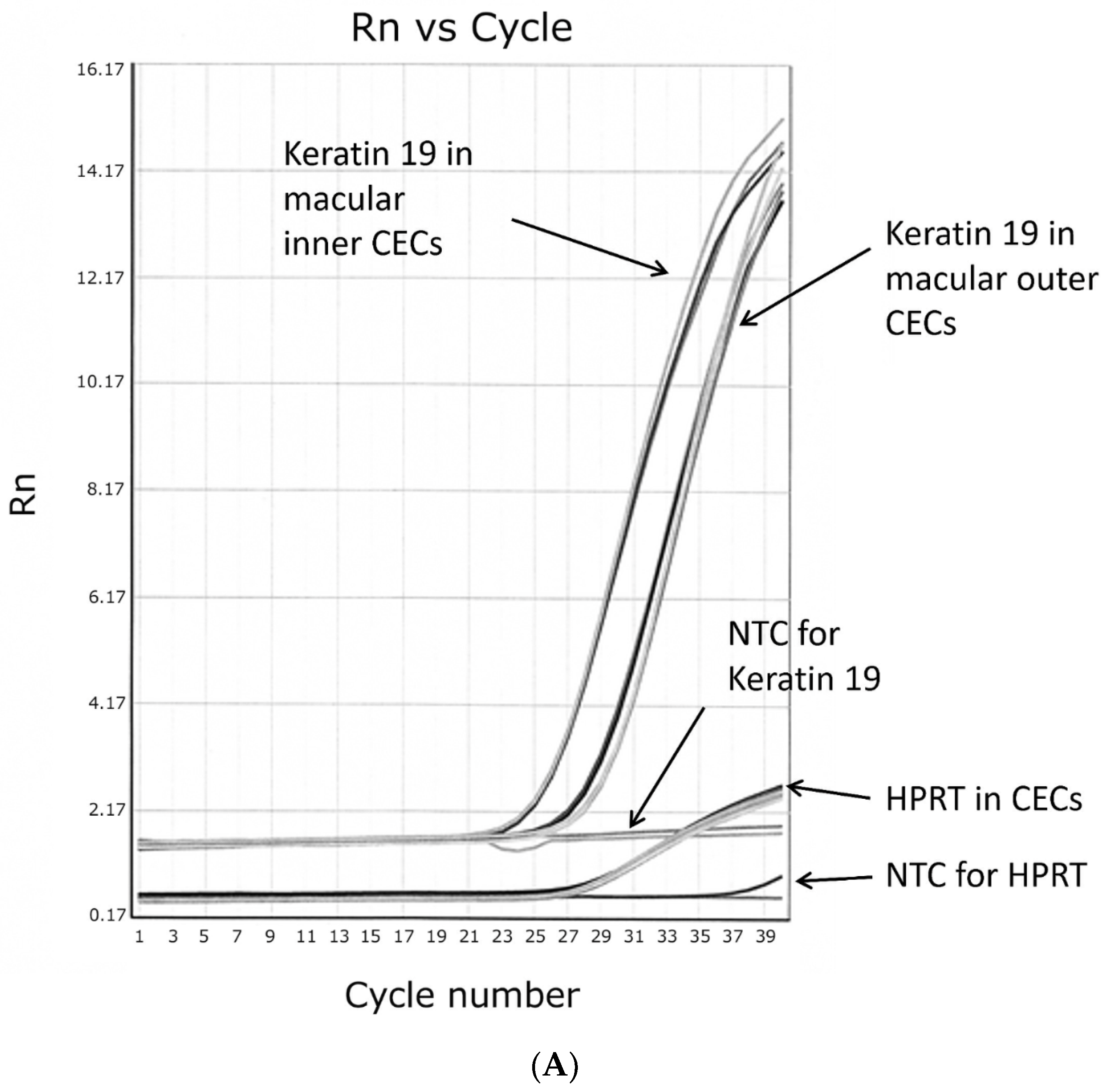
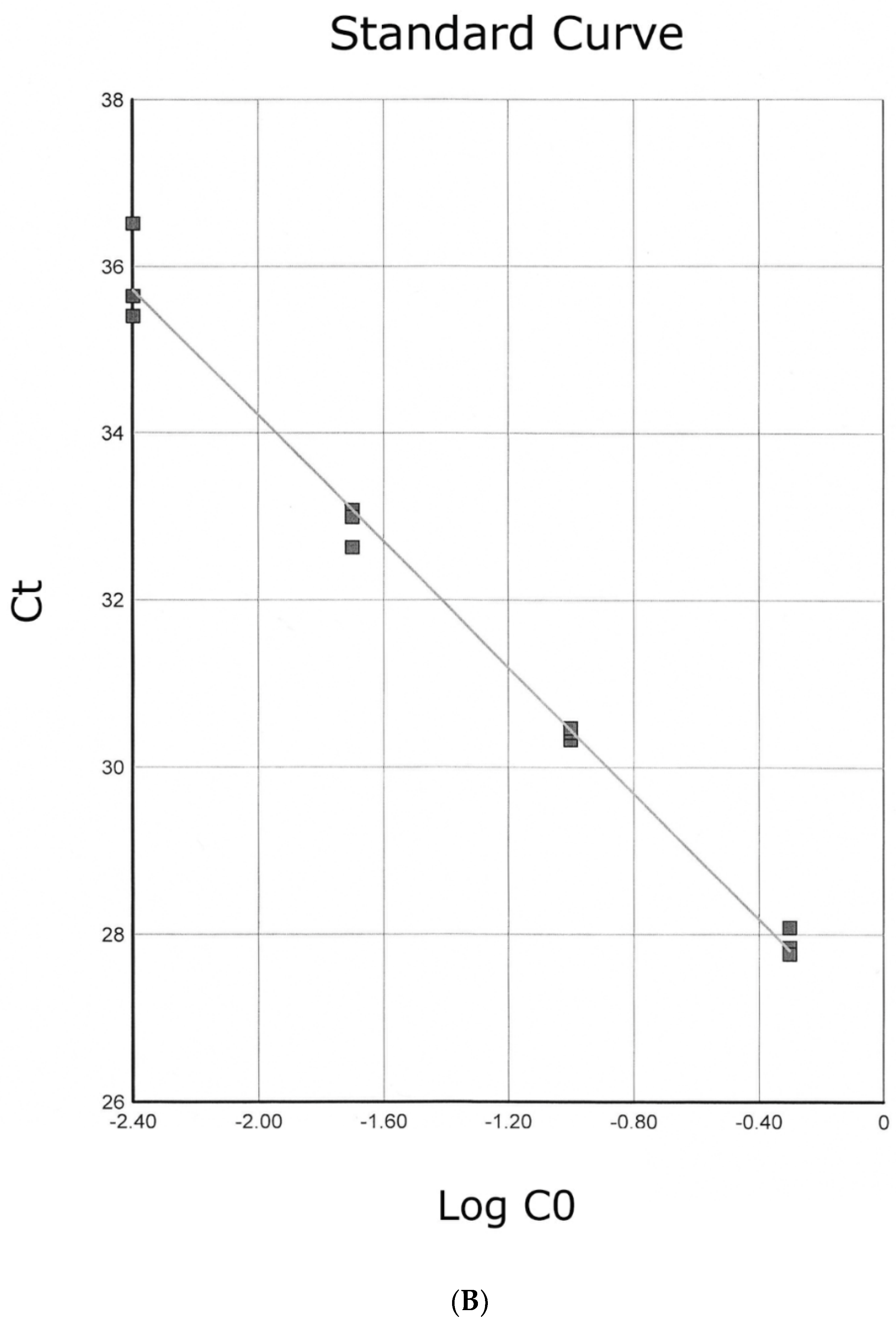
3.5. Real-Time PCR
3.6. Immunohistochemistry
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharon, D.; Blackshaw, S.; Cepko, C.L.; Dryja, T.P. Profile of the genes expressed in the human peripheral retina, macula, and retinal pigment epithelium determined through serial analysis of gene expression (SAGE). Proc. Natl. Acad. Sci. USA 2002, 99, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K.; Tian, J.; Handa, J.T. Similarity of mRNA phenotypes of morphologically normal macular and peripheral retinal pigment epithelial cells in older human eyes. Investig. Ophthalmol. Vis. Sci. 2004, 45, 3291–3301. [Google Scholar] [CrossRef]
- Bowes Rickman, C.; Ebright, J.N.; Zavodni, Z.J.; Yu, L.; Wang, T.; Daiger, S.P.; Wistow, G.; Boon, K.; Hauser, M.A. Defining the human macula transcriptome and candidate retinal disease genes using EyeSAGE. Investig. Ophthalmol. Vis. Sci. 2006, 47, 2305–2316. [Google Scholar] [CrossRef][Green Version]
- Van Soest, S.S.; de Wit, G.M.; Essing, A.H.; ten Brink, J.B.; Kamphuis, W.; de Jong, P.T.; Bergen, A.A. Comparison of human retinal pigment epithelium gene expression in macula and periphery highlights potential topographic differences in Bruch’s membrane. Mol. Vis. 2007, 13, 1608–1617. [Google Scholar]
- Curcio, C.A.; Sloan, K.R.; Kalina, R.E.; Hendrickson, A.E. Human photoreceptor topography. J. Comp. Neurol. 1990, 292, 497–523. [Google Scholar] [CrossRef]
- Hornan, D.M.; Peirson, S.N.; Hardcastle, A.J.; Molday, R.S.; Cheetham, M.E.; Webster, A.R. Novel retinal and cone photoreceptor transcripts revealed by human macular expression profiling. Investig. Ophthalmol. Vis. Sci. 2007, 48, 5388–5396. [Google Scholar] [CrossRef]
- Bernstein, S.L.; Wong, P. Regional expression of disease-related genes in human and monkey retina. Mol. Vis. 1998, 4, 24. [Google Scholar]
- Bron, A.J.; Tripathi, R.C.; Tripathi, B.J. Wolff’s Anatomy of the Eye and Orbit; Chapman and Hall: London, UK, 1997; pp. 460–465. [Google Scholar]
- Keilhauer, C.N.; Delori, F.C. Near-infrared autofluorescence imaging of the fundus: Visualization of ocular melanin. Investig. Ophthalmol. Vis. Sci. 2006, 47, 3556–3564. [Google Scholar] [CrossRef]
- Ramrattan, R.S.; van der Schaft, T.L.; Mooy, C.M.; de Bruijn, W.C.; Mulder, P.G.; de Jong, P.T. Morphometric analysis of Bruch’s membrane, the choriocapillaris, and the choroid in aging. Investig. Ophthalmol. Vis. Sci. 1994, 35, 2857–2864. [Google Scholar]
- Okubo, A.; Rosa, R.H., Jr.; Bunce, C.V.; Alexander, R.A.; Fan, J.T.; Bird, A.C.; Luthert, P.J. The relationships of age changes in retinal pigment epithelium and Bruch’s membrane. Investig. Ophthalmol. Vis. Sci. 1999, 40, 443–449. [Google Scholar]
- Chong, N.H.; Keonin, J.; Luthert, P.J.; Frennesson, C.I.; Weingeist, D.M.; Wolf, R.L.; Mullins, R.F.; Hageman, G.S. Decreased thickness and integrity of the macular elastic layer of Bruch’s membrane correspond to the distribution of lesions associated with age-related macular degeneration. Am. J. Pathol. 2005, 166, 241–251. [Google Scholar] [CrossRef]
- Killingsworth, M.C. Angiogenesis in early choroidal neovascularization secondary to age-related macular degeneration. Graefes Arch. Clin. Exp. Ophthalmol. 1995, 233, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Sarks, J.P.; Sarks, S.H.; Killingsworth, M.C. Morphology of early choroidal neovascularisation in age-related macular degeneration: Correlation with activity. Eye 1997, 11, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Browning, A.C.; Gray, T.; Amoaku, W.M. Isolation, culture, and characterisation of human macular inner choroidal microvascular endothelial cells. Br. J. Ophthalmol. 2005, 89, 1343–1347. [Google Scholar] [CrossRef]
- Browning, A.C.; Halligan, E.P.; Stewart, E.A.; Swan, D.C.; Dove, R.; Samaranayake, G.J.; Amoaku, W.M. Comparative gene expression profiling of human umbilical vein endothelial cells and ocular vascular endothelial cells. Br. J. Ophthalmol. 2012, 96, 128–132. [Google Scholar] [CrossRef]
- Breitling, R.; Armengaud, P.; Amtmann, A.; Herzyk, P. Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004, 573, 83–92. [Google Scholar] [CrossRef]
- Shen, W.Y.; Lee, S.Y.; Yeo, I.; Lai, C.; Mathur, R.; Tan, D.; Constable, I.J.; Rakoczy, P.E. Predilection of the macular region to high incidence of choroidal neovascularization after intense laser photocoagulation in the monkey. Arch. Ophthalmol. 2004, 122, 353–360. [Google Scholar] [CrossRef][Green Version]
- Cruickshanks, K.J.; Klein, R.; Klein, B.E.; Nondahl, D.M. Sunlight and the 5-year incidence of early age-related maculopathy: The beaver dam eye study. Arch. Ophthalmol. 2001, 119, 246–250. [Google Scholar]
- Tomany, S.C.; Cruickshanks, K.J.; Klein, R.; Klein, B.E.; Knudtson, M.D. Sunlight and the 10-year incidence of age-related maculopathy: The Beaver Dam Eye Study. Arch. Ophthalmol. 2004, 122, 750–757. [Google Scholar] [CrossRef]
- Grimm, C.; Wenzel, A.; Williams, T.; Rol, P.; Hafezi, F.; Reme, C. Rhodopsin-mediated blue-light damage to the rat retina: Effect of photoreversal of bleaching. Investig. Ophthalmol. Vis. Sci. 2001, 42, 497–505. [Google Scholar]
- Hafezi, F.; Marti, A.; Munz, K.; Reme, C.E. Light-induced apoptosis: Differential timing in the retina and pigment epithelium. Exp. Eye Res. 1997, 64, 963–970. [Google Scholar] [CrossRef]
- Delcourt, C.; Carriere, I.; Ponton-Sanchez, A.; Fourrey, S.; Lacroux, A.; Papoz, L. Light exposure and the risk of age-related macular degeneration: The Pathologies Oculaires Liees a l’Age (POLA) study. Arch. Ophthalmol. 2001, 119, 1463–1468. [Google Scholar] [CrossRef]
- Darzins, P.; Mitchell, P.; Heller, R.F. Sun exposure and age-related macular degeneration. An Australian case-control study. Ophthalmology 1997, 104, 770–776. [Google Scholar] [CrossRef]
- McCarty, C.A.; Mukesh, B.N.; Fu, C.L.; Mitchell, P.; Wang, J.J.; Taylor, H.R. Risk factors for age-related maculopathy: The Visual Impairment Project. Arch. Ophthalmol. 2001, 119, 1455–1462. [Google Scholar] [CrossRef] [PubMed]
- Lengyel, I.; Tufail, A.; Hosaini, H.A.; Luthert, P.; Bird, A.C.; Jeffery, G. Association of drusen deposition with choroidal intercapillary pillars in the aging human eye. Investig. Ophthalmol. Vis. Sci. 2004, 45, 2886–2892. [Google Scholar] [CrossRef] [PubMed]
- Mullins, R.F.; Skeie, J.M.; Malone, E.A.; Kuehn, M.H. Macular and peripheral distribution of ICAM-1 in the human choriocapillaris and retina. Mol. Vis. 2006, 12, 224–235. [Google Scholar] [PubMed]
- Abi-Hanna, D.; Wakefield, D.; Watkins, S. HLA antigens in ocular tissues. I. In vivo expression in human eyes. Transplantation 1988, 45, 610–613. [Google Scholar] [CrossRef]
- Radeke, M.J.; Peterson, K.E.; Johnson, L.V.; Anderson, D.H. Disease susceptibility of the human macula: Differential gene transcription in the retinal pigmented epithelium/choroid. Exp. Eye Res. 2007, 85, 366–380. [Google Scholar] [CrossRef]
- Whitmore, S.S.; Wagner, A.H.; DeLuca, A.P.; Drack, A.V.; Stone, E.M.; Tucker, B.A.; Zeng, S.; Braun, T.A.; Mullins, R.F.; Scheetz, T.E. Transcriptomic analysis across nasal, temporal, and macular regions of human neural retina and RPE/choroid by RNA-Seq. Exp. Eye Res. 2014, 129, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Skeie, J.M.; Mahajan, V.B. Proteomic landscape of the human choroid—Retinal pigment epithelial complex. JAMA Ophthalmol. 2014, 132, 1271–1281. [Google Scholar] [CrossRef]
- Voigt, A.P.; Mulfaul, K.; Mullin, N.K.; Flamme-Wiese, M.J.; Giacalone, J.C.; Stone, E.M.; Tucker, B.A.; Scheetz, T.E.; Mullins, R.F. Single-cell transcriptomics of the human retinal pigment epithelium and choroid in health and macular degeneration. Proc. Natl. Acad. Sci. USA 2019, 116, 24100–24107. [Google Scholar] [CrossRef]
- Satchell, S.C.; Braet, F. Glomerular endothelial cell fenestrations: An integral component of the glomerular filtration barrier. Am. J. Physiol. Renal Physiol. 2009, 296, F947–F956. [Google Scholar] [CrossRef]
- Kano, T.; Abe, T.; Tomita, H.; Sakata, T.; Ishiguro, S.; Tamai, M. Protective effect against ischemia and light damage of iris pigment epithelial cells transfected with the BDNF gene. Investig. Ophthalmol. Vis. Sci. 2002, 43, 3744–3753. [Google Scholar] [PubMed]
- Nakahashi, T.; Fujimura, H.; Altar, C.A.; Li, J.; Kambayashi, J.; Tandon, N.N.; Sun, B. Vascular endothelial cells synthesize and secrete brain-derived neurotrophic factor. FEBS Lett. 2000, 470, 113–117. [Google Scholar] [CrossRef]
- Amenta, P.S.; Scivoletti, N.A.; Newman, M.D.; Sciancalepore, J.P.; Li, D.; Myers, J.C. Proteoglycan-collagen XV in human tissues is seen linking banded collagen fibers subjacent to the basement membrane. J. Histochem. Cytochem. 2005, 53, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Grierson, I.; Heathcote, L.; Hiscott, P.; Hogg, P.; Briggs, M.; Hagan, S. Hepatocyte growth factor/scatter factor in the eye. Prog. Retin. Eye Res. 2000, 19, 779–802. [Google Scholar] [CrossRef]
- He, P.M.; He, S.; Garner, J.A.; Ryan, S.J.; Hinton, D.R. Retinal pigment epithelial cells secrete and respond to hepatocyte growth factor. Biochem. Biophys. Res. Commun. 1998, 249, 253–257. [Google Scholar] [CrossRef]
- Jin, M.; Barron, E.; He, S.; Ryan, S.J.; Hinton, D.R. Regulation of RPE intercellular junction integrity and function by hepatocyte growth factor. Investig. Ophthalmol. Vis. Sci. 2002, 43, 2782–2790. [Google Scholar]
- Jin, M.; Chen, Y.; He, S.; Ryan, S.J.; Hinton, D.R. Hepatocyte growth factor and its role in the pathogenesis of retinal detachment. Investig. Ophthalmol. Vis. Sci. 2004, 45, 323–329. [Google Scholar] [CrossRef]
- Nishimura, M.; Ikeda, T.; Ushiyama, M.; Nanbu, A.; Kinoshita, S.; Yoshimura, M. Increased vitreous concentrations of human hepatocyte growth factor in proliferative diabetic retinopathy. J. Clin. Endocrinol. Metab. 1999, 84, 659–662. [Google Scholar] [CrossRef]
- Katsura, Y.; Okano, T.; Noritake, M.; Kosano, H.; Nishigori, H.; Kado, S.; Matsuoka, T. Hepatocyte growth factor in vitreous fluid of patients with proliferative diabetic retinopathy and other retinal disorders. Diabetes Care 1998, 21, 1759–1763. [Google Scholar] [CrossRef]
- Hu, W.; Criswell, M.H.; Fong, S.L.; Temm, C.J.; Rajashekhar, G.; Cornell, T.L.; Clauss, M.A. Differences in the temporal expression of regulatory growth factors during choroidal neovascular development. Exp. Eye Res. 2009, 88, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Bhutto, I.A.; McLeod, D.S.; Merges, C.; Hasegawa, T.; Lutty, G.A. Localisation of SDF-1 and its receptor CXCR4 in retina and choroid of aged human eyes and in eyes with age related macular degeneration. Br. J. Ophthalmol. 2006, 90, 906–910. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cai, H.; Fields, M.A.; Hoshino, R.; Del Priore, L.V. Effects of aging and anatomical location on gene expression in human retina. Front. Aging Neurosci. 2012, 4, 1–20. [Google Scholar] [CrossRef]
| Gene Title | Fold Change | Uncorrected p-Value | Corrected p-Value |
|---|---|---|---|
| insulin-like growth factor binding protein 3 | 11.1 | 0 | 0 |
| neurofilament, medium polypeptide | 10.4 | 0 | 0 |
| brain-derived neurotrophic factor | 8.8 | 0 | 0 |
| platelet-derived growth factor receptor, alpha polypeptide | 8.8 | 0 | 0.0029 |
| keratin 19 | 7.3 | 0 | 0.0033 |
| pleckstrin homology-like domain, family A, member 2 | 7.1 | 0 | 0 |
| solute carrier family 6 (neutral amino acid transporter) | 6.6 | 0 | 0 |
| popeye domain containing 3 | 6.0 | 0 | 0.0031 |
| olfactomedin-like 3 | 5.9 | 0 | 0.0038 |
| SIX homeobox 2 | 5.6 | 0 | 0.0035 |
| neurofilament, light polypeptide | 5.5 | 0 | 0.0033 |
| adipocyte-specific adhesion molecule | 5.2 | 0 | 0.0027 |
| CCAAT/enhancer binding protein (C/EBP), beta | 5.2 | 0 | 0.0107 |
| mesoderm specific transcript homolog (mouse) | 5.1 | 0 | 0.0036 |
| ADAM metallopeptidase with thrombospondin type 1 | 5.1 | 0 | 0.005 |
| mannosyl (alpha-1,3-)-glycoprotein | 5.1 | 0 | 0.0029 |
| keratin associated protein 2–4 | 5.0 | 0 | 0.0044 |
| chromosome 5 open reading frame 23 | 4.9 | 0 | 0.004 |
| vascular endothelial growth factor A | 4.9 | 0 | 0.0197 |
| brain expressed, X-linked 1 | 4.8 | 0 | 0.0113 |
| GLI pathogenesis-related 1 | 4.7 | 0 | 0.0096 |
| lysophosphatidic acid receptor 1 | 4.7 | 0 | 0.0177 |
| Aldehyde dehydrogenase 1 family, member L2 | 4.6 | 0 | 0.0264 |
| collagen, type XI, alpha 1 | 4.4 | 1.0 × 10−4 | 0.0388 |
| pleckstrin and Sec7 domain containing 3 | 4.4 | 0 | 0.0037 |
| carbonic anhydrase XII | 4.4 | 1.0 × 10−4 | 0.0393 |
| procollagen C-endopeptidase enhancer 2 | 4.3 | 0 | 0.0272 |
| ABI family, member 3 (NESH) binding protein | 4.3 | 0 | 0.0269 |
| phosphodiesterase 5A, cGMP-specific | 4.2 | 0 | 0.0107 |
| PNMA-like 1 | 4.1 | 0 | 0.0205 |
| tissue factor pathway inhibitor 2 | 4.1 | 1.0 × 10−4 | 0.0408 |
| glycine receptor, beta | 4.0 | 0 | 0.034 |
| fin bud initiation factor homolog (zebrafish) | 4.0 | 0 | 0.013 |
| wingless-type MMTV integration site family, member 5A | 4.0 | 0 | 0.0038 |
| sphingosine-1-phosphate receptor 3 | 4.0 | 0 | 0.0298 |
| carbonic anhydrase XII | 3.9 | 0 | 0.0171 |
| forkhead box F2 | 3.9 | 0 | 0.0321 |
| MSTP150 | 3.9 | 2..0 × 10−4 | 0.0495 |
| Ras-related C3 botulinum toxin substrate 1 | 3.9 | 1.0 × 10−4 | 0.0406 |
| frequenin homolog (Drosophila) | 3.8 | 0 | 0.0207 |
| glycine receptor, beta | 3.8 | 1.0 × 10−4 | 0.0405 |
| vestigial like 3 (Drosophila) | 3.8 | 0 | 0.0259 |
| TP53 regulating kinase | 3.8 | 1.0 × 10−4 | 0.0421 |
| chromosome 6 open reading frame 141 | 3.8 | 0 | 0.0095 |
| calmegin | 3.7 | 1.0 × 10−4 | 0.0359 |
| carbonic anhydrase XII | 3.7 | 0 | 0.0152 |
| hypothetical protein LOC100130506 | 3.7 | 0 | 0.0112 |
| ectonucleotide pyrophosphatase/phosphodiesterase 2 | 3.7 | 1.0 × 10−4 | 0.0437 |
| protocadherin 18 | 3.6 | 0 | 0.0295 |
| leucine rich repeat (in FLII) interacting protein 1 | 3.6 | 0 | 0.0155 |
| hepatocyte growth factor (hepapoietin A; scatter factor) | 3.6 | 0 | 0.0091 |
| protein phosphatase 1, regulatory (inhibitor) subunit 14B | 3.6 | 1.0 × 10−4 | 0.0486 |
| golgi autoantigen, golgin subfamily a, 8A | 3.6 | 0 | 0.0159 |
| lymphoid-restricted membrane protein | 3.5 | 0 | 0.0341 |
| sulfatase 1 | 3.4 | 1.0 × 10−4 | 0.0471 |
| dermatan sulfate epimerase-like | 3.4 | 0 | 0.0116 |
| secretogranin II (chromogranin C) | 3.4 | 1.0 × 10−4 | 0.0408 |
| poliovirus receptor-related 3 | 3.4 | 0 | 0.026 |
| metastasis associated lung adenocarcinoma transcript 1 | 3.4 | 0 | 0.0205 |
| folate receptor 1 (adult) | 3.4 | 0 | 0.021 |
| transforming growth factor, beta receptor 1 | 3.4 | 1.0 × 10−4 | 0.0386 |
| spermatogenesis associated 18 homolog (rat) | 3.4 | 1.0 × 10−4 | 0.0429 |
| tissue factor pathway inhibitor 2 | 3.4 | 0 | 0.0337 |
| emopamil binding protein-like | 3.4 | 1.0 × 10−4 | 0.0354 |
| aldehyde dehydrogenase 1 family, member A3 | 3.4 | 0 | 0.0283 |
| centromere protein V | 3.4 | 0 | 0.0256 |
| interferon-induced protein with tetratricopeptide repeats 1 | 3.4 | 0 | 0.0332 |
| lactamase, beta | 3.4 | 0 | 0.0183 |
| 5’-nucleotidase, ecto (CD73) | 3.3 | 1.0 × 10−4 | 0.0426 |
| tumor necrosis factor (ligand) superfamily, member 15 | 3.3 | 0 | 0.0203 |
| mitogen-activated protein kinase kinase kinase kinase 4 | 3.3 | 1.0 × 10−4 | 0.0448 |
| chromosome 9 open reading frame 40 | 3.3 | 0 | 0.0319 |
| neuropilin (NRP) and tolloid (TLL)-like 2 | 3.3 | 1.0 × 10−4 | 0.0347 |
| family with sequence similarity 13, member B | 3.3 | 1.0 × 10−4 | 0.041 |
| aspartyl-tRNA synthetase | 3.2 | 0 | 0.0195 |
| Chromodomain helicase DNA binding protein 2 | 3.2 | 1.0 × 10−4 | 0.0404 |
| paternally expressed 10 | 3.2 | 0 | 0.0311 |
| versican | 3.1 | 0 | 0.0298 |
| metastasis associated lung adenocarcinoma transcript 1 | 3.1 | 0 | 0.0337 |
| nephronophthisis 3 (adolescent) | 3.1 | 0 | 0.0265 |
| carboxymethylenebutenolidase homolog (Pseudomonas) | 3.1 | 0 | 0.0293 |
| hypothetical LOC100128822 | 3.1 | 1.0 × 10−4 | 0.0434 |
| phosphoglycolate phosphatase | 3.1 | 0 | 0.0262 |
| prostaglandin-endoperoxide synthase 2 | 3.0 | 0 | 0.0227 |
| retinol dehydrogenase 14 (all-trans/9-cis/11-cis) | 3.0 | 0 | 0.0267 |
| carbohydrate sulfotransferase 7 | 3.0 | 1.0 × 10−4 | 0.0444 |
| proline rich 16 | 3.0 | 2.0 × 10−4 | 0.0498 |
| transcription factor Dp-1 | 3.0 | 0 | 0.0107 |
| aryl hydrocarbon receptor | 3.0 | 0 | 0.0346 |
| glycoprotein (transmembrane) nmb | 3.0 | 0 | 0.0295 |
| chromosome 13 open reading frame 15 | 3.0 | 0 | 0.0319 |
| collagen, type I, alpha 2 | 3.0 | 0 | 0.0293 |
| O-linked N-acetylglucosamine (GlcNAc) transferase | 3.0 | 1.0 × 10−4 | 0.0419 |
| B-cell translocation gene 1, anti-proliferative | 3.0 | 1.0 × 10−4 | 0.0424 |
| phosphatidic acid phosphatase type 2B | 3.0 | 0 | 0.0339 |
| cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | 3.0 | 0 | 0.0342 |
| TAF10 RNA polymerase II, TATA box binding protein | 3.0 | 1.0 × 10−4 | 0.0495 |
| serpin peptidase inhibitor, clade E (nexin, | 2.9 | 1.0 × 10−4 | 0.0453 |
| COMM domain containing 2 | 2.9 | 1.0 × 10−4 | 0.0363 |
| transducer of ERBB2, 1 | 2.9 | 1.0 × 10−4 | 0.043 |
| Gene Title | Fold Change | Uncorrected p-Value | Corrected p-Value |
|---|---|---|---|
| histone cluster 1, H3b | 14.7 | 0 | 0 |
| LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) | 10.2 | 0 | 0 |
| translocase of inner mitochondrial membrane 44 homolog (yeast) | 8.9 | 0 | 0 |
| chromosome 6 open reading frame 108 | 8.4 | 0 | 0 |
| fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) | 7.9 | 0 | 0 |
| FXYD domain containing ion transport regulator 6 | 7.8 | 0 | 0 |
| leucine rich repeat containing 15 | 7.6 | 0 | 0 |
| translocase of inner mitochondrial membrane 44 homolog (yeast) | 7.4 | 0 | 0 |
| RAS and EF-hand domain containing | 7.4 | 0 | 0 |
| kinesin light chain 1 | 6.9 | 0 | 0 |
| RAS and EF-hand domain containing | 6.8 | 0 | 0 |
| cyclin K | 6.4 | 0 | 0 |
| polypyrimidine tract binding protein 1 | 6.0 | 0 | 0.001 |
| valyl-tRNA synthetase | 5.9 | 0 | 0.0011 |
| apelin receptor | 5.4 | 0 | 0.0024 |
| fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) | 5.4 | 0 | 0.003 |
| mitogen-activated protein kinase kinase 2 | 5.1 | 0 | 0.0029 |
| cleft lip and palate associated transmembrane protein 1 | 5.1 | 0 | 0.0018 |
| Hypothetical protein LOC339047 | 4.9 | 0 | 0.0006 |
| RNA pseudouridylate synthase domain containing 3 | 4.7 | 0 | 0.0024 |
| histone cluster 1, H2bf | 4.6 | 0 | 0.0025 |
| transforming growth factor beta 1 induced transcript 1 | 4.5 | 0 | 0.0026 |
| mRNA turnover 4 homolog (S. cerevisiae) | 4.5 | 0 | 0.0027 |
| deoxyribonuclease I-like 3 | 4.4 | 0 | 0.0028 |
| RAN binding protein 3 | 4.4 | 0 | 0.0031 |
| histone deacetylase 5 | 4.4 | 0 | 0.0041 |
| cysteine-rich protein 2 | 4.1 | 0 | 0.0023 |
| cell division cycle 34 homolog (S. cerevisiae) | 4.1 | 0 | 0.0103 |
| TAO kinase 1 | 4.1 | 0 | 0.0023 |
| dicarbonyl/L-xylulose reductase | 4.5 | 0 | 0.0037 |
| collagen, type XV, alpha 1 | 3.9 | 0 | 0.009 |
| coronin, actin binding protein, 1B | 3.9 | 0 | 0.0058 |
| mannose receptor, C type 1 /// mannose receptor, C type 1-like 1 | 3.9 | 0 | 0.0076 |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 | 3.9 | 0 | 0.0081 |
| Na+/H+ exchanger domain containing 1 | 3.9 | 0 | 0.0026 |
| sema domain, transmembrane domain (TM) (semaphorin) 6B | 3.9 | 0 | 0.0049 |
| coronin, actin binding protein, 1B | 3.8 | 0 | 0.0076 |
| carboxypeptidase M | 3.8 | 0 | 0.0073 |
| protocadherin 17 | 3.7 | 0 | 0.0086 |
| GINS complex subunit 4 (Sld5 homolog) | 3.7 | 0 | 0.0051 |
| guanine nucleotide binding protein-like 3 (nucleolar)-like | 3.6 | 0 | 0.0058 |
| leucine rich repeat containing 33 | 3.6 | 0 | 0.0087 |
| protein kinase C and casein kinase substrate in neurons 2 | 3.6 | 0 | 0.0048 |
| thimet oligopeptidase 1 | 3.6 | 0 | 0.0088 |
| major histocompatibility complex, class II, DR beta 1 | 3.6 | 0 | 0.0086 |
| hypothetical LOC654433 | 3.6 | 0 | 0.0167 |
| zinc finger protein 688 | 3.6 | 0 | 0.0102 |
| histone cluster 1, H1b | 3.6 | 0 | 0.0089 |
| chromosome 1 open reading frame 93 | 3.5 | 0 | 0.0086 |
| SH3KBP1 binding protein 1 | 3.5 | 0 | 0.0088 |
| cleft lip and palate associated transmembrane protein 1 | 3.5 | 0 | 0.0077 |
| Rho GTPase activating protein 29 | 3.5 | 0 | 0.0094 |
| major histocompatibility complex, class II, DR beta 1 | 3.4 | 0 | 0.0057 |
| MAD1 mitotic arrest deficient-like 1 (yeast) | 3.4 | 0 | 0.0083 |
| exosome component 4 | 3.4 | 0 | 0.0104 |
| YKT6 v-SNARE homolog (S. cerevisiae) | 3.4 | 0 | 0.0087 |
| zinc finger CCCH-type containing 7B | 3.3 | 0 | 0.0087 |
| ATPase type 13A2 | 3.2 | 0 | 0.0117 |
| RAB GTPase binding effector protein 2 | 3.3 | 0 | 0.0085 |
| hepatoma-derived growth factor-related protein 2 | 3.3 | 0 | 0.01 |
| phosphoglucomutase 5 | 3.2 | 0 | 0.0153 |
| matrix metallopeptidase 10 (stromelysin 2) | 3.2 | 0 | 0.009 |
| mitogen-activated protein kinase kinase 2 | 3.1 | 0 | 0.009 |
| endoglin | 3.1 | 1.0 × 10−4 | 0.0241 |
| R-spondin 3 homolog (Xenopus laevis) | 3.1 | 1.0 × 10−4 | 0.0234 |
| F-box and WD repeat domain containing 5 | 3.1 | 1.0 × 10−4 | 0.0285 |
| carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | 3.1 | 0 | 0.009 |
| lipase, endothelial | 3.1 | 0 | 0.0128 |
| Similar to p40 | 3.1 | 0 | 0.0087 |
| spectrin repeat containing, nuclear envelope 2 | 3.1 | 1.0 × 10−4 | 0.0237 |
| kinesin light chain 1 | 3.1 | 0 | 0.0103 |
| homer homolog 3 (Drosophila) | 3.1 | 0 | 0.0102 |
| hypothetical protein LOC286434 | 3.1 | 1.0 × 10−4 | 0.0208 |
| exosome component 4 | 3.1 | 0 | 0.0146 |
| glucocorticoid receptor DNA binding factor 1 | 3.1 | 0 | 0.0085 |
| WD repeat domain 4 | 3.0 | 0 | 0.0091 |
| cytochrome b5 reductase 3 | 3.0 | 0 | 0.0085 |
| chromosome 21 open reading frame 45 | 3.0 | 0 | 0.0165 |
| cytochrome P450, family 1, subfamily B, polypeptide 1 | 3.0 | 0 | 0.0182 |
| peter pan homolog (Drosophila) | 3.0 | 0 | 0.0101 |
| cytochrome P450, family 1, subfamily B, polypeptide 1 | 3.0 | 0 | 0.0167 |
| cyclin D1 | 3.0 | 1.0 × 10−4 | 0.0319 |
| nasal embryonic LHRH factor | 3.0 | 0 | 0.0084 |
| zinc finger protein 160 | 3.0 | 0 | 0.0153 |
| 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 (soluble) | 3.0 | 0 | 0.0102 |
| dipeptidyl-peptidase 9 | 3.0 | 0 | 0.01 |
| small optic lobes homolog (Drosophila) | 3.0 | 0 | 0.0179 |
| splicing factor, arginine/serine-rich 8 | 2.9 | 1.0 × 10−4 | 0.0211 |
| Wolf-Hirschhorn syndrome candidate 1 | 2.9 | 2..0 × 10−4 | 0.0397 |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 | 2.9 | 0 | 0.0135 |
| dedicator of cytokinesis 6 | 2.9 | 0 | 0.015 |
| lysosomal multispanning membrane protein 5 | 2.9 | 0 | 0.0099 |
| FERM domain containing 3 | 2.9 | 1.0 × 10−4 | 0.0191 |
| ankyrin repeat domain 1 (cardiac muscle) | 2.9 | 1.0 × 10−4 | 0.0231 |
| ring finger protein 125 | 2.9 | 0 | 0.0186 |
| Hypothetical protein LOC203274 | 2.9 | 0 | 0.0175 |
| sorbitol dehydrogenase | 2.9 | 0 | 0.0119 |
| ATPase family, AAA domain containing 3A | 2.9 | 0 | 0.01 |
| sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 | 2.9 | 0 | 0.0103 |
| Hypothetical protein LOC100129502 | 2.9 | 1.0 × 10−4 | 0.0209 |
| Difference in Gene Expression (Fold Change) | ||||
|---|---|---|---|---|
| Gene Transcript | Affy ID | Fold Change in Gene Expression Relative to Human Macular Inner CECs | ||
| Microarray (SD) | RT-PCR (SD) | (p Value) | ||
| Keratin 19 | 201650_at | 7.3 (1.8) | 10.5 (2.0) | (0.15) |
| Brain derived Neurotrophic factor | 206382_s_at | 8.9 (0.8) | 7.2 (0.6) | (0.34) |
| CXCL 12 | 203666_at | −2.4 (−1.0) | −3.6 (−1.0) | (0.21) |
| MAPK 11 | 206040_s_at | −2.8 (−0.9) | −3.3 (−0.64) | (0.52) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Browning, A.C.; Halligan, E.P.; Stewart, E.A.; Swan, D.C.; Cockell, S.J.; Amoaku, W.M. Regional Differences in Gene Expression of Proliferating Human Choroidal Endothelial Cells. Int. J. Transl. Med. 2021, 1, 83-100. https://doi.org/10.3390/ijtm1020007
Browning AC, Halligan EP, Stewart EA, Swan DC, Cockell SJ, Amoaku WM. Regional Differences in Gene Expression of Proliferating Human Choroidal Endothelial Cells. International Journal of Translational Medicine. 2021; 1(2):83-100. https://doi.org/10.3390/ijtm1020007
Chicago/Turabian StyleBrowning, Andrew C., Eugene P. Halligan, Elizabeth A. Stewart, Daniel C. Swan, Simon J. Cockell, and Winfried M. Amoaku. 2021. "Regional Differences in Gene Expression of Proliferating Human Choroidal Endothelial Cells" International Journal of Translational Medicine 1, no. 2: 83-100. https://doi.org/10.3390/ijtm1020007
APA StyleBrowning, A. C., Halligan, E. P., Stewart, E. A., Swan, D. C., Cockell, S. J., & Amoaku, W. M. (2021). Regional Differences in Gene Expression of Proliferating Human Choroidal Endothelial Cells. International Journal of Translational Medicine, 1(2), 83-100. https://doi.org/10.3390/ijtm1020007







