Peroxidase-like Activity of G-Quadruplex/Hemin Complexes for Colorimetric Nucleic Acid Analysis: Loop and Flanking Sequences Affect Signal Intensity
Abstract
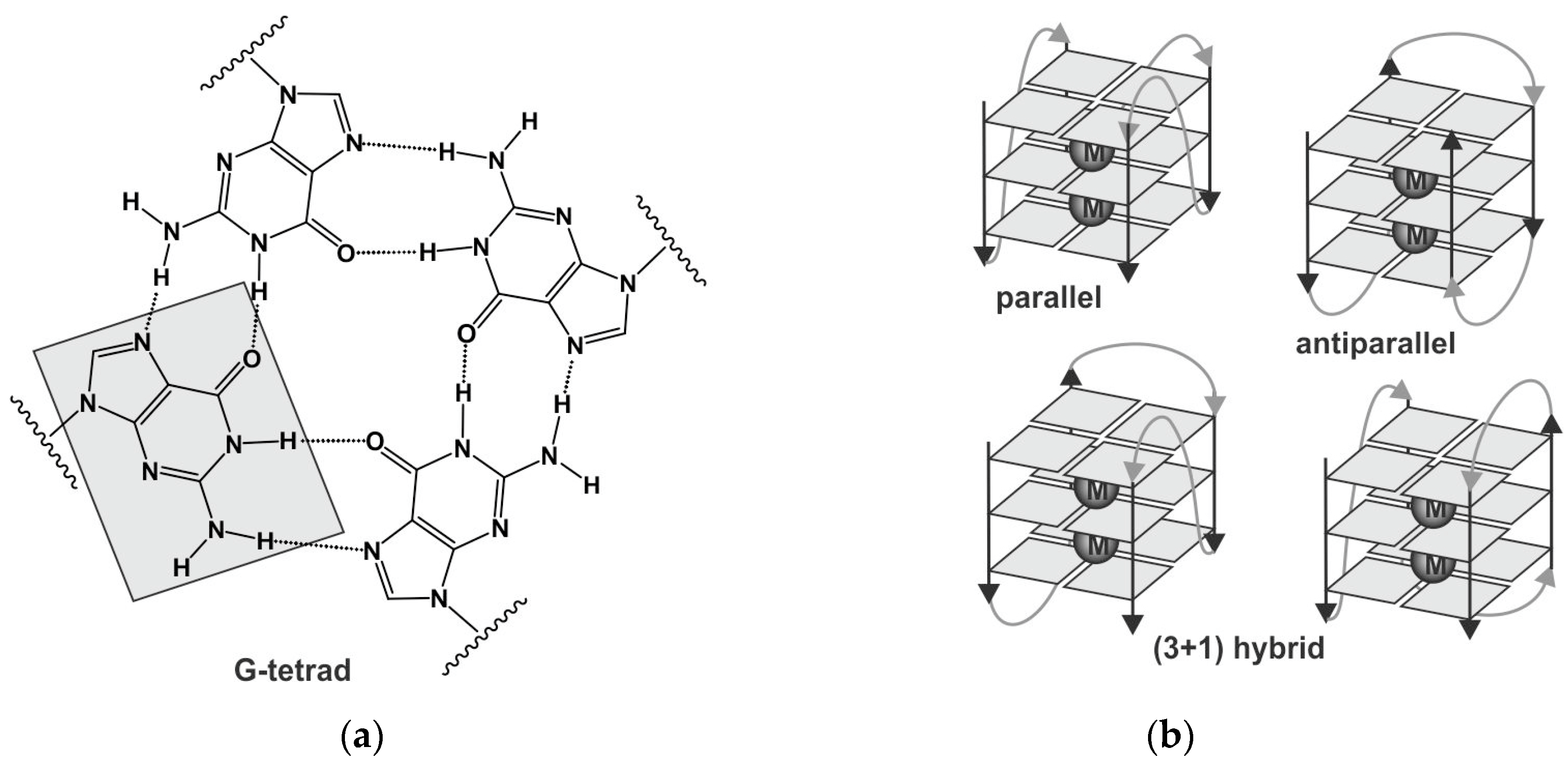
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Circular Dichroism Spectra
2.3. Colorimetric Assay with Unimolecular G-Quadruplex Sequences
2.4. Colorimetric Assay with Split Peroxidase Deoxyribozyme (sPDz) Probes
2.5. Colorimetric Assay with a Cascade System Based on Split Deoxyribozyme (sDz) Probe Releasing G4 Sequences with the PDz Activity
2.6. Statistical Analysis
3. Results
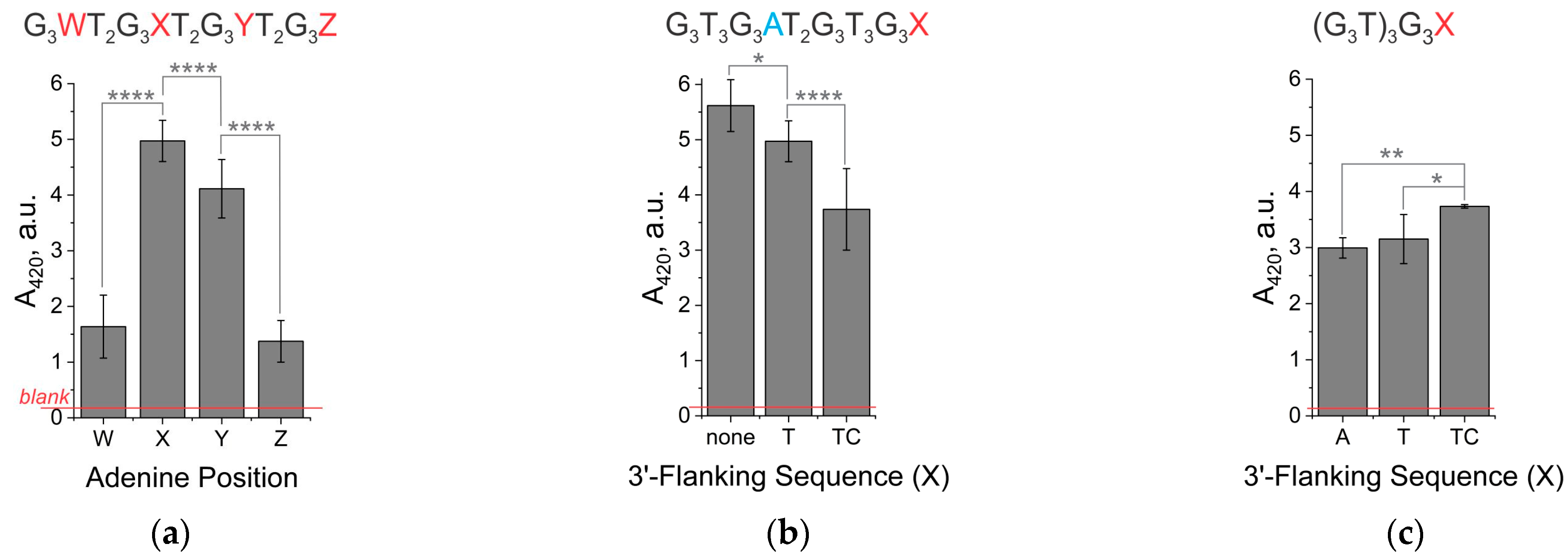
3.1. The Signal Intensity of the Peroxidase-like Activity of Hemin in Complex with Unimolecular G4 Depends on the Presence of Adenine in the Second Loop or 3′-Terminal Nucleotides
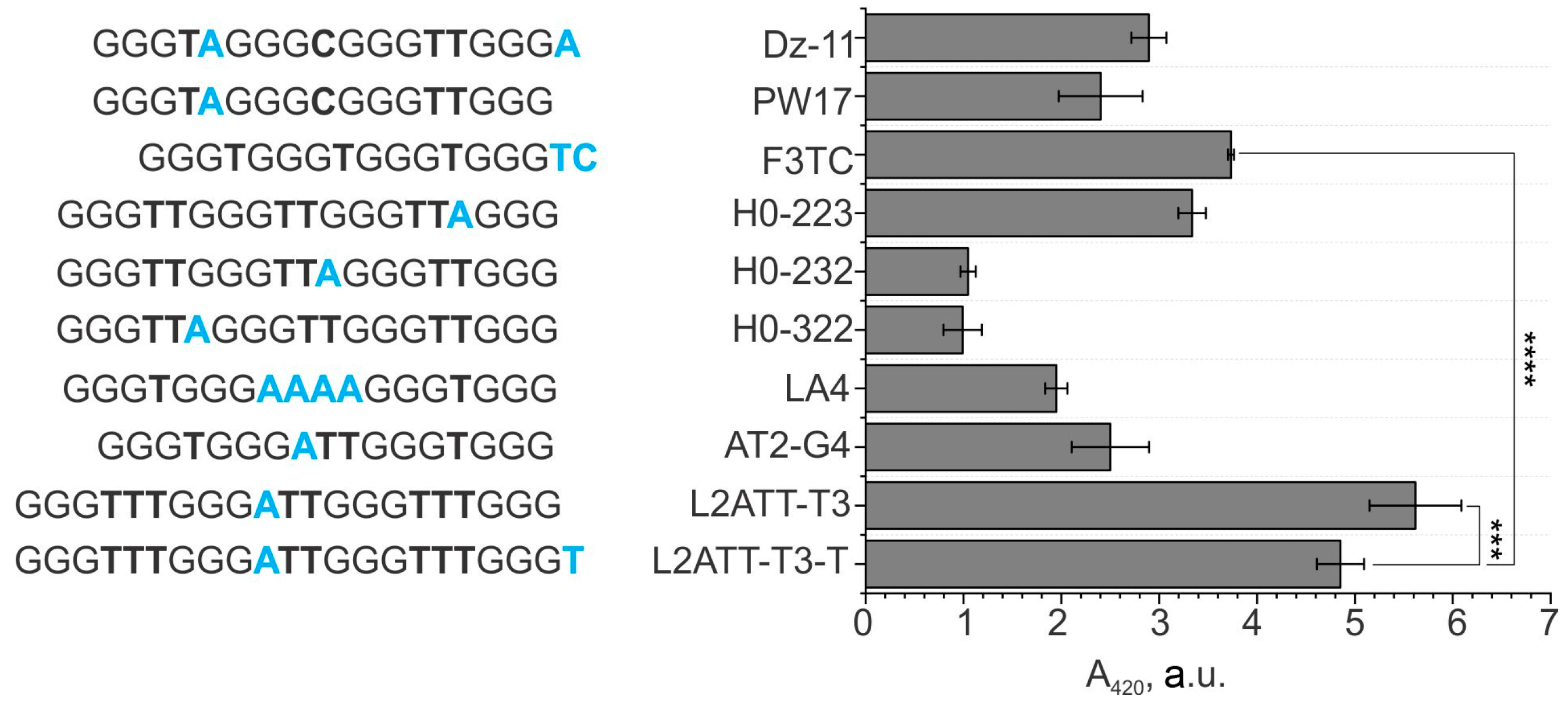
3.2. A Comparison of the Performance of the sPDz Probes Utilizing Different G4 Sequences as a Signal Transducer
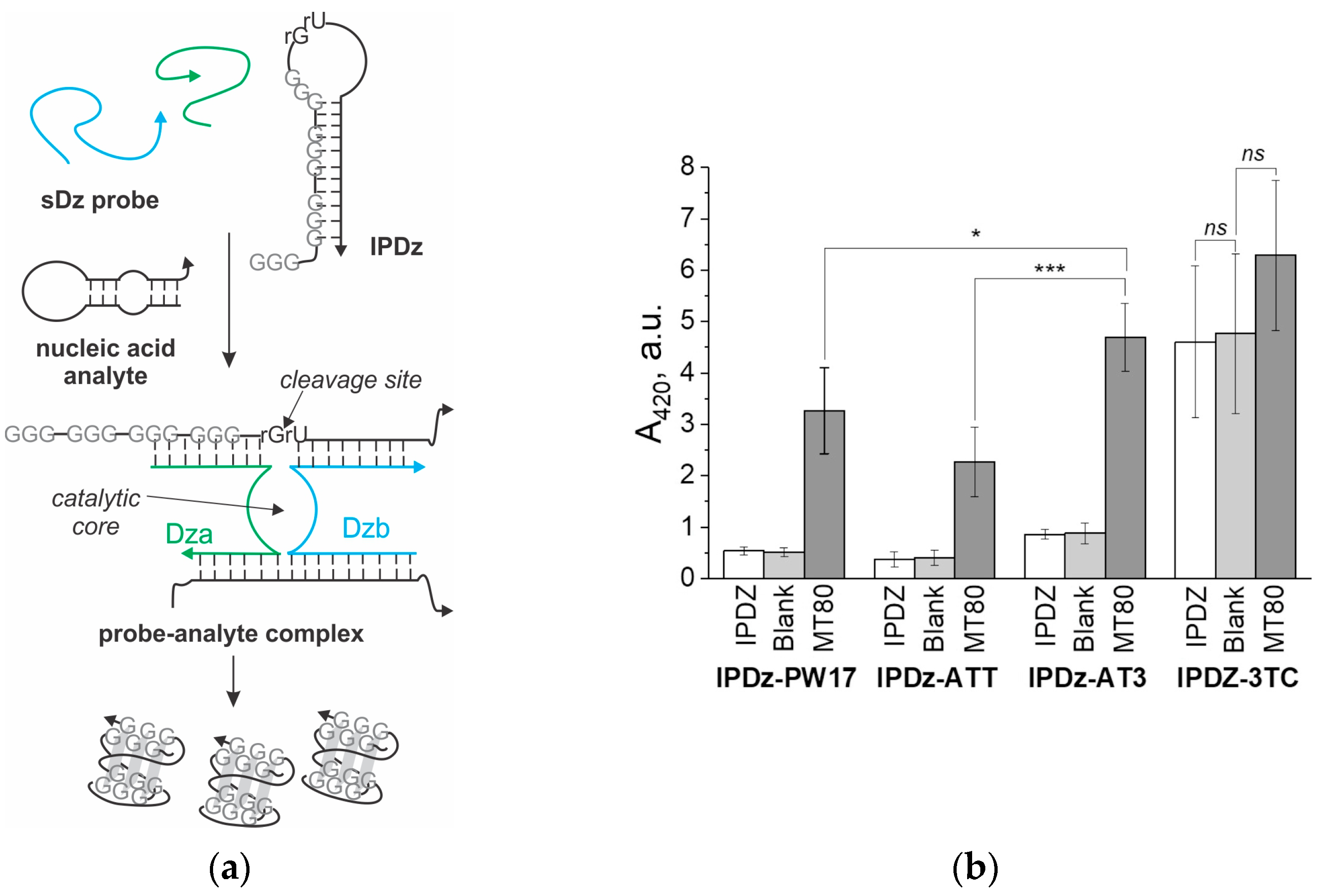
3.3. A Comparison of the Performance of the Cascade sDz/PDz Probes Utilizing Different G4 Sequences as a Signal Reporter
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef]
- Bhattacharyya, D.; Arachchilage, G.M.; Basu, S. Metal cations in G-quadruplex folding and stability. Front. Chem. 2016, 4, 2296–2646. [Google Scholar] [CrossRef] [PubMed]
- Keniry, M. Quadruplex structures in nucleic acids. Biopolymers 2001, 56, 123–146. [Google Scholar] [CrossRef]
- Tippana, R.; Xiao, W.; Myong, S. G-quadruplex conformation and dynamics are determined by loop length and sequence. Nucleic Acids Res. 2014, 42, 8106–8114. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The structure and function of DNA G-quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Roxo, C.; Kotkowiak, W.; Pasternak, A. G-Quadruplex-forming aptamers—Characteristics, applications, and perspectives. Molecules 2019, 24, 3781. [Google Scholar] [CrossRef] [PubMed]
- Travascio, P.; Li, Y.; Sen, D. DNA-enhanced peroxidase activity of a DNA aptamer-hemin complex. Chem. Biol. 1998, 5, 505–517. [Google Scholar] [CrossRef]
- Cheng, X.; Liu, X.; Bing, T.; Cao, Z.; Shangguan, D. General peroxidase activity of G-quadruplex-hemin complexes and its application in ligand screening. Biochemistry 2009, 48, 7817–7823. [Google Scholar] [CrossRef] [PubMed]
- Poon, L.C.-H.; Methot, S.P.; Morabi-Pazooki, W.; Bennet, A.J.; Sen, D. Guanine-rich RNAs and DNAs that bind heme robustly catalyze oxygen transfer reactions. J. Am. Chem. Soc. 2011, 133, 1877–1884. [Google Scholar] [CrossRef] [PubMed]
- Sen, D.; Poon, L.C. RNA and DNA complexes with hemin [Fe (III) heme] are efficient peroxidases and peroxygenases: How do they do it and what does it mean? Crit. Rev. Biochem. Mol. Biol. 2011, 46, 478–492. [Google Scholar] [CrossRef]
- Iwaniuk, E.E.; Adebayo, T.; Coleman, S.; Villaros, C.G.; Nesterova, I.V. Activatable G-quadruplex based catalases for signal transduction in biosensing. Nucleic Acids Res. 2023, 51, 1600–1607. [Google Scholar] [CrossRef] [PubMed]
- Kosman, J.; Juskowiak, B. Peroxidase-mimicking DNAzymes for biosensing applications: A review. Anal. Chim. Acta 2011, 707, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Guo, Z.; Wang, J.; Wang, E. G-quadruplex as signal transducer for biorecognition events. Curr. Pharm. Des. 2012, 18, 2076–2095. [Google Scholar] [CrossRef]
- Neo, J.L.; Kamaladasan, K.; Uttamchandani, M. G-quadruplex based probes for visual detection and sensing. Curr. Pharm. Des. 2012, 18, 2048–2057. [Google Scholar] [CrossRef] [PubMed]
- Ruttkay-Nedecky, B.; Kudr, J.; Nejdl, L.; Maskova, D.; Kizek, R.; Adam, V. G-quadruplexes as sensing probes. Molecules 2013, 18, 14760–14779. [Google Scholar] [CrossRef]
- Ren, J.; Wang, T.; Wang, E.; Wang, J. Versatile G-quadruplex-mediated strategies in label-free biosensors and logic systems. Analyst 2015, 140, 2556–2572. [Google Scholar] [CrossRef]
- Lv, M.; Guo, Y.; Ren, J.; Wang, E. Exploration of intramolecular split G-quadruplex and its analytical applications. Nucleic Acids Res. 2019, 47, 9502–9510. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhou, Y.; Liu, J. G-quadruplex DNA for construction of biosensors. Trends Anal. Chem. 2020, 132, 116060. [Google Scholar] [CrossRef]
- Stefan, L.; Denat, F.; Monchaud, D. Insights into how nucleotide supplements enhance the peroxidase-mimicking DNAzyme activity of the G-quadruplex/hemin system. Nucleic Acids Res. 2012, 40, 8759–8772. [Google Scholar] [CrossRef]
- Travascio, P.; Witting, P.K.; Mauk, A.G.; Sen, D.J. The peroxidase activity of a hemin−DNA oligonucleotide complex: Free radical damage to specific guanine bases of the DNA. J. Am. Chem. Soc. 2001, 123, 1337–1348. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J.; Fang, C.L.; Mei, H.C.; Chang, T.J.; Cao, Z.H.; Shangguan, D.H. Characterization of G-quadruplex/hemin peroxidase: Substrate specificity and inactivation kinetics. Chem. Eur. J. 2011, 17, 14475–14484. [Google Scholar] [CrossRef]
- Li, W.; Li, Y.; Liu, Z.; Lin, B.; Yi, H.; Xu, F.; Nie, Z.; Yao, S. Insight into G-quadruplex-hemin DNAzyme/RNAzyme: Adjacent adenine as the intramolecular species for remarkable enhancement of enzymatic activity. Nucleic Acids Res. 2016, 44, 7373–7384. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, Y.; Cheng, M.; Guo, Y.; Šponer, J.; Monchaud, D.; Mergny, J.-L.; Ju, H.; Zhou, J. How proximal nucleobases regulate the catalytic activity of G-quadruplex/hemin DNAzymes. ACS Catal. 2018, 8, 11352–11361. [Google Scholar] [CrossRef]
- Saito, K.; Tai, H.; Hemmi, H.; Kobayashi, N.; Yamamoto, Y. Interaction between the heme and a G-quartet in a heme-DNA complex. Inorg. Chem. 2012, 51, 8168–8176. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Araki, H.; Shinomiya, R.; Haysaka, K.; Nakayama, Y.; Ochi, K.; Shibata, T.; Momotake, A.; Ohyama, T.; Hagihara, M.; et al. Structures and catalytic activities of complexes between heme and all parallel-stranded monomeric G-quadruplex DNAs. Biochemistry 2018, 57, 5938–5948. [Google Scholar] [CrossRef]
- Stadlbauer, P.; Islam, B.; Otyepka, M.; Chen, J.; Monchaud, D.; Zhou, J.; Mergny, J.-L.; Sponer, J. Insights into G-quadruplex–hemin dynamics using atomistic simulations: Implications for reactivity and folding. J. Chem. Theory Comput. 2021, 17, 1883–1899. [Google Scholar] [CrossRef]
- Kong, D.-M.; Yang, W.; Wu, J.; Li, C.-X.; Shen, H.-X. Structure–function study of peroxidase-like G-quadruplex-hemin complexes. Analyst 2010, 135, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, S.; Wang, J.; Sintim, H.O. DNA-based peroxidation catalyst—What is the exact role of topology on catalysis and is there a special binding site for catalysis? Chem. Eur. J. 2011, 17, 5691–5698. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, S.; Sintim, H.O. Investigating the interactions between cations, peroxidation substrates and G-quadruplex topology in DNAzyme peroxidation reactions using statistical testing. Anal. Chim. Acta 2012, 747, 1–6. [Google Scholar] [CrossRef]
- Nasab, M.G.; Hasani, L.; Nejad, S.M.; Norouzi, D. Interaction of hemin with quadruplex DNA. J. Biol. Phys. 2017, 43, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.-M. Factors influencing the performance of G-quadruplex DNAzyme-based sensors. Methods 2013, 64, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Rachwal, P.A.; Brown, T.; Fox, K.R. Effect of G-tract length on the topology and stability of intramolecular DNA quadruplexes. Biochemistry 2007, 46, 3036–3044. [Google Scholar] [CrossRef]
- Hatzakis, E.; Okamoto, K.; Yang, D. Thermodynamic stability and folding kinetics of the major G-quadruplex and its loop isomers formed in the nuclease hypersensitive element in the human c-Myc promoter: Effect of loops and flanking segments on the stability of parallel-stranded intramolecular G-quadruplexes. Biochemistry 2010, 49, 9152–9160. [Google Scholar] [PubMed]
- Travascio, P.; Sen, D.; Bennet, A.J. DNA and RNA enzymes with peroxidase activity—An investigation into the mechanism of action. Can. J. Chem. 2006, 84, 613. [Google Scholar] [CrossRef]
- Chang, T.; Gong, H.; Ding, P.; Liu, X.; Li, W.; Bing, T.; Cao, Z.; Shangguan, D. Activity enhancement of G-quadruplex/hemin DNAzyme by flanking d(CCC). Chemistry 2016, 12, 4015. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Zhou, J.; Jia, G.; Ai, X.; Mergny, J.L.; Li, C. Relations between the loop transposition of DNA G-quadruplex and the catalytic function of DNAzyme. Biochim. Biophys. Acta 2017, 1861, 1913–1920. [Google Scholar] [CrossRef]
- Chen, J.; Guo, Y.; Zhou, J.; Ju, H. The effect of adenine repeats on G-quadruplex/hemin peroxidase mimicking DNAzyme activity. Chem. Eur. J. 2017, 23, 4210–4215. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Ding, P.; Yang, L.; Li, W.; Luo, Y.; Wang, J. Investigation and improvement of catalytic activity of G-quadruplex/hemin DNAzymes using designed terminal G-tetrads with deoxyadenosine caps. Chem. Sci. 2020, 11, 6896–6906. [Google Scholar] [CrossRef] [PubMed]
- Qui, D.; Mo, J.; Liu, Y.; Zhang, J.; Cheng, Y.; Zhang, X. Effect of distance from catalytic synergy group to iron porphyrin center on activity of G-quadruplex/hemin DNAzyme. Molecules 2020, 25, 3425. [Google Scholar] [CrossRef]
- Kong, D.-M.; Wu, J.; Wang, N.; Yang, W.; Shen, H.-X. Peroxidase activity–structure relationship of the intermolecular four-stranded G-quadruplex–hemin complexes and their application in Hg2+ ion detection. Talanta 2009, 80, 459–465. [Google Scholar] [CrossRef]
- Del Villar-Guerra, R.; Trent, J.O.; Chaires, J.B. G-quadruplex secondary structure obtained from circular dichroism spectroscopy. Angew. Chem. Int. Ed. Engl. 2018, 57, 7171–7175. [Google Scholar] [CrossRef] [PubMed]
- da Silva, M.W. Geometric formalism for DNA quadruplex folding. Chem. Eur. J. 2007, 13, 9738–9745. [Google Scholar] [CrossRef]
- Nakayama, S.; Sintim, H.O. Biomolecule detection with peroxidase-mimicking DNAzymes; expanding detection modality with fluorogenic compounds. Mol. BioSyst. 2009, 6, 95–97. [Google Scholar] [CrossRef] [PubMed]
- Kolpashchikov, D.M. Split DNA enzyme for visual single nucleotide polymorphism typing. J. Am. Chem. Soc. 2008, 130, 2934–2935. [Google Scholar] [CrossRef]
- Deng, M.; Zhang, D.; Zhou, Y.; Zhou, X. Highly effective colorimetric and visual detection of nucleic acids using an asymmetrically split peroxidase DNAzyme. J. Am. Chem. Soc. 2008, 130, 13095–13102. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Feng, S.; Luo, F.; Wang, S.; Sun, X.; Zhou, X.; Zhang, X.-L. Visual detection of rpoB mutations in rifampin-resistant Mycobacterium tuberculosis strains by use of an asymmetrically split peroxidase DNAzyme. J. Clin. Microbiol. 2012, 50, 3443–3450. [Google Scholar] [CrossRef] [PubMed]
- Darius, A.K.L.; Ling, N.J.; Mahesh, U. Visual detection of DNA from salmonella and mycobacterium using split DNAzymes. Mol. BioSyst. 2010, 6, 792–794. [Google Scholar] [CrossRef] [PubMed]
- Connelly, R.P.; Morozkin, E.S.; Gerasimova, Y.V. Alphanumerical visual display made of DNA logic gates for drug susceptibility testing of pathogens. ChemBioChem 2018, 19, 203–206. [Google Scholar] [CrossRef]
- Connelly, R.P.; Verduzco, S.; Farnell, S.; Yishay, T.; Gerasimova, Y.V. Toward a rational approach to design split G-quadruplex probes. ACS Chem. Biol. 2019, 14, 2701–2712. [Google Scholar] [CrossRef]
- Batule, B.S.; Kim, S.U.; Mun, H.; Choi, C.; Shim, W.-B.; Kim, M.-G. Colorimetric detection of norovirus in oyster samples through DNAzyme as a signaling probe. J. Agric. Food Chem. 2018, 66, 3003–3008. [Google Scholar] [CrossRef]
- Lu, X.; Shi, X.; Wu, G.; Wu, T.; Qin, R.; Wang, Y. Visual detection and differentiation of Classic Swine Fever Virus strains using nucleic acid sequence based amplification (NASBA) and G-quadruplex DNAzyme assay. Sci. Rep. 2017, 7, 44211. [Google Scholar] [CrossRef]
- Gerasimova, Y.V.; Cornett, E.M.; Edwards, E.; Su, X.; Rohde, K.H.; Kolpashchikov, D.M. Deoxyribozyme cascade for visual detection of bacterial RNA. Chembiochem 2013, 14, 2087–2090. [Google Scholar] [CrossRef]
- Reed, A.J.; Connelly, R.P.; Williams, A.; Tran, M.; Shim, B.S.; Choe, H.; Gerasimova, Y.V. Label-free pathogen detection by a deoxyribozyme cascade with visual signal readout. Sens. Actuators B Chem. 2019, 282, 945–951. [Google Scholar] [CrossRef] [PubMed]
- Wood, H.N.; Sidders, A.E.; Brumsey, L.E.; Morozkin, E.S.; Gerasimova, Y.V.; Rohde, K.H. Species typing of nontuberculous Mycobacteria by use of deoxyribozyme sensors. Clin. Chem. 2019, 65, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Dhar, B.C.; Reed, A.J.; Mitra, S.; Rodrigues Sanchez, P.; Nedorezova, D.D.; Connelly, R.P.; Rohde, K.H.; Gerasimova, Y.V. Cascade of deoxyribozymes for the colorimetric analysis of drug resistance in Mycobacterium tuberculosis. Biosens. Bioelecton. 2020, 165, 112385. [Google Scholar] [CrossRef] [PubMed]
- Zadeh, J.N.; Steenberg, C.D.; Bois, J.S.; Wolfe, B.R.; Pierce, M.B.; Khan, A.R.; Dirks, R.M.; Pierce, N.A. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2011, 32, 170–173. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Connelly, R.P.; Fonseca, V.; Gerasimova, Y.V. Peroxidase-like Activity of G-Quadruplex/Hemin Complexes for Colorimetric Nucleic Acid Analysis: Loop and Flanking Sequences Affect Signal Intensity. DNA 2025, 5, 12. https://doi.org/10.3390/dna5010012
Connelly RP, Fonseca V, Gerasimova YV. Peroxidase-like Activity of G-Quadruplex/Hemin Complexes for Colorimetric Nucleic Acid Analysis: Loop and Flanking Sequences Affect Signal Intensity. DNA. 2025; 5(1):12. https://doi.org/10.3390/dna5010012
Chicago/Turabian StyleConnelly, Ryan P., Valentina Fonseca, and Yulia V. Gerasimova. 2025. "Peroxidase-like Activity of G-Quadruplex/Hemin Complexes for Colorimetric Nucleic Acid Analysis: Loop and Flanking Sequences Affect Signal Intensity" DNA 5, no. 1: 12. https://doi.org/10.3390/dna5010012
APA StyleConnelly, R. P., Fonseca, V., & Gerasimova, Y. V. (2025). Peroxidase-like Activity of G-Quadruplex/Hemin Complexes for Colorimetric Nucleic Acid Analysis: Loop and Flanking Sequences Affect Signal Intensity. DNA, 5(1), 12. https://doi.org/10.3390/dna5010012






