Inka Child Mummy Found in Cerro Aconcagua (Argentina) Traced Back to Populations of the Northern Peruvian Coast through Y-Chromosome Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement and Sampling
2.2. Genotyping the Y-Chromosome SNPs and STRs
2.3. Y-STR Data Analysis
2.4. Y-SNP Data Analysis
3. Results
3.1. Y-STR Results
3.2. Y-SNP Results
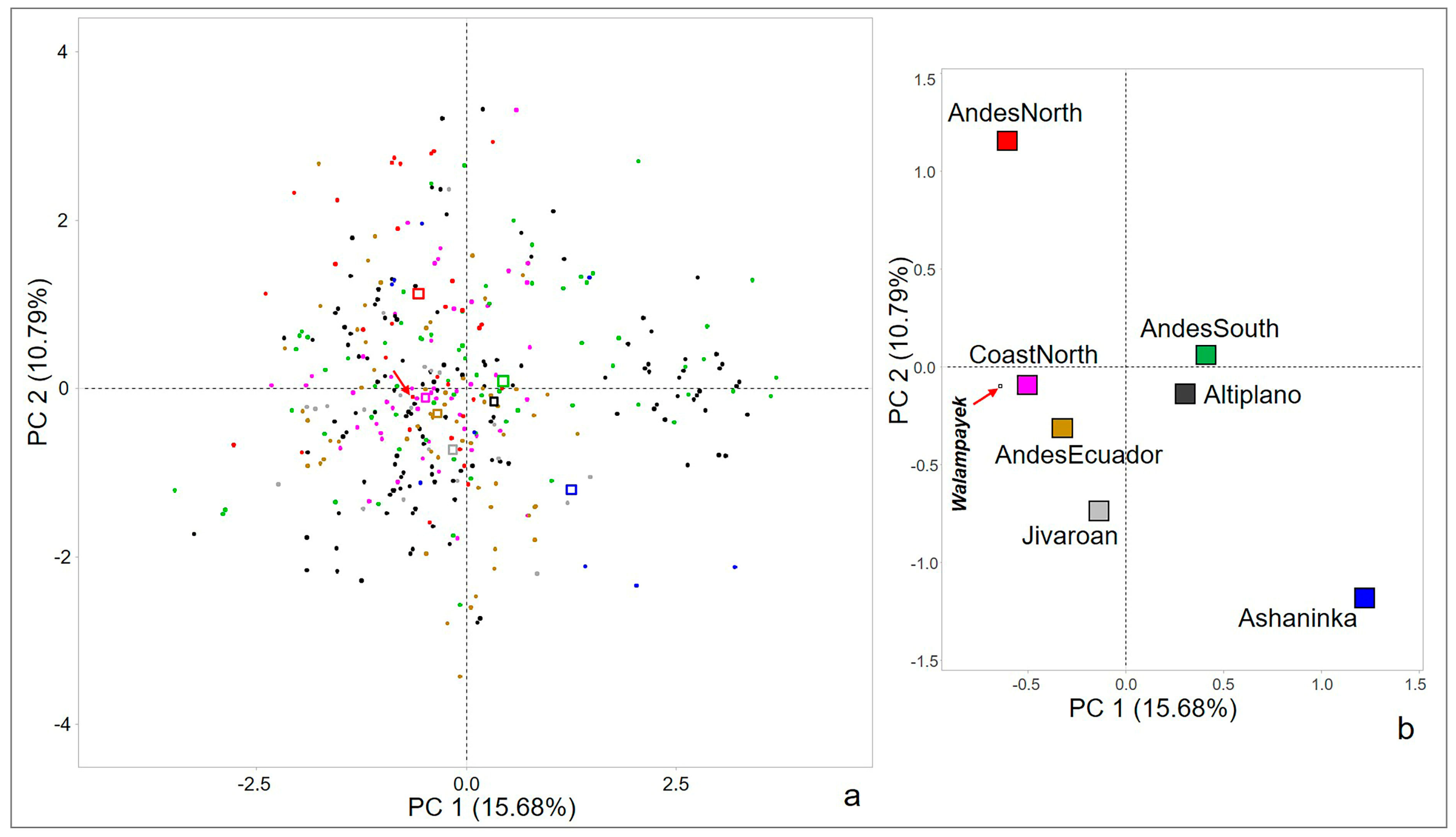
3.3. Principal Component Analysis Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Reinhard, J.; Constanza-Ceruti, M. Inca Rituals and Sacred Mountains. A Study of the World’s Highest Archaeological Sites; Cotsen Institute of Archaeology, University of California: Los Angeles, CA, USA, 2010. [Google Scholar]
- Schobinger, J. Los santuarios de altura incaicos y el Aconcagua: Aspectos generales e interpretativos. Rel. Soc. Argent. Antropol. 1999, 24, 7–27. Available online: http://sedici.unlp.edu.ar/handle/10915/20077 (accessed on 22 November 2022).
- Moreno-Mayar, J.V.; Vinner, L.; de Barros Damgaard, P.; de la Fuente, C.; Chan, J.; Spence, J.P.; Allentoft, M.E.; Vimala, T.; Racimo, F.; Pinotti, T.; et al. Early human dispersals within the Americas. Science 2018, 362, eaav2621. [Google Scholar] [CrossRef] [PubMed]
- Roberto-Bárcena, J. Pigmentos en el ritual funerario de la momia del Co. Aconcagua (Provincia de Mendoza, República Argentina). Xama 1989, 2, 61–116. [Google Scholar]
- Roberto-Bárcena, J. Estudios Sobre el Santuario Incaico del Cerro Aconcagua. In El Santuario Incaico del Cerro Aconcagua; EDIUNC (Juan Schobinger, compilador); Universidad Nacional del Cuyo: Mendoza, Argentina, 2001. [Google Scholar]
- Pinotti, T.; Bergström, A.; Geppert, M.; Bawn, M.; Ohasi, D.; Shi, W.; Lacerda, D.R.; Solli, A.; Norstedt, J.; Reed, K.; et al. Y chromosome sequences reveal a short Beringian Standstill, rapid expansion, and early population structure of Native American founders. Curr. Biol. 2019, 29, 149–157. [Google Scholar] [CrossRef]
- Gómez-Carballa, A.; Catelli, L.; Pardo-Seco, J.; Martinón-Torres, F.; Roewer, L.; Vullo, C.; Salas, A. The complete mitogenome of a 500-year-old Inca child mummy. Sci. Rep. 2015, 5, 16462. [Google Scholar] [CrossRef]
- Salas, A.; Catelli, L.; Pardo-Seco, J.; Gómez-Carballa, A.; Martinón-Torres, F.; Roberto-Barcena, J.; Vullo, C. Y-chromosome Peruvian origin of the 500-year-old Inca child mummy sacrificed in Cerro Aconcagua (Argentina). Sci. Bull. 2018, 63, 1457–1459. [Google Scholar] [CrossRef]
- Tamm, E.; Kivisild, T.; Reidla, M.; Metspalu, M.; Smith, D.G.; Mulligan, C.J.; Bravi, C.M.; Rickards, O.; Martinez-Labarga, C.; Khusnutdinova, E.K.; et al. Beringian standstill and spread of Native American founders. PLoS ONE 2007, 2, e829. [Google Scholar] [CrossRef]
- Derenko, M.; Malyarchuk, B.; Grzybowski, T.; Denisova, G.; Rogalla, U.; Perkova, M.; Dambueva, I.; Zakharov, I. Origin and post-glacial dispersal of mitochondrial DNA haplogroups C and D in northern Asia. PLoS ONE 2010, 5, e15214. [Google Scholar] [CrossRef]
- Nakatsuka, N.; Lazaridis, I.; Barbieri, C.; Skoglund, P.; Rohland, N.; Mallick, S.; Harkins-Kinkaid, K.; Ferry, M.; Harney, É.; Michel, M.; et al. A paleogenomic reconstruction of the deep population history of the Andes. Cell 2020, 181, 1131–1145.e21. [Google Scholar] [CrossRef]
- Bongers, J.L.; Nakatsuka, N.; O’Shea, C.; Harper, T.K.; Tantaleán, H.; Stanish, C.; Fehren-Schmitz, L. Integration of ancient DNA with transdisciplinary dataset finds strong support for Inca resettlement in the south Peruvian coast. Proc. Natl. Acad. Sci. USA 2020, 117, 18359–18368. [Google Scholar] [CrossRef]
- Uceda, S.; Mujica, E. Hacia el Final del Milenio. Actas del Segundo Coloquio Sobre la Cultura Moche, 2nd ed.; Trujillo 1 al 7 de Agosto de 1999; Universidad Nacional de Trujillo y PUCP: Trujillo, Perú, 2003. [Google Scholar]
- Barbieri, C.; Barquera, R.; Arias, L.; Sandoval, J.R.; Acosta, O.; Zurita, C.; Aguilar-Campos, A.; Tito-Álvarez, A.M.; Serrano-Osuna, R.; Gray, R.D.; et al. The current genomic landscape of western South America: Andes, Amazonia and Pacific Coast. Mol. Biol. Evol. 2019, 36, 2698–2713. [Google Scholar] [CrossRef] [PubMed]
- Sandoval, J.R.; Lacerda, D.R.; Jota, M.S.A.; Salazar-Granara, A.; Vieira, P.P.R.; Acosta, O.; Cuellar, C.; Revollo, S.; Fujita, R.; Santos, F.R. The Genographic Project Consortium, The genetic history of indigenous populations of the Peruvian and Bolivian Altiplano: The legacy of the Uros. PLoS ONE 2013, 8, e73006. [Google Scholar] [CrossRef] [PubMed]
- Sandoval, J.R.; Lacerda, D.R.; Acosta, O.; Jota, M.S.; Robles-Ruiz, P.; Salazar-Granara, A.; Vieira, P.P.R.; Paz-y-Miño, C.; Fujita, R.; Santos, F.R. The Genographic Project Consortium, The genetic history of Peruvian Quechua-Lamistas and Chankas: Uniparental DNA patterns among autochthonous Amazonian and Andean populations. Ann. Hum. Genet. 2016, 80, 88–101. [Google Scholar] [CrossRef] [PubMed]
- Sandoval, J.R.; Lacerda, D.R.; Jota, M.S.; Elward, R.; Acosta, O.; Pinedo, D.; Danos, P.; Cuellar, C.; Revollo, S.; Santos, F.R.; et al. Genetic ancestry of families of putative Inka descent. Mol. Genet. Genom. 2018, 293, 873–881. [Google Scholar] [CrossRef]
- Sandoval, J.R.; Lacerda, D.R.; Jota, M.S.; Robles-Ruiz, P.; Danos, P.; Paz-y-Miño, C.; Wells, S.; Santos, F.R.; Fujita, R. Tracing the genetic history of the ‘Cañaris’ from Ecuador and Peru using uniparental DNA markers. BMC Genom. 2020, 21 (Suppl. S7), 413. [Google Scholar] [CrossRef]
- Sandoval, J.R.; Revollo, S.; Cuellar, C.; Lacerda, D.R.; Jota, M.S.; Fujita, R.; Santos, F.R. Genetic portrait of the Amazonian communities of Peru and Bolivia: The legacy of the Takanan-speaking people. Ann. Hum. Genet. 2023, 87, 210–221. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-Joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Polzin, T.; Daneshmand, S.V. On Steiner trees and minimum spanning trees in hypergraphs. Oper. Res. Lett. 2003, 31, 12–20. [Google Scholar] [CrossRef]
- Forster, P.; Harding, R.; Torroni, A.; Bandelt, H.J. Origin and evolution of Native American mtDNA variation: A reappraisal. Am. J. Hum. Genet. 1996, 59, 935–945. [Google Scholar]
- Zhivotovsky, L.A.; Underhill, P.A.; Cinnioğlu, C.; Kayser, M.; Morar, B.; Kivisild, T.; Scozzari, R.; Cruciani, F.; Destro-Bisol, G.; Spedini, G.; et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population divergence time. Am. J. Hum. Genet. 2004, 74, 50–61. [Google Scholar] [CrossRef]
- Athey, W.T. Haplogroup prediction from Y-STR values using a Bayesian-allele frequency approach. J. Genet. Geneal. 2006, 2, 34–39. [Google Scholar][Green Version]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Poznik, G.D.; Henn, B.M.; Yee, M.C.; Sliwerska, E.; Euskirchen, G.M.; Lin, A.A.; Snyder, M.; Quintana-Murci, L.; Kidd, J.M.; Underhill, P.A.; et al. Sequencing Y chromosomes resolves discrepancy in time to common ancestor of males versus females. Science 2013, 341, 562–565. [Google Scholar] [CrossRef]
- Jota, M.S.; Lacerda, D.R.; Sandoval, J.R.; Vieira, P.P.R.; Ohasi, D.; Santos-Júnior, J.E.; Acosta, O.; Cuellar, C.; Revollo, S.; Paz-y-Miño, C.; et al. New native South American Y chromosome lineages. J. Hum. Genet. 2016, 61, 593–603. [Google Scholar] [CrossRef]
- Roewer, L.; Nothnagel, M.; Gusmão, L.; Gomes, V.; Gonzáles, M.; Corach, D.; Sala, A.; Alechine, E.; Palha, T.; Santos, N.; et al. Continent-wide decoupling of Y chromosomal genetic variation from language and geography in Native South Americans. PLoS Genet. 2013, 9, e1003460. [Google Scholar] [CrossRef]
- Wilson, A.S.; Brown, E.L.; Villa, C.; Lynnerup, N.; Healey, A.; Constanza Ceruti, M.; Reinhard, J.; Previgliano, C.H.; Arias Araoz, F.; Gonzales Diez, J.; et al. Archaeological, radiological, and biological evidence offer insight into Inca child sacrifice. Proc. Natl. Acad. Sci. USA 2013, 10, 13322–13327. [Google Scholar] [CrossRef] [PubMed]
- Socha, D.M.; Reinhard, J.; Chávez Perea, R. Inca human sacrifices on Misti Volcano (Peru). Lat. Am. Antiq. 2021, 32, 138–153. [Google Scholar] [CrossRef]
- Prieto, G.; Verano, J.W.; Goepfert, N.; Kennett, D.; Quilter, J.; LeBlanc, S.; Fehren-Schmitz, L.; Forst, J.; Lund, M.; Dement, B.; et al. A mass sacrifice of children and camelids at the Huanchaquito-Las Llamas site, Moche Valley, Peru. PLoS ONE 2019, 14, e0211691. [Google Scholar] [CrossRef]
- Llamas, B.; Fehren-Schmitz, L.; Valverde, G.; Soubrier, J.; Mallick, S.; Rohland, N.; Nordenfelt, S.; Valdiosera, C.; Richards, S.M.; Rohrlach, A.; et al. Ancient mitochondrial DNA provides high-resolution time scale of the peopling of the Americas. Sci. Adv. 2016, 2, e1501385. [Google Scholar] [CrossRef]
- Cabello de Balboa, M. Miscelánea Antártica (1586). In Una Historia del Perú Antiguo; Instituto de Etnología, Facultad de Letras, UNMSM: Lima, Perú, 1951. [Google Scholar]
- Espinoza-Soriano, W. El valle de Jayanca y el reino de los mochica, Siglos XV y XVI. Bull. Inst. Fr. Et. And. 1975, 3–4, 243–274. [Google Scholar] [CrossRef]
- Guevara, E.K.; Palo, J.U.; Guillén, S.; Sajantila, A. MtDNA and Y-chromosomal diversity in the Chachapoya, a population from the northeast Peruvian Andes-Amazon divide. Am. J. Hum. Biol. 2016, 28, 857–867. [Google Scholar] [CrossRef] [PubMed]
- Toscanini, U.; Gaviria, A.; Pardo-Seco, J.; Gómez-Carballa, A.; Moscoso, F.; Vela, M.; Cobos, S.; Lupero, A.; Zambrano, A.K.; Martinón-Torres, F.; et al. The geographic mosaic of Ecuadorian Y-chromosome ancestry. Forensic Sci. Int. Genet. 2018, 33, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Regan, J. Mito y Rito. Una comparación entre algunas imágenes mochicas y jibaras. Investig. Soc. 1999, 3, 27–46. [Google Scholar] [CrossRef]
- Salazar, L.; Burger, R.; Forst, J.; Barquera, R.; Nesbitt, J.; Calero, J.; Washburn, E.; Verano, J.; Zhu, K.; Sop, K.; et al. Insights into the genetic histories and lifeways of Machu Picchu´s occupants. Sci. Adv. 2023, 9, eadg3377. [Google Scholar] [CrossRef] [PubMed]

| Sample/Locality | 17 Y-STR DYS19-DYS385a-DYS385b-DYS389a-DYS389b-DYS390-DYS391-DYS392-DYS393-DYS437-DYS438-DYS439-DYS448-DYS456-DYS458-DYS635-YGATAH4 | Reference |
|---|---|---|
| Child mummy of Aconcagua (Mendoza, Argentina) | 14-16-16-14-17-25-10-14-13-14-11-12-20-15-16- 22-12 | Salas et al. [8] |
| MAL10 (Catacaos, Piura) | 14-15-16-14-17-25-10-14-13-14-11-12-20-15-16- 22-12 | This study |
| VICUS2, VICUS4, VICUS7, NARI6 (Chulucanas, La Arena, Catacaos, Narihuala; Piura) | 14-14-16-14-17-25-10-14-13-14-11-12-20-15-16- 22-12 | This study |
| WASI36 (Cajamarca) | 14-15-15-14-17-25-10-14-13-14-11-12-20-15-16- 22-12 | Sandoval et al. [18] |
| CHO-12 (Chotuna, Lambayeque) | 14-14-16-14-17-25-10-14-13-14-12-12-20-15-16- 22-12 | This study |
| Pomac02, Pomac05 (Lambayeque) | 14-14-16-14-17-25-10-14-13-14-11-12-20-15-15- 22-12 | This study |
| POMAC-12, Q_Caj15, CAX25 (Lambayeque, Cajamarca) | 14-14-15-14-17-25-10-14-13-14-11-12-20-15-16- 22-12 | This study, Sandoval et al. [15,18] |
| Q_Caj17 (Cajamarca) | 14-14-15-14-17-25-10-14-14-14-11-12-20-15-16- 22-12 | Sandoval et al. [15] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sandoval, J.R.; Fujita, R.; Jota, M.S.; Pinotti, T.; Santos, F.R. Inka Child Mummy Found in Cerro Aconcagua (Argentina) Traced Back to Populations of the Northern Peruvian Coast through Y-Chromosome Analysis. DNA 2023, 3, 137-147. https://doi.org/10.3390/dna3040012
Sandoval JR, Fujita R, Jota MS, Pinotti T, Santos FR. Inka Child Mummy Found in Cerro Aconcagua (Argentina) Traced Back to Populations of the Northern Peruvian Coast through Y-Chromosome Analysis. DNA. 2023; 3(4):137-147. https://doi.org/10.3390/dna3040012
Chicago/Turabian StyleSandoval, José R., Ricardo Fujita, Marilza S. Jota, Thomaz Pinotti, and Fabrício R. Santos. 2023. "Inka Child Mummy Found in Cerro Aconcagua (Argentina) Traced Back to Populations of the Northern Peruvian Coast through Y-Chromosome Analysis" DNA 3, no. 4: 137-147. https://doi.org/10.3390/dna3040012
APA StyleSandoval, J. R., Fujita, R., Jota, M. S., Pinotti, T., & Santos, F. R. (2023). Inka Child Mummy Found in Cerro Aconcagua (Argentina) Traced Back to Populations of the Northern Peruvian Coast through Y-Chromosome Analysis. DNA, 3(4), 137-147. https://doi.org/10.3390/dna3040012









