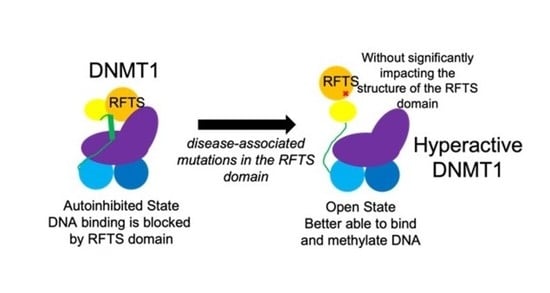
Disease-Associated Mutation A554V Disrupts Normal Autoinhibition of DNMT1
Abstract
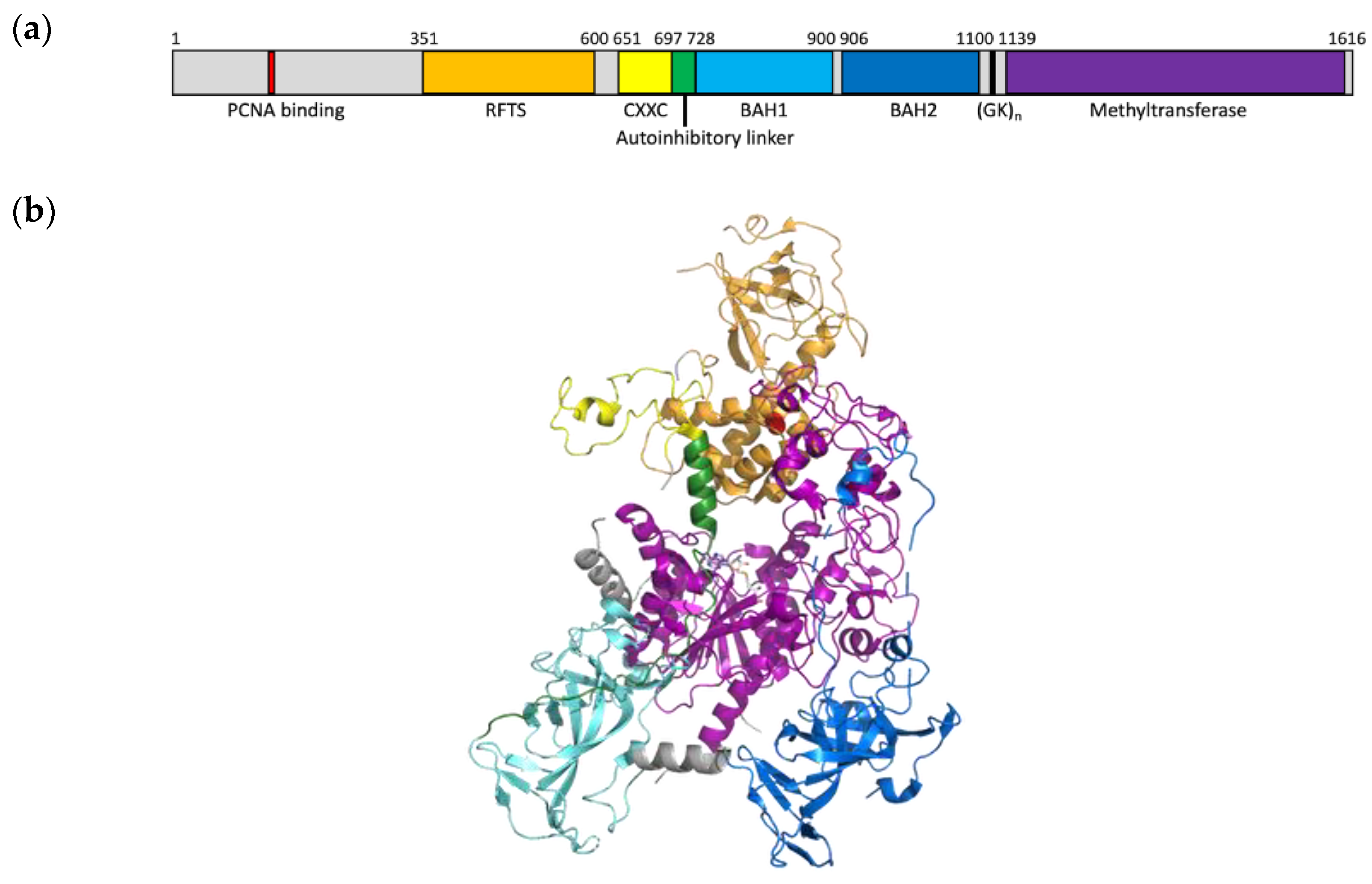
1. Introduction
2. Materials and Methods
2.1. Expression Constructs and Site-Directed Mutagenesis
2.2. Protein Expression and Purification
2.3. Fluorescence Polarization
2.4. DNA Methylation Assay
2.5. Differential Scanning Fluorimetry (DSF)
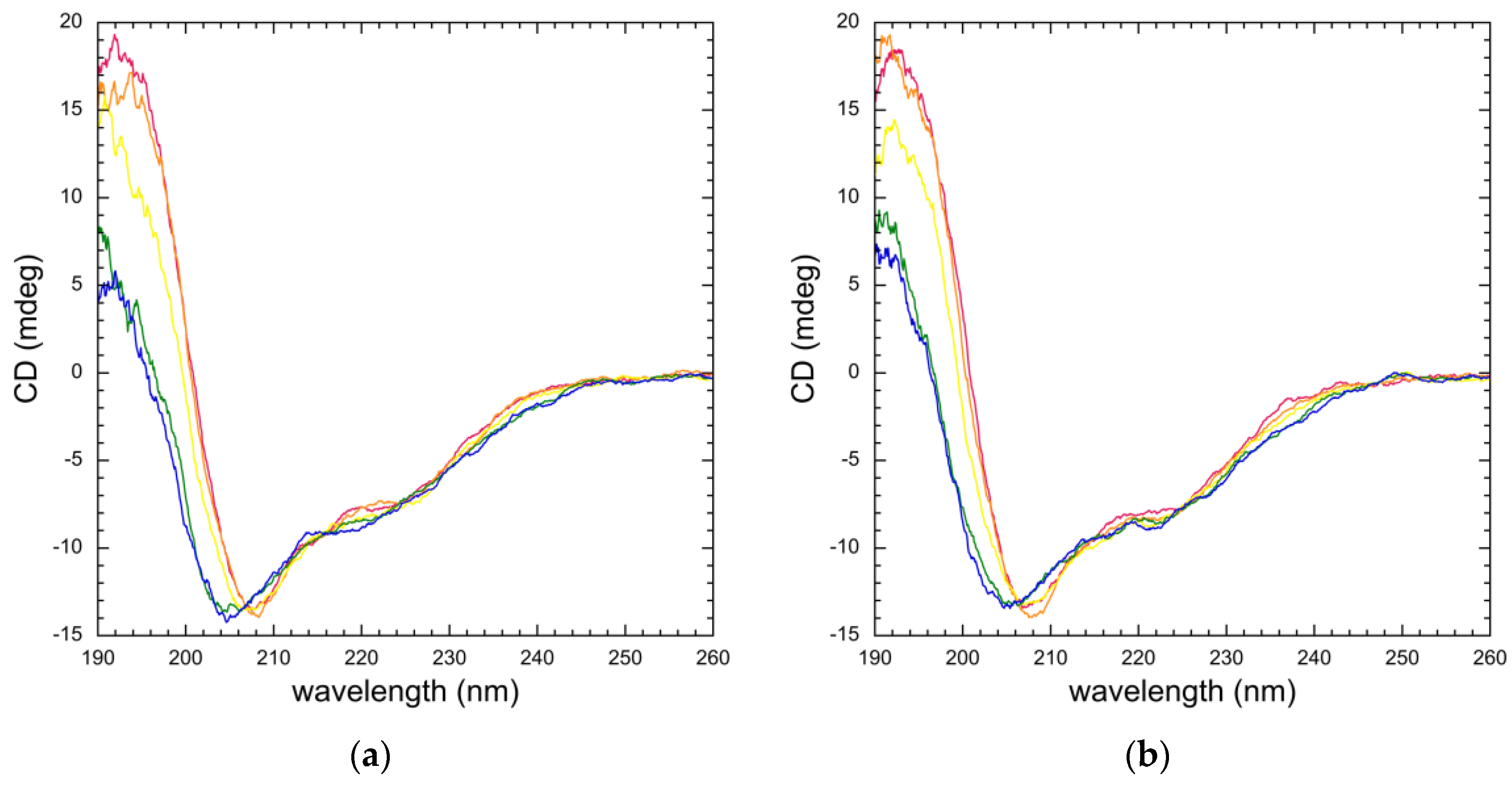
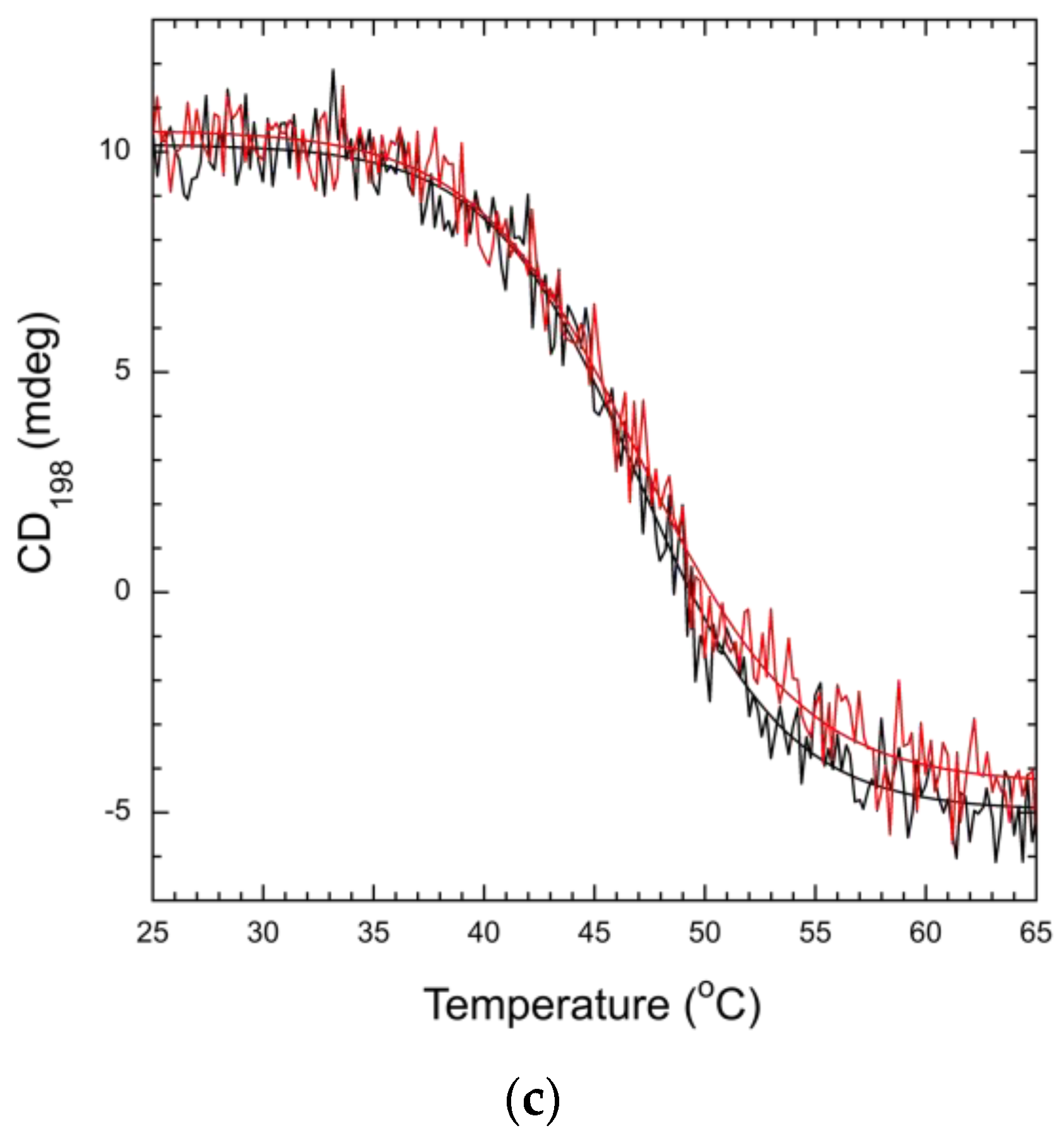
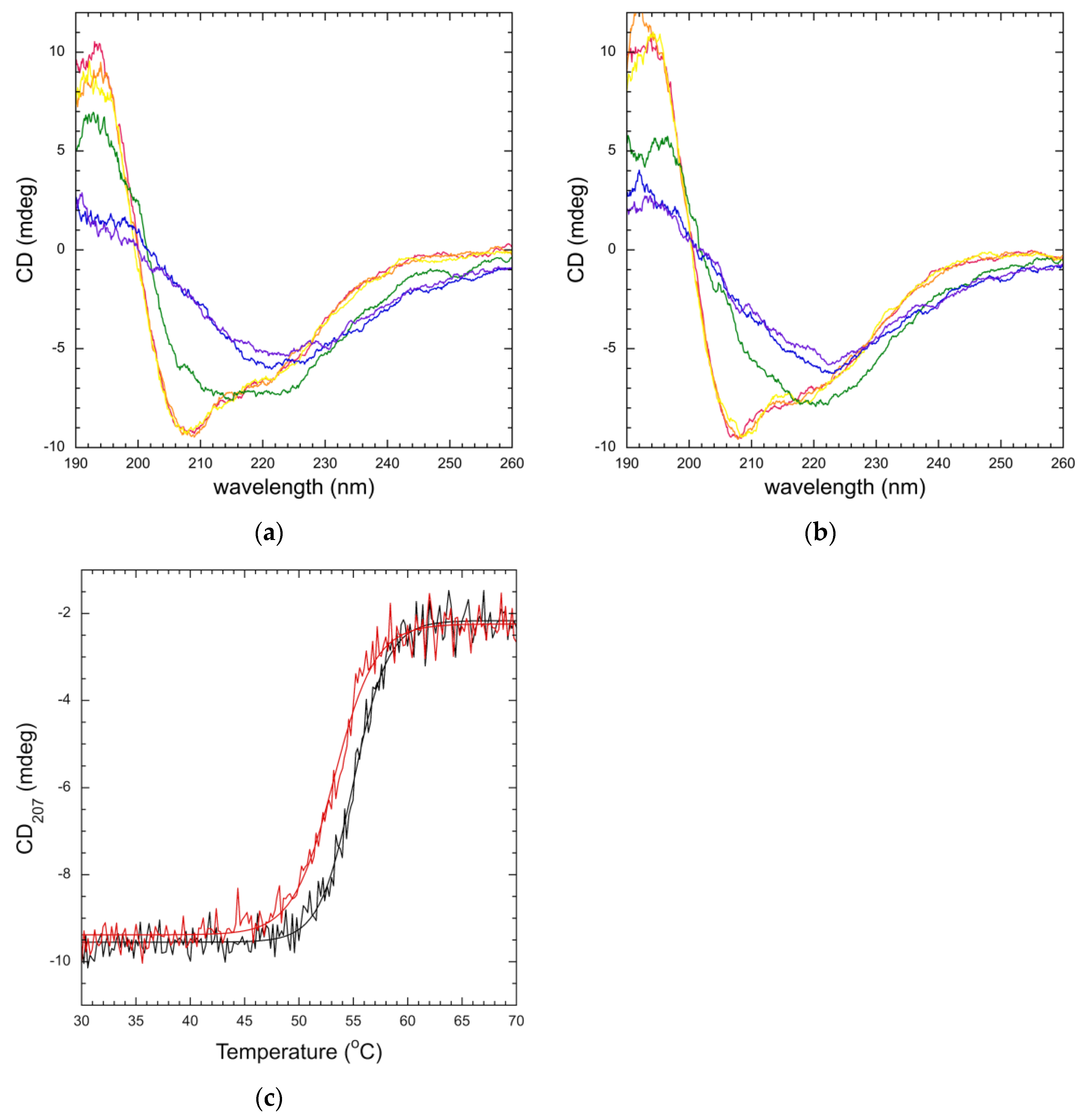
2.6. Circular Dichroism (CD) Spectroscopy
3. Results
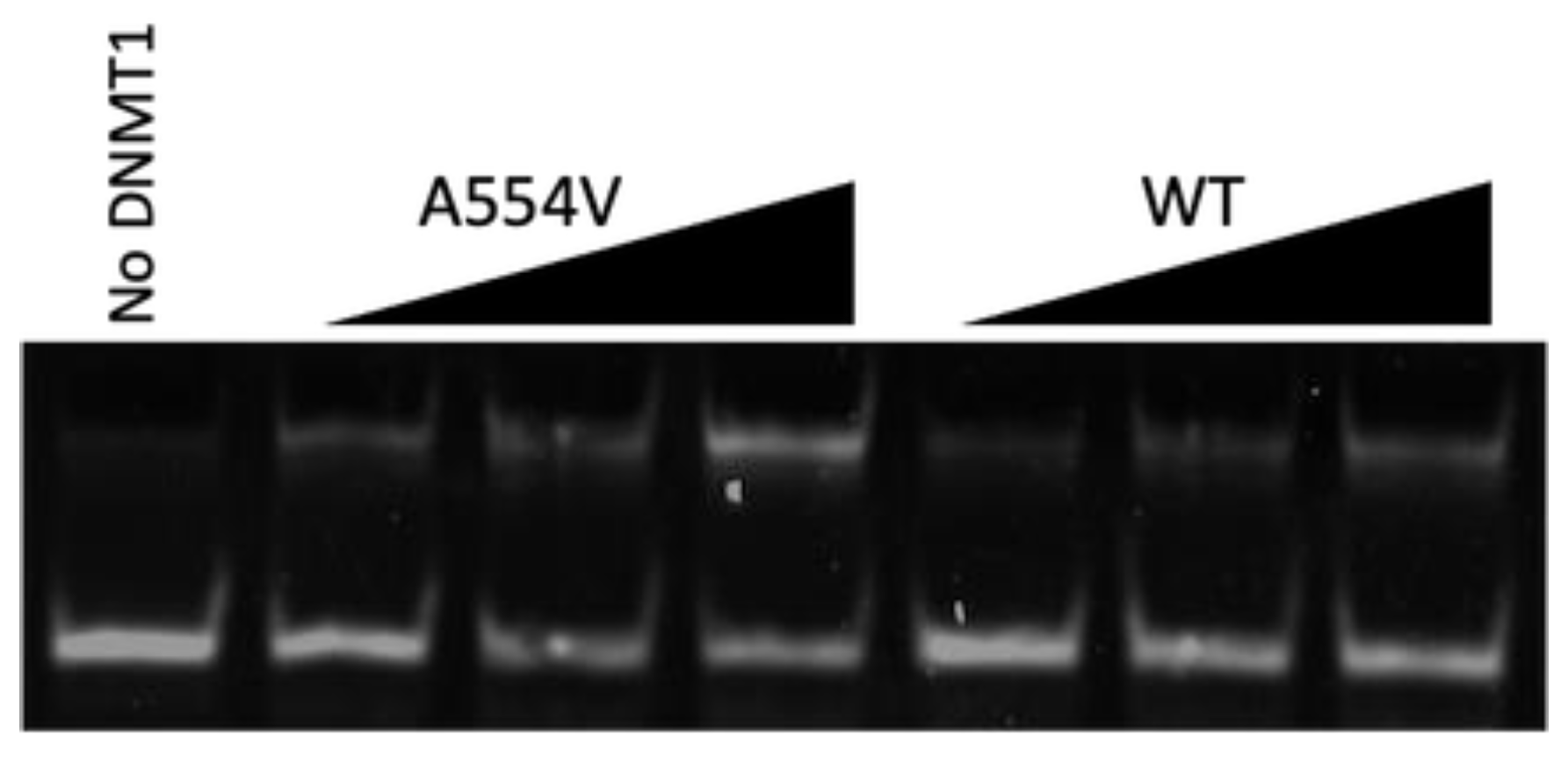
3.1. Mutation Increases DNA Binding Affinity
3.2. Mutation Increases DNA Methylation Activity
3.3. Mutation Impacts Structure and Stability
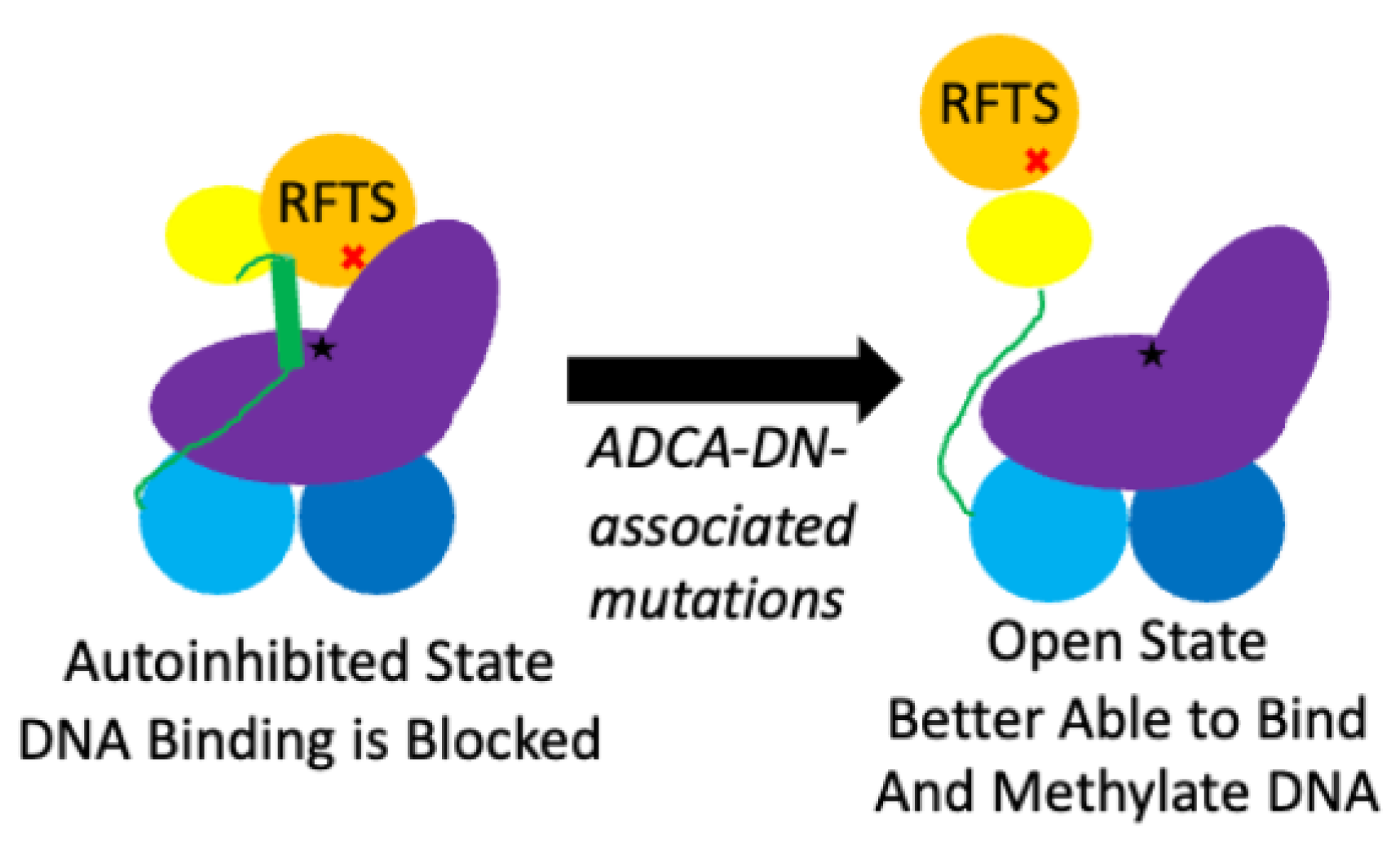
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Norvil, A.B.; Saha, D.; Saleem Dar, M.; Gowher, H. Effect of Disease-Associated Germline Mutations on Structure Function Relationship of DNA Methyltransferases. Genes 2019, 10, 369. [Google Scholar] [CrossRef]
- Hamidi, T.; Singh, A.K.; Chen, T. Genetic Alterations of DNA Methylation Machinery in Human Diseases. Epigenomics 2015, 7, 247–265. [Google Scholar] [CrossRef]
- Ren, W.; Gao, L.; Song, J. Structural Basis of DNMT1 and DNMT3A-Mediated DNA Methylation. Genes 2018, 9, 620. [Google Scholar] [CrossRef] [PubMed]
- Jeltsch, A.; Jurkowska, R.Z. New Concepts in DNA Methylation. Trends Biochem. Sci. 2014, 39, 310–318. [Google Scholar] [CrossRef]
- Jair, K.-W.; Bachman, K.E.; Suzuki, H.; Ting, A.H.; Rhee, I.; Yen, R.-W.C.; Baylin, S.B.; Schuebel, K.E. De Novo CpG Island Methylation in Human Cancer Cells. Cancer Res. 2006, 66, 682–692. [Google Scholar] [CrossRef] [PubMed]
- Inano, K.; Suetake, I.; Ueda, T.; Miyake, Y.; Nakamura, M.; Okada, M.; Tajima, S. Maintenance-Type DNA Methyltransferase Is Highly Expressed in Post-Mitotic Neurons and Localized in the Cytoplasmic Compartment. J. Biochem. 2000, 128, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Leonhardt, H.; Page, A.W.; Weier, H.-U.; Bestor, T.H. A Targeting Sequence Directs DNA Methyltransferase to Sites of DNA Replication in Mammalian Nuclei. Cell 1992, 71, 865–873. [Google Scholar] [CrossRef]
- Syeda, F.; Fagan, R.L.; Wean, M.; Avvakumov, G.V.; Walker, J.R.; Xue, S.; Dhe-Paganon, S.; Brenner, C. The Replication Focus Targeting Sequence (RFTS) Domain Is a DNA-Competitive Inhibitor of Dnmt1. J. Biol. Chem. 2011, 286, 15344–15351. [Google Scholar] [CrossRef]
- Takeshita, K.; Suetake, I.; Yamashita, E.; Suga, M.; Narita, H.; Nakagawa, A.; Tajima, S. Structural Insight into Maintenance Methylation by Mouse DNA Methyltransferase 1 (Dnmt1). Proc. Natl. Acad. Sci. USA 2011, 108, 9055–9059. [Google Scholar] [CrossRef]
- Zhang, Z.-M.; Liu, S.; Lin, K.; Luo, Y.; Perry, J.J.; Wang, Y.; Song, J. Crystal Structure of Human DNA Methyltransferase 1. J. Mol. Biol. 2015, 427, 2520–2531. [Google Scholar] [CrossRef]
- Bashtrykov, P.; Rajavelu, A.; Hackner, B.; Ragozin, S.; Carell, T.; Jeltsch, A. Targeted Mutagenesis Results in an Activation of DNA Methyltransferase 1 and Confirms an Autoinhibitory Role of Its RFTS Domain. ChemBioChem 2014, 15, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Bashtrykov, P.; Jankevicius, G.; Jurkowska, R.Z.; Ragozin, S.; Jeltsch, A. The UHRF1 Protein Stimulates the Activity and Specificity of the Maintenance DNA Methyltransferase DNMT1 by an Allosteric Mechanism. J. Biol. Chem. 2014, 289, 4106–4115. [Google Scholar] [CrossRef]
- Berkyurek, A.C.; Suetake, I.; Arita, K.; Takeshita, K.; Nakagawa, A.; Shirakawa, M.; Tajima, S. The DNA Methyltransferase Dnmt1 Directly Interacts with the SET and RING Finger-Associated (SRA) Domain of the Multifunctional Protein Uhrf1 to Facilitate Accession of the Catalytic Center to Hemi-Methylated DNA. J. Biol. Chem. 2014, 289, 379–386. [Google Scholar] [CrossRef]
- Ren, W.; Fan, H.; Grimm, S.A.; Guo, Y.; Kim, J.J.; Yin, J.; Li, L.; Petell, C.J.; Tan, X.-F.; Zhang, Z.-M.; et al. Direct Readout of Heterochromatic H3K9me3 Regulates DNMT1-Mediated Maintenance DNA Methylation. Proc. Natl. Acad. Sci. USA 2020, 117, 18439–18447. [Google Scholar] [CrossRef] [PubMed]
- Ishiyama, S.; Nishiyama, A.; Saeki, Y.; Moritsugu, K.; Morimoto, D.; Yamaguchi, L.; Arai, N.; Matsumura, R.; Kawakami, T.; Mishima, Y.; et al. Structure of the Dnmt1 Reader Module Complexed with a Unique Two-Mono-Ubiquitin Mark on Histone H3 Reveals the Basis for DNA Methylation Maintenance. Mol. Cell 2017, 68, 350–360.e7. [Google Scholar] [CrossRef] [PubMed]
- Misaki, T.; Yamaguchi, L.; Sun, J.; Orii, M.; Nishiyama, A.; Nakanishi, M. The Replication Foci Targeting Sequence (RFTS) of DNMT1 Functions as a Potent Histone H3 Binding Domain Regulated by Autoinhibition. Biochem. Biophys. Res. Commun. 2016, 470, 741–747. [Google Scholar] [CrossRef]
- Klein, C.J.; Botuyan, M.-V.; Wu, Y.; Ward, C.J.; Nicholson, G.A.; Hammans, S.; Hojo, K.; Yamanishi, H.; Karpf, A.R.; Wallace, D.C.; et al. Mutations in DNMT1 Cause Hereditary Sensory Neuropathy with Dementia and Hearing Loss. Nat. Genet. 2011, 43, 595–600. [Google Scholar] [CrossRef]
- Klein, C.J.; Bird, T.; Ertekin-Taner, N.; Lincoln, S.; Hjorth, R.; Wu, Y.; Kwok, J.; Mer, G.; Dyck, P.J.; Nicholson, G.A. DNMT1 Mutation Hot Spot Causes Varied Phenotypes of HSAN1 with Dementia and Hearing Loss. Neurology 2013, 80, 824–828. [Google Scholar] [CrossRef]
- Winkelmann, J.; Lin, L.; Schormair, B.; Kornum, B.R.; Faraco, J.; Plazzi, G.; Melberg, A.; Cornelio, F.; Urban, A.E.; Pizza, F.; et al. Mutations in DNMT1 Cause Autosomal Dominant Cerebellar Ataxia, Deafness and Narcolepsy. Hum. Mol. Genet. 2012, 21, 2205–2210. [Google Scholar] [CrossRef]
- Moghadam, K.K.; Pizza, F.; La Morgia, C.; Franceschini, C.; Tonon, C.; Lodi, R.; Barboni, P.; Seri, M.; Ferrari, S.; Liguori, R.; et al. Narcolepsy Is a Common Phenotype in HSAN IE and ADCA-DN. Brain 2014, 137, 1643–1655. [Google Scholar] [CrossRef]
- Yuan, J.; Higuchi, Y.; Nagado, T.; Nozuma, S.; Nakamura, T.; Matsuura, E.; Hashiguchi, A.; Sakiyama, Y.; Yoshimura, A.; Takashima, H. Novel Mutation in the Replication Focus Targeting Sequence Domain of DNMT1 Causes Hereditary Sensory and Autonomic Neuropathy IE: Yuan. J. Peripher. Nerv. Syst. 2013, 18, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Baets, J.; Duan, X.; Wu, Y.; Smith, G.; Seeley, W.W.; Mademan, I.; McGrath, N.M.; Beadell, N.C.; Khoury, J.; Botuyan, M.-V.; et al. Defects of Mutant DNMT1 Are Linked to a Spectrum of Neurological Disorders. Brain 2015, 138, 845–861. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Wu, Y.; Ordog, T.; Baheti, S.; Nie, J.; Duan, X.; Hojo, K.; Kocher, J.-P.; Dyck, P.J.; Klein, C.J. Aberrant Signature Methylome by DNMT1 Hot Spot Mutation in Hereditary Sensory and Autonomic Neuropathy 1E. Epigenetics 2014, 9, 1184–1193. [Google Scholar] [CrossRef]
- Kernohan, K.D.; Cigana Schenkel, L.; Huang, L.; Smith, A.; Pare, G.; Ainsworth, P.; Boycott, K.M.; Warman-Chardon, J.; Sadikovic, B. Identification of a Methylation Profile for DNMT1-Associated Autosomal Dominant Cerebellar Ataxia, Deafness, and Narcolepsy. Clin. Epigenetics 2016, 8, 91. [Google Scholar] [CrossRef]
- Dolen, E.K.; McGinnis, J.H.; Tavory, R.N.; Weiss, J.A.; Switzer, R.L. Disease-Associated Mutations G589A and V590F Relieve Replication Focus Targeting Sequence-Mediated Autoinhibition of DNA Methyltransferase 1. Biochemistry 2019, 58, 5151–5159. [Google Scholar] [CrossRef] [PubMed]
- Switzer, R.L.; Medrano, J.; Reedel, D.A.; Weiss, J. Substituted Anthraquinones Represent a Potential Scaffold for DNA Methyltransferase 1-Specific Inhibitors. PLoS ONE 2019, 14, e0219830. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.-H.; Goto, Y.; Réfrégiers, M.; Kardos, J. Accurate Secondary Structure Prediction and Fold Recognition for Circular Dichroism Spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, E3095–E3103. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Bulyáki, É.; Kun, J.; Moussong, É.; Lee, Y.-H.; Goto, Y.; Réfrégiers, M.; Kardos, J. BeStSel: A Web Server for Accurate Protein Secondary Structure Prediction and Fold Recognition from the Circular Dichroism Spectra. Nucleic Acids Res. 2018, 46, W315–W322. [Google Scholar] [CrossRef]
- Vilkaitis, G.; Suetake, I.; Klimašauskas, S.; Tajima, S. Processive Methylation of Hemimethylated CpG Sites by Mouse Dnmt1 DNA Methyltransferase. J. Biol. Chem. 2005, 280, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Mishima, Y.; Brueckner, L.; Takahashi, S.; Kawakami, T.; Arita, K.; Oka, S.; Otani, J.; Hojo, H.; Shirakawa, M.; Shinohara, A.; et al. RFTS-Dependent Negative Regulation of Dnmt1 by Nucleosome Structure and Histone Tails. FEBS J. 2017, 284, 3455–3469. [Google Scholar] [CrossRef]
- Palfey, B.A.; Switzer, R.L. Kinetics of Enzyme Catalysis; ACS In Focus; American Chemical Society: Washington, DC, USA, 2022; ISBN 978-0-8412-9939-9. [Google Scholar]
- Micsonai, A.; Bulyáki, É.; Kardos, J. BeStSel: From Secondary Structure Analysis to Protein Fold Prediction by Circular Dichroism Spectroscopy. In Structural Genomics; Chen, Y.W., Yiu, C.-P.B., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2021; Volume 2199, pp. 175–189. ISBN 978-1-07-160891-3. [Google Scholar]
- Greenfield, N.J. Using Circular Dichroism Spectra to Estimate Protein Secondary Structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef]
- Benjwal, S. Monitoring Protein Aggregation during Thermal Unfolding in Circular Dichroism Experiments. Protein Sci. 2006, 15, 635–639. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using Circular Dichroism Collected as a Function of Temperature to Determine the Thermodynamics of Protein Unfolding and Binding Interactions. Nat. Protoc. 2006, 1, 2527–2535. [Google Scholar] [CrossRef]
- Niklasson, M.; Andresen, C.; Helander, S.; Roth, M.G.L.; Zimdahl Kahlin, A.; Lindqvist Appell, M.; Mårtensson, L.; Lundström, P. Robust and Convenient Analysis of Protein Thermal and Chemical Stability. Protein Sci. 2015, 24, 2055–2062. [Google Scholar] [CrossRef]
- Fagan, R.L.; Wu, M.; Chédin, F.; Brenner, C. An Ultrasensitive High Throughput Screen for DNA Methyltransferase 1-Targeted Molecular Probes. PLoS ONE 2013, 8, e78752. [Google Scholar] [CrossRef]
- Kanada, K.; Takeshita, K.; Suetake, I.; Tajima, S.; Nakagawa, A. Conserved Threonine 1505 in the Catalytic Domain Stabilizes Mouse DNA Methyltransferase 1. J. Biochem. 2017, 162, 178–271. [Google Scholar] [CrossRef] [PubMed]
- Goto, K.; Numata, M.; Komura, J.-I.; Ono, T.; Bestor, T.H.; Kondo, H. Expression of DNA Methyltransferase Gene in Mature and Immature Neurons as Well as Proliferating Cells in Mice. Differentiation 1994, 56, 39–44. [Google Scholar] [CrossRef]
- Jobe, E.M.; Zhao, X. DNA Methylation and Adult Neurogenesis. Brain Plast. 2017, 3, 5–26. [Google Scholar] [CrossRef] [PubMed]
- Smets, M.; Link, S.; Wolf, P.; Schneider, K.; Solis, V.; Ryan, J.; Meilinger, D.; Qin, W.; Leonhardt, H. DNMT1 Mutations Found in HSANIE Patients Affect Interaction with UHRF1 and Neuronal Differentiation. Hum. Mol. Genet. 2017, 26, 1522–1534. [Google Scholar] [CrossRef] [PubMed]
- Ngan, K.C.-H.; Hoenig, S.M.; Kwok, H.S.; Lue, N.Z.; Gosavi, P.M.; Tanner, D.A.; Garcia, E.M.; Lee, C.; Liau, B.B. Activity-Based CRISPR Scanning Uncovers Allostery in DNA Methylation Maintenance Machinery. eLife 2023, 12, e80640. [Google Scholar] [CrossRef]

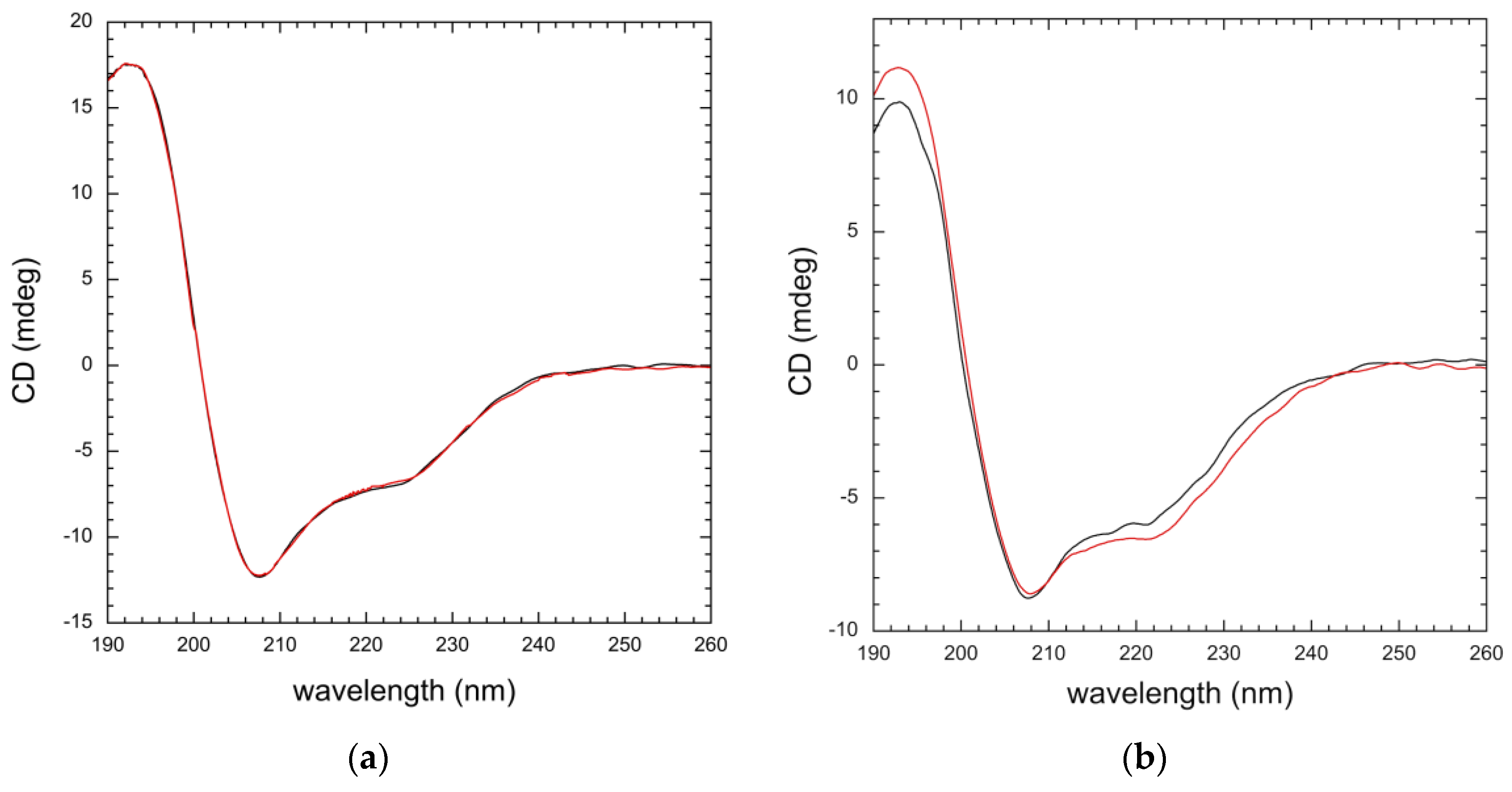
| Domain | Helix 1 | Antiparallel | Parallel | Turn | Other |
|---|---|---|---|---|---|
| Wild type | 17.5 | 23.3 | 0 | 14.9 | 44.3 |
| A554V | 17.3 | 22.3 | 0 | 14.9 | 45.2 |
| Protein | Helix 1 | Antiparallel | Parallel | Turn | Other |
|---|---|---|---|---|---|
| Wild-type | 16.1 | 26.4 | 0 | 14.1 | 43.4 |
| A554V | 14.4 | 26.5 | 0 | 14.4 | 44.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Switzer, R.L.; Hartman, Z.J.; Hewett, G.R.; Carroll, C.F. Disease-Associated Mutation A554V Disrupts Normal Autoinhibition of DNMT1. DNA 2023, 3, 119-133. https://doi.org/10.3390/dna3030010
Switzer RL, Hartman ZJ, Hewett GR, Carroll CF. Disease-Associated Mutation A554V Disrupts Normal Autoinhibition of DNMT1. DNA. 2023; 3(3):119-133. https://doi.org/10.3390/dna3030010
Chicago/Turabian StyleSwitzer, Rebecca L., Zach J. Hartman, Geoffrey R. Hewett, and Clara F. Carroll. 2023. "Disease-Associated Mutation A554V Disrupts Normal Autoinhibition of DNMT1" DNA 3, no. 3: 119-133. https://doi.org/10.3390/dna3030010
APA StyleSwitzer, R. L., Hartman, Z. J., Hewett, G. R., & Carroll, C. F. (2023). Disease-Associated Mutation A554V Disrupts Normal Autoinhibition of DNMT1. DNA, 3(3), 119-133. https://doi.org/10.3390/dna3030010