Dynamics Differences between Minimal Models of Second and First-Order Chemical Self-Replication
Abstract
1. Introduction
2. Minimal Models
2.1. Templator Model of First-Order Self-Replication
2.2. Minimal Dimer Model of Second-Order Self-Replication
2.3. The Extended Model of Second-Order Self-Replication
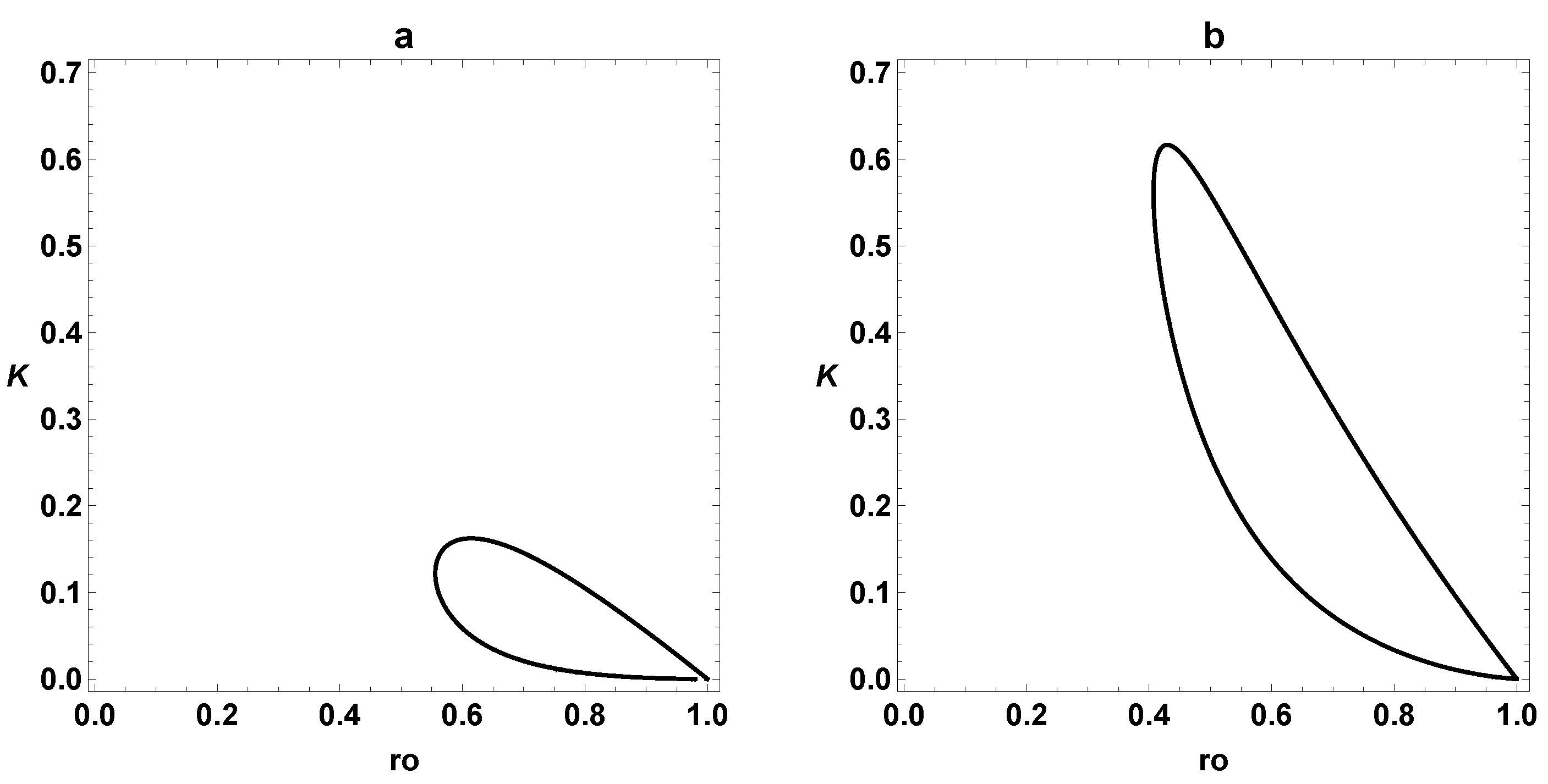
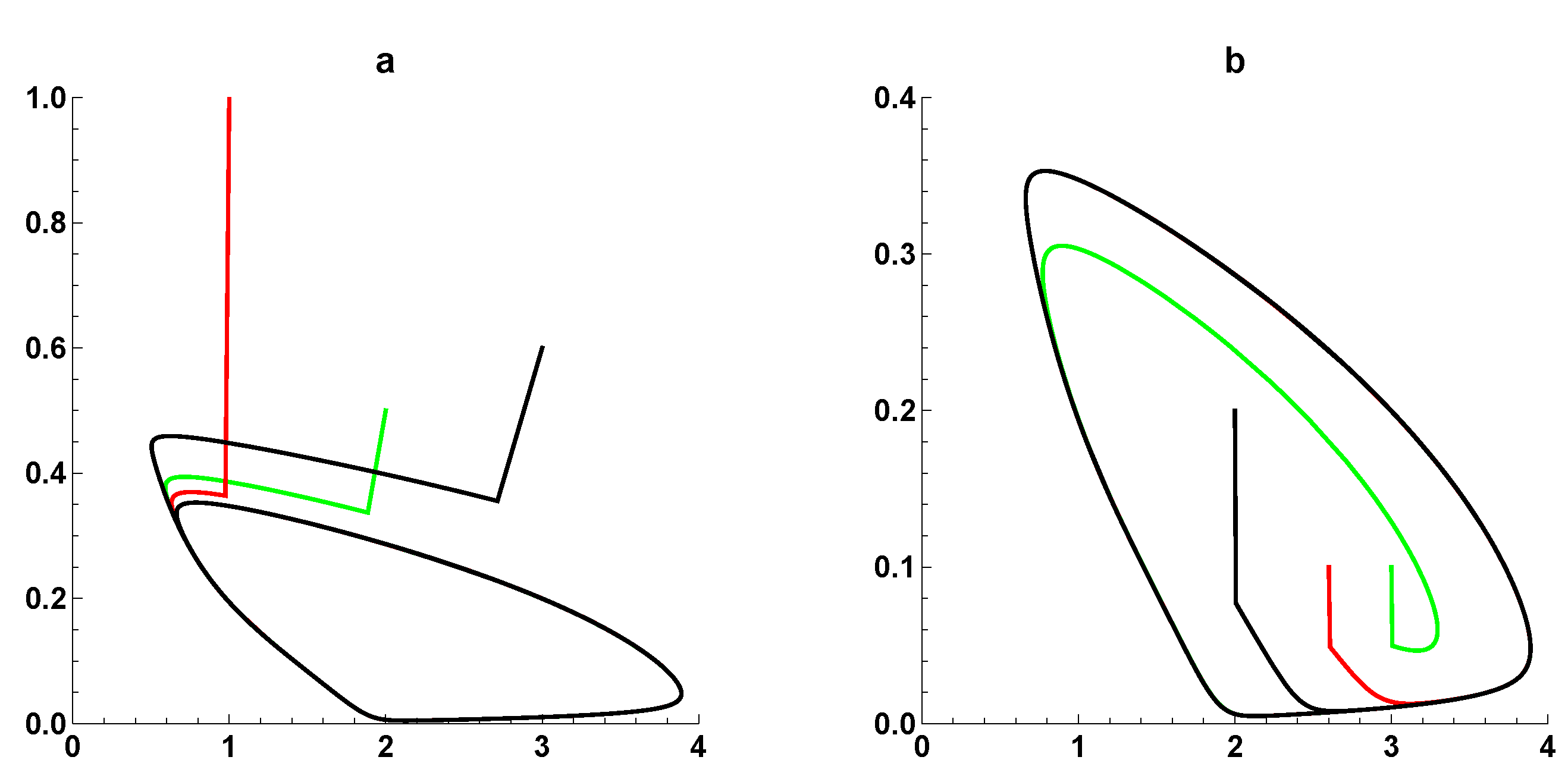
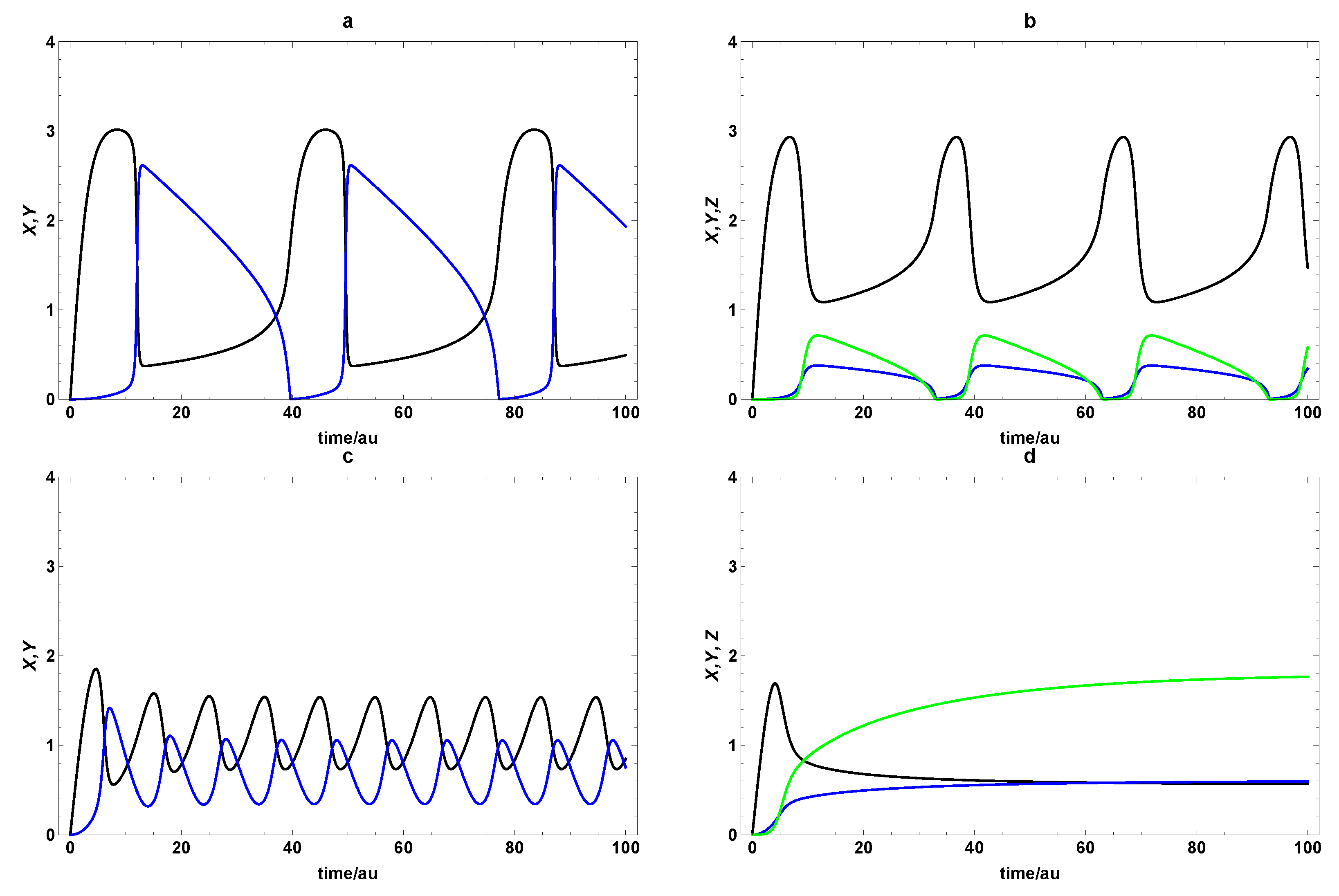
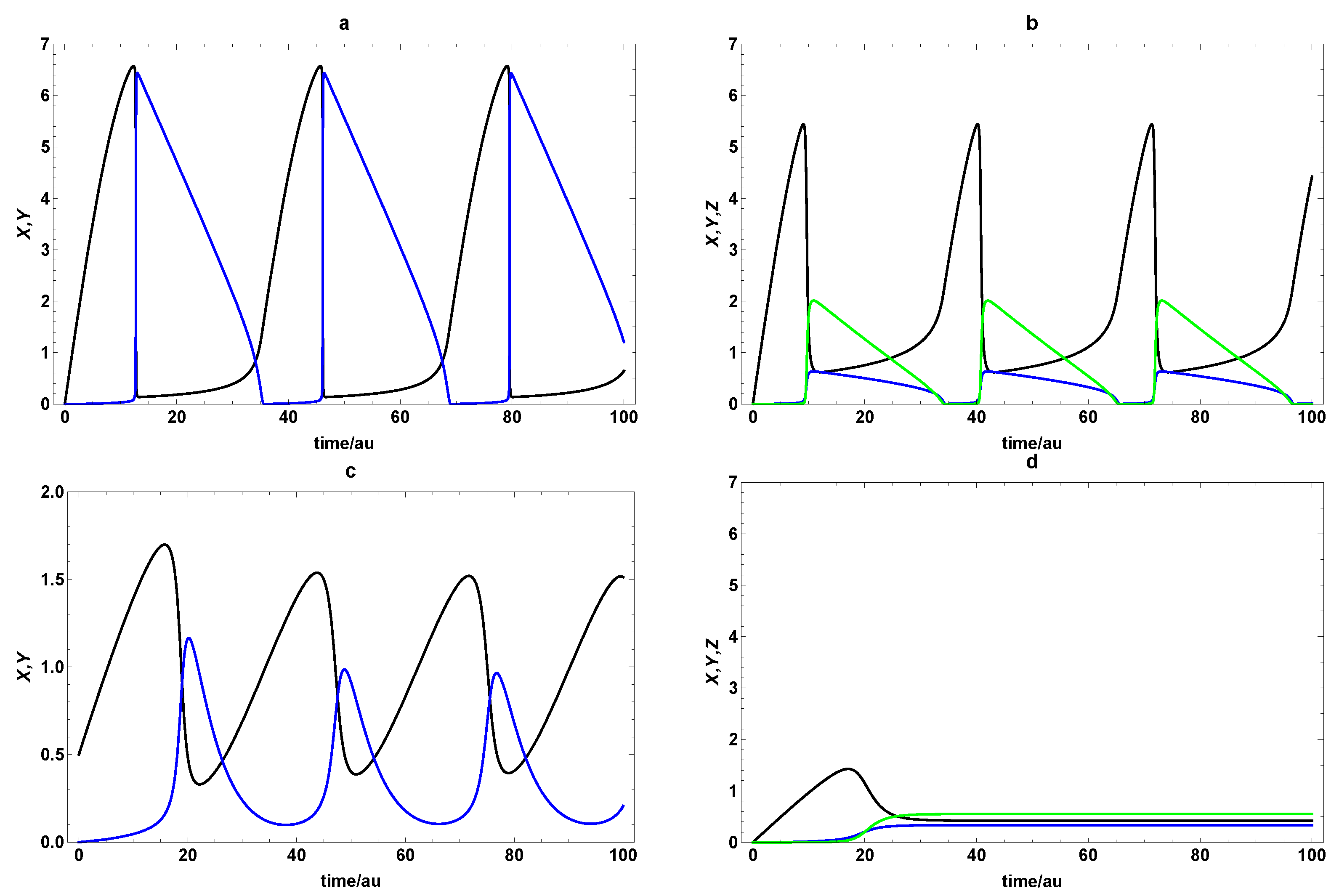
3. Dynamic Characterization
3.1. First- and Second-Order Minimal Models of Self-Replication
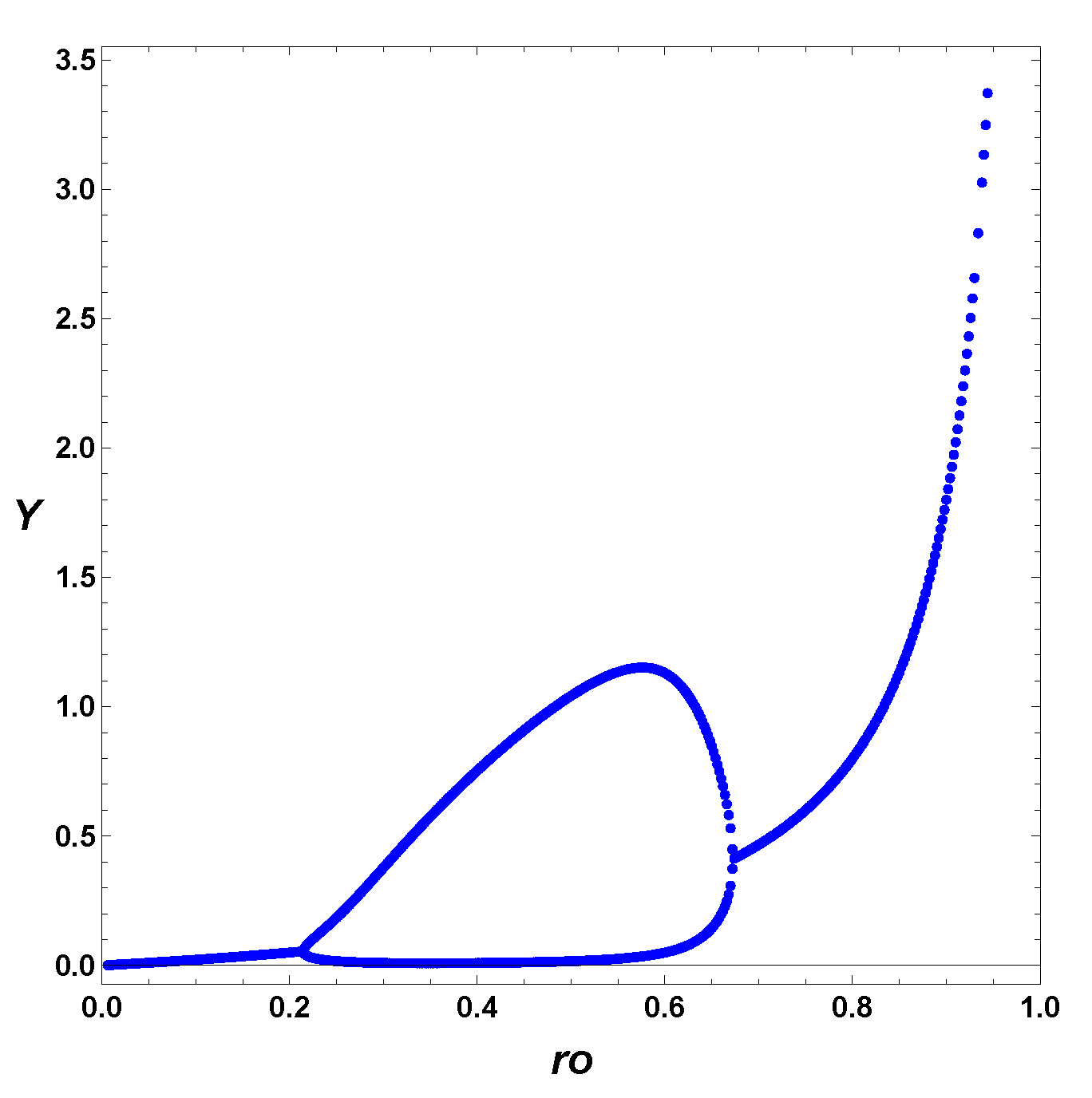
3.2. Extended Second-Order Self-Replication
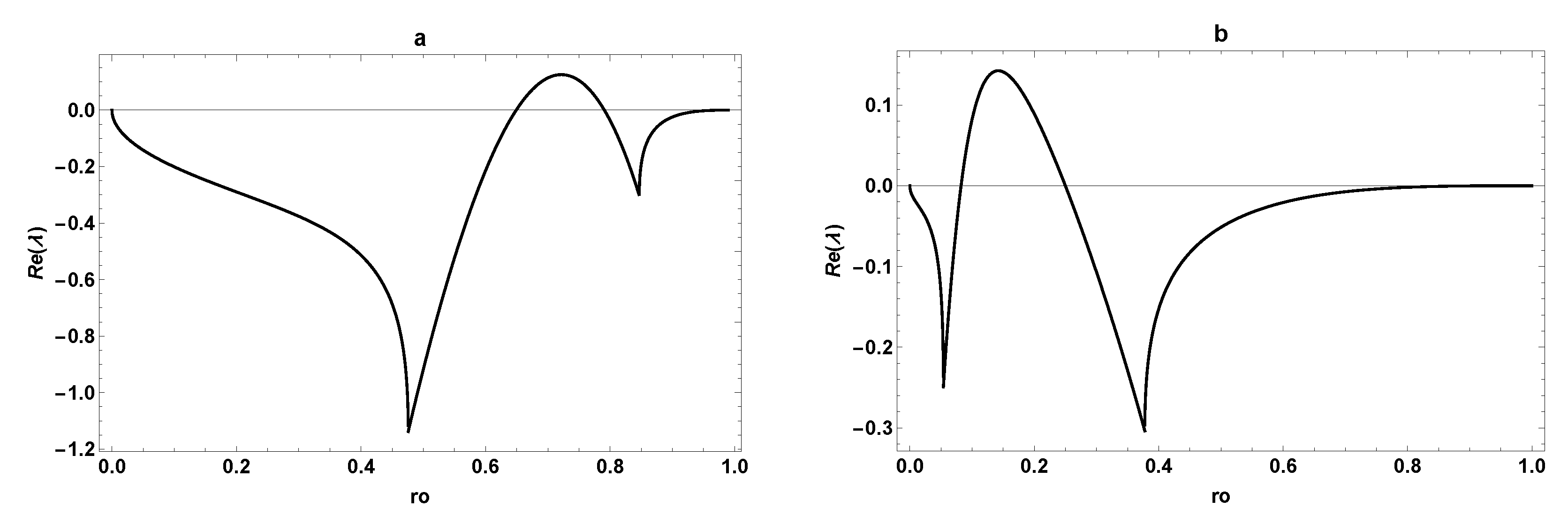
3.3. Comparison between SOM and ESOM
4. Dynamic Characterization
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zielinski, W.S.; Orgel, L.E. Autocatalytic synthesis of a tetranucleotide analog. Nature 1987, 327, 346–347. [Google Scholar] [CrossRef]
- Orgel, L.E. Molecular replication. Nature 1992, 358, 203–209. [Google Scholar] [CrossRef]
- Orgel, L.E. Unnatural Selection in Chemical Systems. Acc. Chem. Res. 1995, 28, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Von Kiedrowski, G. A self-replicating hexadeoxynucleotide. Amgew. Chem. Int. Ed. 1986, 25, 932–935. [Google Scholar] [CrossRef]
- Von Kiedrowski, G.; Wlotzka, B.; Helbing, J.; Matzen, M.; Jordan, S. Parabolic Growth of a Self-Replicating Hexadeoxynucleotide Bearing a 3′-5′-Phosphoamidate Linkage. Angew. Chem. Int. Ed. Engl. 1991, 30, 423–426. [Google Scholar] [CrossRef]
- Bag, B.; von Kiedrowski, G. Templates, autocatalytic and molecular replication. Pure Appl. Chem. 1996, 68, 2145–2152. [Google Scholar] [CrossRef]
- Luther, A.; Brandsch, R.; von Kiedrowski, G. Surface-promoted replication and exponential amplification of DNA analogues. Nature 1998, 386, 245–248. [Google Scholar] [CrossRef]
- Li, T.; Nicolaou, K.C. Chemical self-replication of palindromic duplex DNA. Nature 1994, 369, 218–221. [Google Scholar] [CrossRef]
- Tjivikua, T.; Ballester, P.; Rebek, J., Jr. A Self-replicating system. J. Am. Chem. Soc. 1990, 112, 1249–1250. [Google Scholar] [CrossRef]
- Rebek, J., Jr. Molecular Recognition with Model Systems. Angew. Chem. Int. Ed. Engl. 1990, 29, 245–255. [Google Scholar] [CrossRef]
- Nowick, J.S.; Feng, Q.; Tjivikua, T.; Ballester, P.; Rebek, J., Jr. Kinetic Studies and Modeling of a Self-Replicating System. J. Am. Chem. Soc. 1991, 113, 8831–8839. [Google Scholar] [CrossRef]
- Hong, I.-I.; Fing, Q.; Rotello, V.; Rebek, J., Jr. Competition, Cooperation, and Mutation: Improving a Synthetic Replicator by Light Irradiation. Science 1992, 255, 848–850. [Google Scholar] [CrossRef]
- Wintner, E.A.; Conn, M.M.; Rebek, J., Jr. Studies in Molecular Replication. Acc. Chem. Res. 1994, 27, 198–203. [Google Scholar] [CrossRef]
- Wintner, E.A.; Conn, M.M.; Rebek, J., Jr. Self-Replicating Molecules: A Second Generation. J. Am. Chem. Soc. 1994, 116, 8877–8884. [Google Scholar] [CrossRef]
- Rebek, J., Jr. Synthetic Self-replicating Molecules. Sci. Am. 1994, 271, 48–55. [Google Scholar] [CrossRef]
- Rebek, J., Jr. A template for Life, surface, and linear rate. Chem. Br. 1994, 30, 286–290. [Google Scholar]
- Szathmary, E. Sub-Exponential Growth and Coexistence of Non-Enzymatically Replicating Templates. J. Theor. Biol. 1989, 138, 55–58. [Google Scholar] [CrossRef]
- Lee, D.H.; Granja, J.R.; Martinez, J.A.; Severin, K.; Ghadiri, M.R. A self-replicating peptide. Nature 1996, 382, 525–528. [Google Scholar] [CrossRef]
- Severin, K.; Lee, D.H.; Martinez, J.A.; Ghadiri, M.R. Peptide self-replication via template-directed ligation. Chem. Eur. J. 1997, 3, 1017–1024. [Google Scholar] [CrossRef]
- Lee, D.H.; Severin, K.; Ghadiri, M.R. Autocatalytic networks: The transition from molecular self-replication to molecular ecosystems. Curr. Opin. Chem. Biol. 1997, 1, 491–496. [Google Scholar] [CrossRef]
- Lee, D.H.; Severin, K.; Yokobayashi, Y.; Ghadiri, M.R. Emergence of symbiosis in peptide self-replication through a hypercyclic network. Nature 1997, 390, 591–594. [Google Scholar] [CrossRef]
- Saghatalias, A.K.; Yokobayashi, Y.; Soltani, K.; Ghadiri, M.R. A chiroselective peptide replicator. Nature 2001, 409, 797–801. [Google Scholar] [CrossRef]
- Yao, S.; Gosh, I.; Zutshi, R.; Chmielewski, J. A pH-Modulated, Self-Replicating Peptide. J. Am. Chem. Soc. 1997, 119, 10559–10560. [Google Scholar] [CrossRef]
- Yao, S.; Gosh, I.; Zutshi, R.; Chmielewski, J. Selective amplification by auto- and cross-catalysis in a replicating peptide system. Nature 1998, 396, 447–450. [Google Scholar] [CrossRef] [PubMed]
- Issac, R.; Ham, Y.-W.; Chmielewski, J. The design of self-replicating helical peptides. Curr. Opin. Struct. Biol. 2001, 11, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Issac, R.; Chmielewski, J. Approaching Exponential Growth with a Self-Replicating Peptide. J. Am. Chem. Soc. 2001, 124, 6808–6809. [Google Scholar] [CrossRef] [PubMed]
- Paul, N.; Joyce, G.F. A self-replicating ligase ribozyme. Proc. Nat. Acad. Sci. USA 2002, 99, 12733–12740. [Google Scholar] [CrossRef]
- Paul, N.; Joyce, G.F. Minimal self-replicating systems. Curr. Opin. Chem. Biol. 2004, 8, 634–639. [Google Scholar] [CrossRef]
- Lincolm, T.A.; Joyce, G.F. Self-Sustained Replication of an RNA Enzyme. Science 2009, 323, 1229–1232. [Google Scholar] [CrossRef]
- Peacock-López, E.; Radov, D.B.; Flesner, C.S. Mixed-mode oscillations in a template mechanism. Biophys. Chem. 1997, 65, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Ashkenasy, G.; Ghadiri, M.R. Boolean Logic Functions of Synthetic Peptide Network. J. Am. Chem. Soc. 2004, 126, 11140–11141. [Google Scholar] [CrossRef]
- Dadon, Z.; Wagner, N.; Ashkenasy, G. The Road to Nonenzymatic Molecular Networks. Angew. Chem. Int. Ed. Engl. 2008, 47, 6128–6136. [Google Scholar] [CrossRef]
- Alasibi, S.; Peacock-López, E.; Ashkenasy, G. Coupled Oscillations and Circadian Rhythms in Molecular Replication Networks. J. Phys. Chem. Lett. 2015, 15, 60–65. [Google Scholar]
- Dadon, Z.; Wagner, N.; Alasibi, S.; Samiappan, M.; Mukherjee, R.; Ashkenasy, G. Competition and Cooperation in Dynamic Replication Networks. Chem. Eur. J. 2015, 21, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Wagner, N.; Mukherjee, R.; Maity, I.; Peacock-López, E.; Ashkenasy, G. Bistability and Bifurcation in Minimal Self-Replication and Nonenzymatic Catalytic Network. ChemPhysChem 2017, 18, 1842–1850. [Google Scholar] [CrossRef]
- Wagner, N.; Hochberg, D.; Peacock-López, E.; Maity, I.; Ashkenasy, G. Open Prebiotic Environment Drive Emergent Phenomena and Complex Behavior. Life 2019, 9, 45. [Google Scholar] [CrossRef]
- Maity, I.; Wagner, N.; Mukherjee, R.; Dev, D.; Peacock-López, E.; Ashkenasy, G. A chemically fuelled nonenzymatic bistable network. Nat. Commun. 2019, 10, 4636. [Google Scholar] [CrossRef] [PubMed]
- Poincaré, H. Le Méthodes Nouvelles de la Mécanique Célest; Gauthier-Villars Et Filles: Paris, France, 1892; Volume I. [Google Scholar]
- Andronov, A.A.; Witt, A. Sur la théorie mathématique des auto-oscillations. CR Acad. Sci. Paris 1930, 190, 256. [Google Scholar]
- Kuznetsov, Y.A. Elements of Applied Bifurcation Theory, 3rd ed.; Springer: New York, NY, USA, 2010. [Google Scholar]
- Hopf, E. Sachsische Akademie der Wissenschaften Leipzig. Math. Phys. 1942, 94, 1. [Google Scholar]
- Gijima, D.T.; Peacock-López, E. A dynamic study of biochemical self-replication. Mathematics 2020, 8, 1042. [Google Scholar] [CrossRef]
- Shah, A.K.; Peacock- López, E. Complex dynamics of competitive first-order self-replication. In Chaos Theory—Recent Advances, New Perspective, and Applications; Andriychu, M., Ed.; IntechOpen: London, UK, 2022. [Google Scholar]
- Beutel, K.M.; Peacock-López, E. Complex dynamics in a cross-catalytic mechanism of chemical self-replication. J. Chem. Phys. 2007, 126, 125104. [Google Scholar] [CrossRef]
- Eigen, M.; Schuster, P. The hypercycle. A principle of natural self-organization. Part A: Emergence of the hypercycle. Naturwissenschaften 1977, 64, 541–565. [Google Scholar] [CrossRef]
- Eigen, M.; Schuster, P. The Hypercycle. Naturwissenschaften 1978, 65, 7–41. [Google Scholar] [CrossRef]
- Nakajima, M.; Imai, K.; Ito, H.; Nishiwaki, T.; Murayama, Y.; Iwasaki, H.; Oyama, T.; Kondo, T. Reconstitution of Circadian Oscillations of Cyanobacterial KaiC Phosphorylation in Vitro. Science 2005, 308, 414–415. [Google Scholar] [CrossRef]
- Rust, M.J.; Markson, J.S.; Lane, W.S.; Fischer, D.S.; O’Shea, E.K. Ordered Phosphorylation Governs Oscillation of the Three-Protein Circadian Clock. Science 2007, 318, 809–812. [Google Scholar] [CrossRef] [PubMed]
- Ludlow, R.F.; Otto, S. System Chemistry. Chem. Soc. Rev. 2008, 37, 101–108. [Google Scholar] [CrossRef]
- Liu, B.; Wu, J.; Geerts, M.; Markovitch, O.; Pappas, C.G.; Liu, K.; Otto, S. Out-of-Equilibrium Self-Replication Allows Selection for Dynamic Kinetic Stability in a System of Competing Replicators. Angew. Chem. 2022, 134, e202117605. [Google Scholar]
- Howlett, M.G.; Engwerda, A.H.J.; Scanes, R.J.H.; Fletcher, S.P. An autonomous oscillating supramolecular self-replicator. Nat. Chem. 2022, 14, 805–810. [Google Scholar] [CrossRef]
- Bandela, A.K.; Wagner, N.; Sadihov, H.; Morales-Reina, S.; Chotera-Ouda, A.; Basu, K.; Cohen-Luria, R.; del la Escosura, A.; Ashkenasy, G. Primitive selection of the fittest emerging through functional synergy in nucleopeptides networks. Proc. Nat. Acad. Sci. USA 2021, 118, e2015285118. [Google Scholar] [CrossRef]










Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moseley, L.A.; Peacock-López, E. Dynamics Differences between Minimal Models of Second and First-Order Chemical Self-Replication. Dynamics 2023, 3, 425-443. https://doi.org/10.3390/dynamics3030023
Moseley LA, Peacock-López E. Dynamics Differences between Minimal Models of Second and First-Order Chemical Self-Replication. Dynamics. 2023; 3(3):425-443. https://doi.org/10.3390/dynamics3030023
Chicago/Turabian StyleMoseley, Lauren A., and Enrique Peacock-López. 2023. "Dynamics Differences between Minimal Models of Second and First-Order Chemical Self-Replication" Dynamics 3, no. 3: 425-443. https://doi.org/10.3390/dynamics3030023
APA StyleMoseley, L. A., & Peacock-López, E. (2023). Dynamics Differences between Minimal Models of Second and First-Order Chemical Self-Replication. Dynamics, 3(3), 425-443. https://doi.org/10.3390/dynamics3030023






