Multiobjective Optimization of a Fed-Batch Bienzymatic Reactor for Mannitol Production
Abstract
1. Introduction
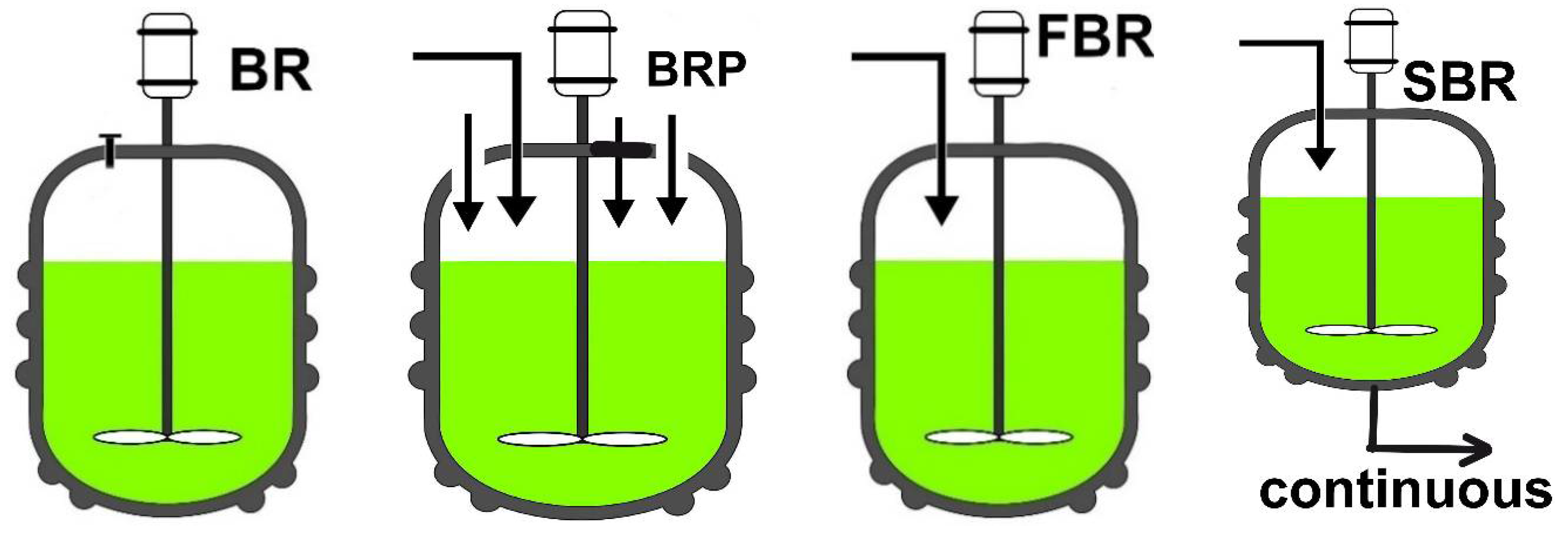
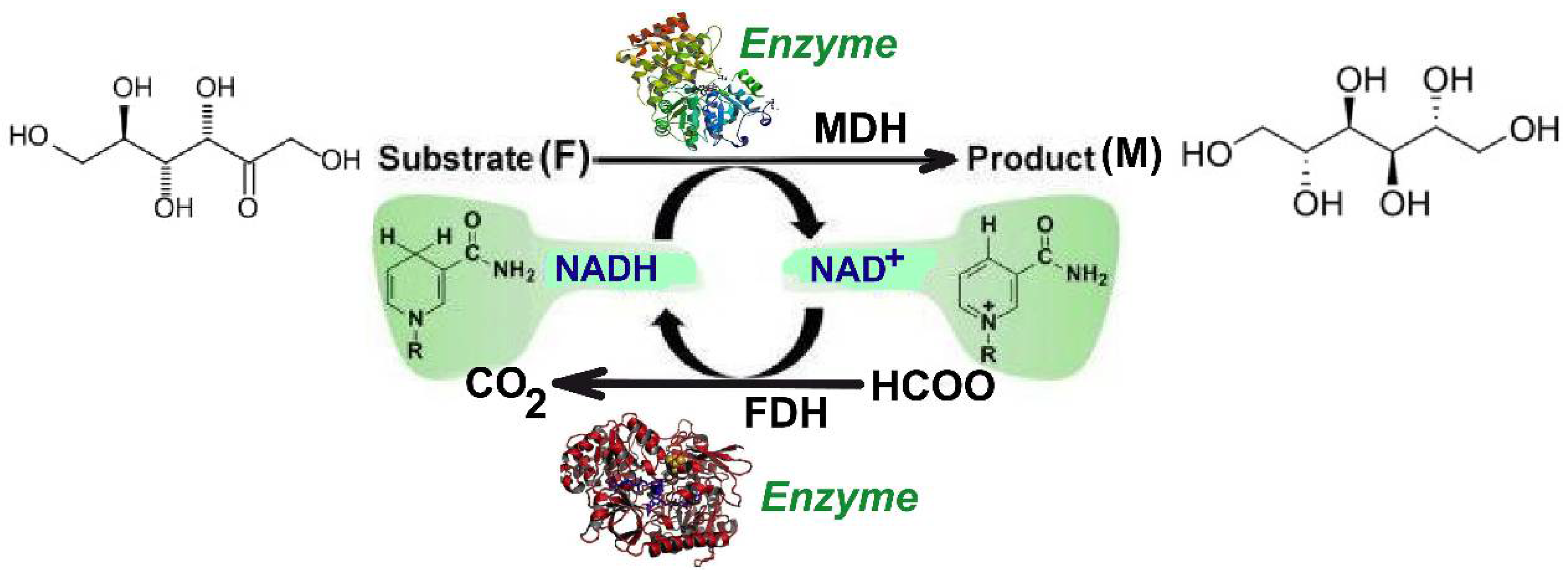
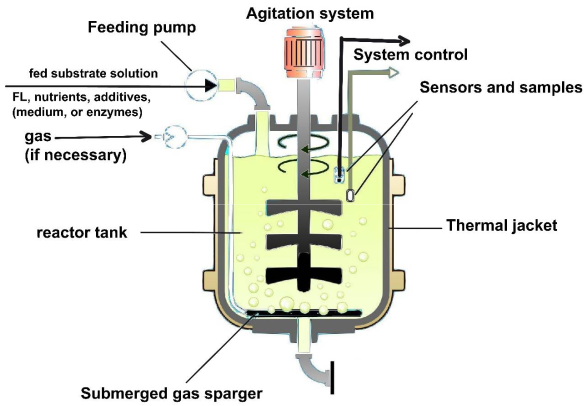
2. Dynamic Models for the Bienzymatic Process and FBR
| Parameter | Values and Remarks [13] |
|---|---|
| Temperature/Pressure/pH (buffer solution) | 25 °C/Normal (1 atm.)/7 |
| Molar initial concentrations | |
| Fructose, [F]o | 0.1–1 M ([13]) (0.1–4 M) (this paper, in the range of kinetic model validity); (a) |
| [NADH]o | 0.008–0.5 M ([13]) (0.1–0.5 M) (this paper); 0.1 M (initial guess for optimal FBR) |
| [NAD+]o | 0.0005 M [13] |
| Formate initial [HCOO]o | Identical to initial [F]o |
| Others: [M]o = [CO2]o = 0 | None |
| = CO2 saturation level at 25 °C and pH = 7 ± 0.1 | 0.0313 M [54,55] pH control by an “automatic addition of HCl” [13] |
| ) | 48 h (this paper), and for an optimal single BR [27] 40 h for each BR in the SeqBR series of NBR = 10 BR [27] |
| ) | 0.5 L (adopted); variable due to the continuous feeding of the FBR. Max. 3 L |
| ) | 0.02 L h−1 (initial). Its time stepwise dynamics are to be optimized in the adopted range of 0.01–0.04 L h−1 (limited by the reactor volume) |
| FDH enzyme (referred to the reactor liquid) | Initial value = 0. Its time stepwise additions are to be optimized in the range of 0.1–2 kU/L. |
| MDH enzyme (referred to the reactor liquid) | Initial value = 0. Its time stepwise additions are to be optimized in the range of 0.1–2 kU/L. Some other compounds (dithiothreitol) are added by [13] to stabilize the MDH. |
| Reactions: | Rate Expressions: |
|---|---|
| Rate constants = 2 × 10−3; = 8.3259 × 10−3; 1/h//(U/L); = 7.2367 × 10−2 M; = 8.8047 × 10−2 M; = 1; = 1; = 5.0061 × 10−2; = 90.181 [27] | |
| Process stoichiometry:; ; ; | |
| Species | Remarks |
|---|---|
| Substrate (fructose, F) and cofactor (NADH) mass balances: time stepwise unknown values to be determined from the FBR optimization; time stepwise unknown values to be determined from the FBR optimization; | For the optimal FBR with adopted Ndiv = 10, the feeding policy is (a): |
| Formate (HCOO) and exhausted cofactor (NAD+) mass balances: j = 1,.., time stepwise values | M [13] |
| Mannitol (M) and CO2 mass balances: j = 1,.., time stepwise values | (t = 0) = 0. (t = 0) = 0. ; (b) |
| Enzymes MDH and FDH mass balances (c): time stepwise unknown values to be determined from the FBR optimization; = 0. time stepwise unknown values to be determined from the FBR optimization; = 0. | For the optimal FBR with adopted Ndiv = 10, the feeding policy is (a): |
| Liquid volume in the reactor (d): time stepwise unknown values to be determined from the FBR optimization. The unknown values from the FBR optimization. | For the optimal FBR with adopted Ndiv =10, the feeding policy is (a): |
3. Optimization Problem Formulation
3.1. The Control Variable Choice
- -
- The fed enzymes (;),
- -
- The fed substrate (), and the fed cofactor ();
- -
- The feed flow rate () of the solution containing the substrates, enzymes, and the cofactor.
3.2. Optimization Objectives and the Composite Objective Function (W) Choice
- (i)
- maximum FBR productivity in mannitol;
- (ii)
- minimum consumption of the costly enzymes MDH and FDH;
- (iii)
- (iv)
- an adjustable substrate consumption (within defined limits) to ensure its maximum conversion to mannitol.
3.3. Optimization Problem Constraints
3.4. Time Stepwise Divisions Choice
- (a)
- “by choosing unequal time-arcs, of lengths to be determined by the optimization rule;
- (b)
- by considering the whole batch time as an optimization variable;
- (c)
- by increasing the number of equal time-arcs (Ndiv) to obtain a more refined and adaptable FBR operating policy, but keeping the same non-uniform feeding policy for the chosen control variables.
- (d)
- by extend the search min/max limits of the control variables, or even by considering them as unknown (to be determined).
- (e)
- by feeding the bioreactor with a variable feed flow-rate, but with a solution of an uniform concentration for the all other control variables, over a small or large number (Ndiv) of time-arcs.
- Alternatives (a–c) are not good options, because as (Ndiv) increases, the necessary computational effort to solve the NLP problem {Equation (1C) + Equation (2i–v)} grows significantly due to considerable increase in the number of searching variables. Thus, the quick (real-time) implementation of the derived FBR operating policy becomes questionable. Additionally, multiple optimal operating policies can exist for the resulted over-parameterized constrained NLP optimization problem, thus increasing the difficulty to quickly locate a feasible globally optimal solution of the FBR optimization problem.”
- The alternative (d) is unlikely because it cannot limit the excessive consumption of substrates and cannot prevent the hydrodynamic stress or the excessive product dilution. Moreover, such an alternative might indicate unrealistic results outside the validity of the used kinetic model [36], whose plausibility is strictly restricted to the investigated experimental domain (Table 1). “In our numerical analysis, carefully documented upper bounds of control variables Equation (2iii) were tested to ensure realistic results for the FBR optimal operating policy.”
- “The alternative (e) is also not feasible, even if a larger (Ndiv) will be used (see (c)). That is because, it is well-known that the variability of the FBR feeding over the batch time-arcs is the main degree-of-freedom used to obtain FBR optimal operating policies of superior quality” [25,28,45,59]. By giving up to the variable feed flow rate, the enzymes, substrate, and cofactor concentrations, suboptimal FBR operating policies will be obtained and of low performance.
3.5. The Used Solver and the Problem Solution Particularities
3.6. Optimized BR and SEQBR Policies
- (a)
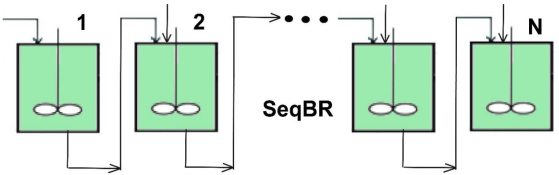
- The optimization of the initial load of a simple single BR of Maria [27,36]. The optimal BR policies have been derived by using an equivalent composite multiobjective function seeking the minimum enzyme (MDH,FDH) consumption simultaneously with realizing a maximum mannitol production under the multiple constraints similar to Equation (2i–v). To realize the equivalence in the direct comparison terms of the present FBR with (Ndiv) = 10 equal time-arcs, a N = 10× times repeated (cyclic) BR (of the same size) and an optimized series SeqBR of NBR = 10 equal BR-s (of the same size) are considered below in the discussion (Section 4).
- (b)
- The optimization of a SeqBR with a series of NBR = 10 BR-s (Figure 2) [27]. At the end of each batch, the content of the BR from the series is passed to the next BR but adjusting its initial load (that is [F]o, [NADH]o, [MDH]o, [FDH]o) according to the optimal operating strategy derived by using an equivalent composite multiobjective function realizing the minimum enzyme (MDH,FDH) consumption simultaneously with the maximum mannitol production under multiple constraints similar to Equation (2i–v). For a direct comparison to the analyzed FBR with (Ndiv) = 10 equal time-arcs, NBR = 10 equal BR-s (of the same size) are considered below in the discussion (Section 4).
4. FBR Optimization Results and Discussion
4.1. The Resulted FBR Optimal Operating Policy
- (a)
- In Figure 4A, the simulated key species concentrations dynamics, that is in Equation (2a).
- (b)
- In Figure 4B, the variable policy of the feed flow rate (, j = 1,…, Ndiv) and the liquid volume dynamics.
- (c)
- In Figure 4C, the variable feeding policy with the substrate (, j = 1,…,Ndiv), and with NADH (, j = 1,…, Ndiv),
- (d)
- In Figure 4D, the variable feeding policy with MDH enzyme (, j=1,…,Ndiv).
- (e)
- In Figure 4E, the variable feeding policy with FDH enzyme (, j=1,…,Ndiv).


- (i)
- (ii)
4.2. Comparative Discussion of the Optimization Results
- (i)
- “There is a close connection between the coupling reactions, enzyme concentrations, and the supra-unitary NADH/NAD+ ratio over the batch (Figure 4A). This means that the two reactions are well coupled, [NADH] being higher than [NAD+] for most of the time, that is a NADH/NAD+ ratio higher than 1 most of the time, thus maintaining a process efficiency [36].
- (ii)
- (iii)
- “As revealed by repeated simulations of Maria [36] (not presented here), the BR performances are more sensitive to [MDH] and [NADH] than to [FDH].”
- (iv)
- Bulk [MDH] grows continuously due to its continuous feeding (Figure 4D). By contrast, [FDH] evolution in the bulk phase is more special (Figure 4E). It grows continuously due to the continuous feeding with FDH until a certain time (10 h). After this time, [FDH] in bulk declines because its reduced feeding rate cannot compensate the reactor content dilution rate.
- (v)
- As a general conclusion, the optimally operated FBR reported better performance compared to the optimal SeqBR, which, in turn, reported better performance than the repeated optimally operated BR. Thus, for a comparable mannitol (M) production: (a) the MDH enzyme consumption of FBR is much smaller, that is 10x (vs. SeqBR) and 130x (vs. BR); (b) the FDH enzyme consumption in FBR is also smaller at 9x (vs. SeqBR) and 15–50x (vs. BR); (c) the NADH consumption in FBR is smaller by 2–9x (vs. SeqBR) and 1.5–7x (vs. BR); (d) compared to the experimental (nonoptimal) BR trials of Slatner et al. [13] (within the range of [F]o = 0.1–1 M, [NADH]o = 0.25–1 mM; [NAD+]o = 0.0005 M; [HCOO]o = [F]o; [;] ∈ [0.1–2] kU/L), the optimal FBR reported much smaller enzyme consumptions, that is, 10x for MDH and 4.5x for FDH.
- (vi)
- As revealed by Figure 4D,E, it is not necessary to keep the enzymes feeding at a high level over the whole batch. On the contrary, an initial high addition over the first 1–2 time-arcs, followed by a continuous addition of a low level, are enough to ensure the necessary enzyme concentrations so to result in the required high reaction rates. The same observation is also valid for the NADH feeding policy.
- (vii)
- One of the better FBR performance costs is the final 2× dilution of the reactor content (Figure 4B). However, balancing the product dilution with the considerable savings in terms of the expensive enzymes and the regenerated cofactor, the use of FBR is worth the investment in its (model-based) offline optimal control.
- (viii)
- The relatively large time-arcs (of ca. 5 h) of the FBR allow an easy operation control over each time-arc, but also when switching between them.
- (ix)
- As proved by the present numerical analysis, “solving this engineering problem by using an experimental procedure, as tried by Slatner et al. [13], may not be the best choice because it involves high costs due to the required large number of batch tests”, even if at a small lab-scale.
- (x)
- In the FBR variable operating case, conversion is more difficult to interpret because the substrate is continuously added to the initial one. Due to this FBR operation particularity, the substrate consumption cannot be referred solely to its initial concentration but rather to the added substrate during the batch, following the variable concentration of Figure 4C. This is why the substrate F consumption is computed with the Equation (3) formula. In the present case, the formal conversion is high, that is: (transformed F in M)/(totally fed F) = 72.5%. The lack of substrate near total conversion is not a critical issue as long as there are lot of solutions to solve the product separations problems, that is, the use: (i) of immobilized enzymes and (ii) of efficient technologies to separate saccharides, as is well-presented in the literature [63,64,65].
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Nomenclature
| “Species j concentration | |
| Species j saturation level | |
| FL | Feed flow rate of the liquid |
| kj, k, Kj, , , KMj | Rate constants |
| Number of reactions | |
| Ndiv | Number of time-arcs of equal lengths in which the batch time is uniformly divided; on every time-arc, the control variables are kept constant |
| , , R1, R2 | Species j reaction rate, or the i-th reaction rate |
| Time | |
| Final batch time | |
| VL | The BR liquid volume |
| W | The overall (composite) objective function of the optimization problem Equation (1A,C) |
| Greek Symbols | |
| Δt | Time interval |
| stoichiometric coefficient of species j in the reaction i | |
| Index | |
| o, 0 | Initial |
| inlet | Referring to the FBR feeding |
| F, final | Final |
| Abbreviations | |
| BR | “batch reactor |
| BRP | BR with intermittent addition of enzymes/substrate(s)/additives |
| E | enzyme |
| CSTR | Continuous stirred tank reactor |
| F | D-Fructose |
| FBR | Fed-batch reactor |
| FDH | Formate dehydrogenase |
| FXBR | Fixed-bed solid-liquid continuous reactor |
| Fobj1-Fobj4 | Objective functions defined in eqn.(1B) |
| HFCS | fructose/glucose syrup |
| HCOO− | Formate ion |
| M | Mannitol, or molar concentration |
| MA(S)CR | Mechanically agitated solid-liquid (semi-)continuous reactor |
| MDH | Mannitol dehydrogenase |
| MMA | The numerical algorithm of Maria [51,52] to solve optimization problems |
| Min/Max | minimum/maximum |
| NAD(P)H | nicotinamide adenine dinucleotide (phosphate) |
| NAD, NAD+ | Nicotinamide adenine dinucleotide (oxidized form) » |
| NLP | Nonlinear optimization problem, subjected to nonlinear constraints |
| NBR | Number of equal BR included in the SeqBR series |
| SeqBR | Sequential batch-to-batch reactor |
| SBR | Semi-batch reactor” |
| T1 …T10 | Switching times between time-arcs |
| [X] | Concentration of the species X |
| Λ | « and » (simultaneously), in the math sense. |
References
- Liese, A.; Seelbach, K.; Wandrey, C. Industrial Biotransformations; Wiley-VCH: Weinheim, Germany, 2006. [Google Scholar]
- Straathof, A.J.J.; Adlercreutz, P. Applied Biocatalysis; Harwood Academic Publ.: Amsterdam, The Netherlands, 2005. [Google Scholar]
- Wikipedia, “Mannitol”. 2021. Available online: https://en.wikipedia.org/wiki/Mannitol (accessed on 7 August 2021).
- Saha, B.C.; Racine, F.M. Biotechnological production of mannitol and its application. Appl. Microbiol. Biotechnol. 2011, 89, 879–891. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, S.M.; Mohan, A.; Srinivastava, S.K. Challenges in enzymatic route of mannitol production. ISRN Biotechnol. 2013, 2013, 914187. [Google Scholar] [CrossRef] [PubMed]
- Von Weymarn, N. Process Development for Mannitol Production by Lactic acid Bacteria. Ph.D. Thesis, Helsinki University of Technology, Laboratory of Bioprocess Engineering, Beijing, China, 2002. Available online: http://lib.tkk.fi/Diss/2002/isbn9512258854/ (accessed on 7 August 2021).
- Ghoreishi, S.M.; Shahrestani, R.G. Innovative strategies for engineering mannitol production. Trends Food Sci. Technol. 2009, 20, 263–268. [Google Scholar] [CrossRef]
- Makkee, M.; Kieboom, A.P.G.; Van Bekkum, H. Production methods of D-mannitol. Starch/Staerke 1985, 37, 136–141. [Google Scholar] [CrossRef]
- Khan, A.; Bhide, A.; Gadre, R. Mannitol production from glycerol by resting cells of Candida magnoliae. Bioresour. Technol. 2009, 100, 4911–4913. [Google Scholar] [CrossRef] [PubMed]
- Song, K.H.; Lee, J.K.; Song, J.Y.; Hong, S.G.; Baek, H.; Kim, S.Y.; Hyun, H.H. Production of mannitol by a novel strain of Candida magnoliae. Biotechnol. Lett. 2002, 24, 9–12. [Google Scholar] [CrossRef]
- Loesche, W.J.; Kornman, K.S. Production of mannitol by Streptococcus mutans. Arch. Oral Biol. 1976, 21, 551–553. [Google Scholar] [CrossRef]
- Bäumchen, C.; Roth, A.H.F.J.; Biedendieck, R.; Malten, M.; Follmann, M.; Sahm, H.; Bringer-Meyer, S.; Jahn, D. D-Mannitol production by resting state whole cell biotransformation of D-fructose by heterologous mannitol and formate dehydrogenase gene expression in Bacillus megentarium. Biotechnol. J. 2007, 2, 1408–1416. [Google Scholar] [CrossRef]
- Slatner, M.; Nagl, G.; Haltrich, D.; Kulbe, K.D.; Nidetzky, B. Enzymatic production of pure D-mannitol at high productivity. Biocatal. Biotransform. 1998, 16, 351–363. [Google Scholar] [CrossRef]
- Chenault, H.K.; Whitesides, G.M. Regeneration of nicotinamide cofactors for use in organic synthesis. Appl. Biochem. Biotechnol. 1987, 14, 147–197. [Google Scholar] [CrossRef]
- Maria, G.; Ene, M.D. Modelling enzymatic reduction of 2-keto-D-glucose by suspended aldose reductase. Chem. Biochem. Eng. Q. 2013, 27, 385–395. Available online: http://silverstripe.fkit.hr/cabeq/assets/Uploads/Cabeq-2013-04-01.pdf (accessed on 30 December 2021).
- Peacock, D.; Boulter, D. Kinetic studies of formate dehydrogenase. Biochem. J. 1970, 120, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.; Michalik, C.; Zavrel, M.; Spiess, A.; Marquardt, W.; Ansorge-Schumacher, M.B. Mechanistic model for prediction of formate dehydrogenase kinetics under industrially relevant conditions. Biotechnol. Prog. 2010, 26, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Moulijn, J.A.; Makkee, M.; van Diepen, A. Chemical Process Technology; Wiley: New York, NY, USA, 2001. [Google Scholar]
- Buchholz, K.; Hempel, D.C. From Gene to Product (Editorial). Eng. Life Sci. 2006, 6, 437. [Google Scholar] [CrossRef]
- Hempel, D.C. Development of biotechnological processes by integrating genetic and engineering methods. Eng. Life Sci. 2006, 6, 443–447. [Google Scholar] [CrossRef]
- Xue, R.; Woodley, J.M. Process technology for multi-enzymatic reaction systems. Bioresour. Technol. 2012, 115, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Blanch, H.W.; Clark, D.S. Biochemical Engineering; Marcel Dekker: New York, NY, USA, 1997. [Google Scholar]
- Hames, B.D.; Hooper, N.M.; Houghton, J.D. Instant Notes in Biochemistry; Springer Verlag: New York, NY, USA, 1997. [Google Scholar]
- Maria, G.; Ene, M.D.; Jipa, I. Modelling enzymatic oxidation of D-glucose with pyranose 2-oxidase in the presence of catalase. J. Mol. Catal. B Enzym. 2012, 74, 209–218. [Google Scholar] [CrossRef]
- Mendes, R.; Rocha, I.; Pinto, J.P.; Ferreira, E.C.; Rocha, M. Differential Evolution for the Offline and Online Optimization of Fed-Batch Fermentation Processes. In Advances in Differential Evolution. Studies in Computational Intelligence, Chakraborty; Chakraborty, U.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 299–317. [Google Scholar]
- Maria, G.; Crisan, M. Operation of a mechanically agitated semi-continuous multi-enzymatic reactor by using the Pareto-optimal multiple front method. J. Process Control. 2017, 53, 95–105. [Google Scholar] [CrossRef]
- Maria, G.; Peptănaru, I.M. Model-based optimization of mannitol production by using a sequence of batch reactors for a coupled bi-enzymatic process–A dynamic approach. Dyn.-Basel 2021, 1, 134–154. [Google Scholar] [CrossRef]
- Maria, G. Model-based optimization of a fed-batch bioreactor for mAb production using a hybridoma cell culture. Mol.-Basel–Org. Chem. 2020, 25, 5648. [Google Scholar] [CrossRef]
- Bonvin, D. Optimal operation of batch reactors—A personal view. J. Process Control. 1998, 8, 355–368. [Google Scholar] [CrossRef]
- Smets, I.Y.; Claes, J.E.; November, E.J.; Bastin, G.P.; van Impe, J.F. Optimal adaptive control of (bio)chemical reactors: Past, present and future. J. Process Control. 2004, 14, 795–805. [Google Scholar] [CrossRef]
- Srinivasan, B.; Bonvin, D.; Visser, E.; Palanki, S. Dynamic optimization of batch processes: II. Role of measurements in handling uncertainty. Comput. Chem. Eng. 2003, 27, 27–44. [Google Scholar] [CrossRef]
- Martinez, E. Batch-to-batch optimization of batch processes using the STATSIMPLEX search method. In Proceedings of the 2nd Mercosur Congress on Chemical Engineering, Rio de Janeiro, Costa Verde, Brasil, 14–18 August 2005; p. 20. [Google Scholar]
- DiBiasio, D. Introduction to the control of biological reactors. In Chemical Engineering Problems in Biotechnology; Shuler, M.L., Ed.; American Institute of Chemical Engineers: New York, NY, USA, 1989; pp. 351–391. [Google Scholar]
- Rao, M.; Qiu, H. Process Control Engineering: A Textbook for Chemical, Mechanical and Electrical Engineers; Gordon and Breach Science Publ.: Amsterdam, The Netherlands, 1993. [Google Scholar]
- Hartig, F.; Keil, F.J.; Luus, R. Comparison of optimization methods for a fed-batch reactor. Hung. J. Ind. Chem. 1995, 23, 81–160. [Google Scholar]
- Maria, G. Model-based optimisation of a batch reactor with a coupled bi-enzymatic process for mannitol production. Comput. Chem. Eng. 2020, 133, 106628–106635. [Google Scholar] [CrossRef]
- Koller, M. A review on established and emerging fermentation schemes for microbial production of polyhydroxyalkanoate (PHA) biopolyesters. Fermentation 2018, 4, 30. [Google Scholar] [CrossRef]
- Irvine, R.L.; Busch, A.W. Sequencing batch biological reactors–An overview. J. Water Pollut. Control. Fed. 1979, 51, 235–243. Available online: http://www.jstor.org/stable/25039819 (accessed on 20 January 2022).
- Shacham, M. Semicontinuous fed-batch and cyclic-fed batch operation. In Proceedings of the Workshop “Modern Problem Solving Techniques in Engineering with POLYMATH, Excel and MATLAB”, Tel-Aviv, Israel, 23 September 2008; pp. 1439–1441, Chapter 14.13. Available online: http://www.eng.tau.ac.il/~brauner/Workshop08Participant/Example-7/Prob-14-13.pdf (accessed on 23 February 2021).
- US EPA. Wastewater Technology Fact Sheet: Sequencing Batch Reactors; OWM, Water Permits Division, Municipal Branch: Washington, WC, USA, 2012. Available online: https://www3.epa.gov/npdes/pubs/sbr_new.pdf (accessed on 23 February 2021).
- Rätze, K.H.G.; Jokiel, M.; Sundmacher, K. Cyclic operation of a semibatch reactor for the hydroformylation of long-chain olefins and integration in a continuous production process. Chem. Eng. J. 2019, 377, 120453. [Google Scholar] [CrossRef]
- Sawatzki, A.; Hans, S.; Narayanan, H.; Haby, B.; Krausch, N.; Sokolov, M.; Glauche, F.; Riedel, S.L.; Neubauer, P.; Bournazou, M.N.C. Accelerated bioprocess development of endopolygalacturonase-production with Saccharomyces cerevisiae using multivariate prediction in a 48 mini-bioreactor automated platform. Bioengineering 2018, 5, 101. [Google Scholar] [CrossRef]
- Hans, S.; Ulmer, C.; Narayanan, H.; Brautaset, T.; Krausch, N.; Neubauer, P.; Schäffl, I.; Sokolov, M.; Bournazou, M.N.C. Monitoring parallel robotic cultivations with online multivariate analysis. Processes 2020, 8, 582. [Google Scholar] [CrossRef]
- Maria, G.; Crisan, M. Evaluation of optimal operation alternatives of reactors used for D-glucose oxidation in a bi-enzymatic system with a complex deactivation kinetics. Asia-Pac. J. Chem. Eng. 2014, 10, 22–44. [Google Scholar] [CrossRef]
- Avili, M.G.; Fazaelipoor, M.H.; Jafari, S.A.; Ataei, S.A. Comparison between batch and fed-batch production of rhamnolipid by Pseudomonas aeruginosa. Iran. Jl. Biotechnol. 2012, 10, 263–269. Available online: https://www.researchgate.net/publication/288966207_Comparison_between_batch_and_fed-batch_production_of_rhamnolipid_by_Pseudomonas_aeruginosa (accessed on 9 January 2022).
- Amribt, Z.; Dewasme, L.; Wouwer, A.V.; Bogaerts, P. Optimization and robustness analysis of hybridoma cell fed-batch cultures using the overflow metabolism model. Bioprocess Biosyst. Eng. 2014, 37, 1637–1652. [Google Scholar] [CrossRef] [PubMed]
- Froment, G.F.; Bischoff, K.B. Chemical Reactor Analysis and Design; Wiley: New York, NY, USA, 1990. [Google Scholar]
- Franco-Lara, E.; Weuster-Botz, D. Estimation of optimal feeding strategies for fed-batch bioprocesses. Bioprocess Biosyst. Eng. 2005, 28, 71. [Google Scholar] [CrossRef][Green Version]
- Maria, G. Model-based heuristic optimised operating policies for D-glucose oxidation in a batch reactor with pulsate addition of enzyme. Comput. Chem. Eng. 2007, 31, 1231–1241. [Google Scholar] [CrossRef]
- Wikipedia, “Bioreactor”. 2021. Available online: https://en.wikipedia.org/wiki/Bioreactor (accessed on 30 December 2021).
- Hamza, M.H. (Ed.) Modelling, Identification and Control; IASTED/ACTA Press: Anaheim, CA, USA, 2003; pp. 112–118. Available online: https://www.actapress.com/Content_of_Proceeding.aspx?proceedingID=213 (accessed on 7 March 2021).
- Maria, G. A review of algorithms and trends in kinetic model identification for chemical and biochemical systems. Chem. Biochem. Eng. Q. 2004, 18, 195–222. [Google Scholar] [CrossRef]
- Moser, A. Bioprocess Technology: Kinetics and Reactors; Springer: New York, NY, USA, 1988. [Google Scholar]
- Carroll, J.J.; Mather, A.E. The system carbon dioxide-water and the Krichevsky-Kasarnovsky equation. J. Solut. Chem. 1992, 21, 607–621. [Google Scholar] [CrossRef]
- Reid, R.C.; Prausnitz, J.M.; Poling, B.E. The Properties of Gases and Liquids, 4th ed.; McGraw-Hill: Boston, MA, USA, 1987. [Google Scholar]
- Dutta, R. Fundamentals of Biochemical Engineering; Springer: Berlin/Heidelberg, Germany, 2008. [Google Scholar]
- Rao, S.S. Engineering Optimization—Theory and Practice; Wiley: New York, NY, USA, 2009; Chapter 14.10. [Google Scholar]
- Scoban, A.G.; Maria, G. Model-based optimization of the feeding policy of a fluidized bed bioreactor for mercury uptake by immobilized P. putida cells. Asia-Pac. J. Chem. Eng. 2016, 11, 721–734. [Google Scholar] [CrossRef]
- Maria, G.; Renea, L. Tryptophan production maximization in a fed-batch bioreactor with modified E. coli cells, by optimizing its operating policy based on an extended structured cell kinetic model. Bioeng.-Basel 2021, 8, 210. [Google Scholar] [CrossRef]
- Bharat, A.; Process Analytical Technology (PAT). Msc. Diss., P.D.V.V.P.F.S. College of pharmacy, AhmedNagar, India. 2013. Available online: https://www.slideshare.net/anjalibharat19/process-analytical-tchnology (accessed on 21 October 2021).
- Loeblein, C.; Perkins, J.; Srinivasan, B.; Bonvin, D. Performance analysis of on-line batch optimization systems. Comput. Chem. Eng. 1997, 21, S867–S872. [Google Scholar] [CrossRef]
- Maria, G. Adaptive random search and short-cut techniques for process model identification and monitoring. In Proceedings of the FOCAPO98 International Conference on Foundations of Computer Aided Process Operations, Snowbird, UT, USA, 5–10 July 1998; pp. 351–359. [Google Scholar]
- Gillarová, S.; Henke, S.; Svoboda, T.; Kadlec, P.; Hinková, A.; Bubník, Z.; Pour, V.; Sluková, M. Chromatographic separation of mannitol from mixtures of other carbohydrates in aqueous solutions. Czech J. Food Sci. 2021, 39, 281–288. [Google Scholar] [CrossRef]
- Blignaut, J.; Albataineh, K.; Banat, Y.; Abu El-Rub, Z. Comparison of Adsorbent Behavior in Glucose/Fructose Separation by Simulated Moving Bed (SMB) Chromatography, Res. Report; University of Twente: Enschede, The Netherlands, 2001. [Google Scholar]
- Lee, K.N. Continuous Separation of Glucose and Fructose at High Concentration Using Two-Section Simulated Moving Bed Process. Korean J. Chem. Eng. 2003, 20, 532–537. [Google Scholar] [CrossRef]
- Rapaport, A.; Dochain, D. Minimal time control of fed-batch processes with growth functions having several maxima. IEEE Trans. Autom. Control. 2011, 56, 2671–2676. Available online: https://hal.archives-ouvertes.fr/hal-00529001/document (accessed on 15 January 2022). [CrossRef]
- Gajardo, P.; Ramirez, H.C.; Rapaport, A.E. Minimal time sequential batch reactors with bounded and impulse controls for one or more species. SIAM J. Control. Optim. 2008, 47, 2827–2856. [Google Scholar] [CrossRef][Green Version]
- Flores, V.H.R. Optimal Control Problems for Bioremediation of Water Resources. Ph.D. Thesis, Universidad de Chile, Santiago, Chile, 2016. Available online: https://repositorio.uchile.cl/bitstream/handle/2250/141336/Problemas-de-control-optimo-para-la-biorremediacion-de-recursos-acuiferos.pdf?sequence=1&isAllowed=y (accessed on 15 January 2022).
| Reactor Type | F consumption (Moles/h) | M Production (M/h) | MDH Consumption (kU/L) | FDH Consumption (kU/L) | NADH Consumption (moles/h) | Remarks |
|---|---|---|---|---|---|---|
| Optimal BR (a,c) | 0.021–0.062 (c) | 0.017–0.054 (c) | 1–13.88 (c) | 3–10 (c) | 0.002–0.010 (c) | 10 BR cycles of 48 h each |
| Optimal SeqBR (b) | 0.025–0.075 (b) | 0.012–0.044 (b) | 1.112 | 1.755 | 0.0025–0.0125 (b) | Series of 10 BR, 40 h run for each BR |
| Optimal FBR (this paper) | 0.0362 | 0.0262 | 0.0882 kU/(0.5–1.2 L FBR) | 0.1686 kU/(0.5–1.2 L FBR) | 0.0014 | Single batch of 48 h |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maria, G.; Renea, L.; Maria, C. Multiobjective Optimization of a Fed-Batch Bienzymatic Reactor for Mannitol Production. Dynamics 2022, 2, 270-294. https://doi.org/10.3390/dynamics2030014
Maria G, Renea L, Maria C. Multiobjective Optimization of a Fed-Batch Bienzymatic Reactor for Mannitol Production. Dynamics. 2022; 2(3):270-294. https://doi.org/10.3390/dynamics2030014
Chicago/Turabian StyleMaria, Gheorghe, Laura Renea, and Cristina Maria. 2022. "Multiobjective Optimization of a Fed-Batch Bienzymatic Reactor for Mannitol Production" Dynamics 2, no. 3: 270-294. https://doi.org/10.3390/dynamics2030014
APA StyleMaria, G., Renea, L., & Maria, C. (2022). Multiobjective Optimization of a Fed-Batch Bienzymatic Reactor for Mannitol Production. Dynamics, 2(3), 270-294. https://doi.org/10.3390/dynamics2030014






