Selection and Characterization of Antibodies Recognizing Unnatural Base Pairs
Abstract
1. Introduction
2. Materials and Methods
2.1. Hirao’s Pair Synthesis
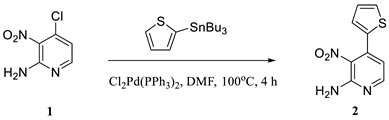
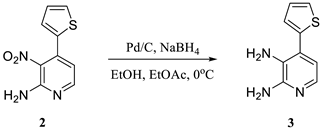
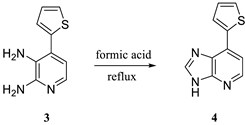
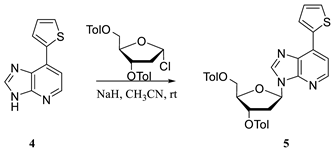
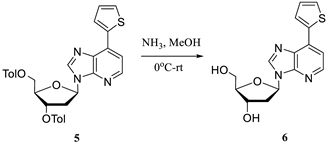

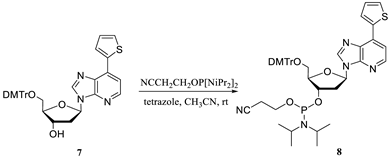
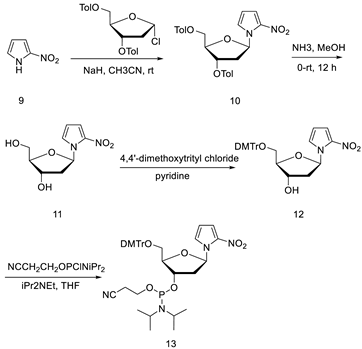
2.2. Antibody Selection
2.2.1. Antigen Design
2.2.2. Phage Display Selection
2.2.3. Yeast Display Selection
2.2.4. Yeast Plasmid Preparation and scFv Gene Sequencing
2.2.5. Specificity of Binding and Kinetic Study of Yeast-Displayed scFvs
2.2.6. Conversion of scFvs to scFv-Fc (Minibody) and IgGs
2.3. Soluble Antibody Characterization Assays
2.3.1. Preparation of Plasmid Containing Unnatural or Natural Oligos (pU or pN)
2.3.2. Common Steps in ELISA/FLISA
2.3.3. Electrophoretic Mobility Shift Assay
3. Results and Discussion
3.1. Antigen Choice and Preparation
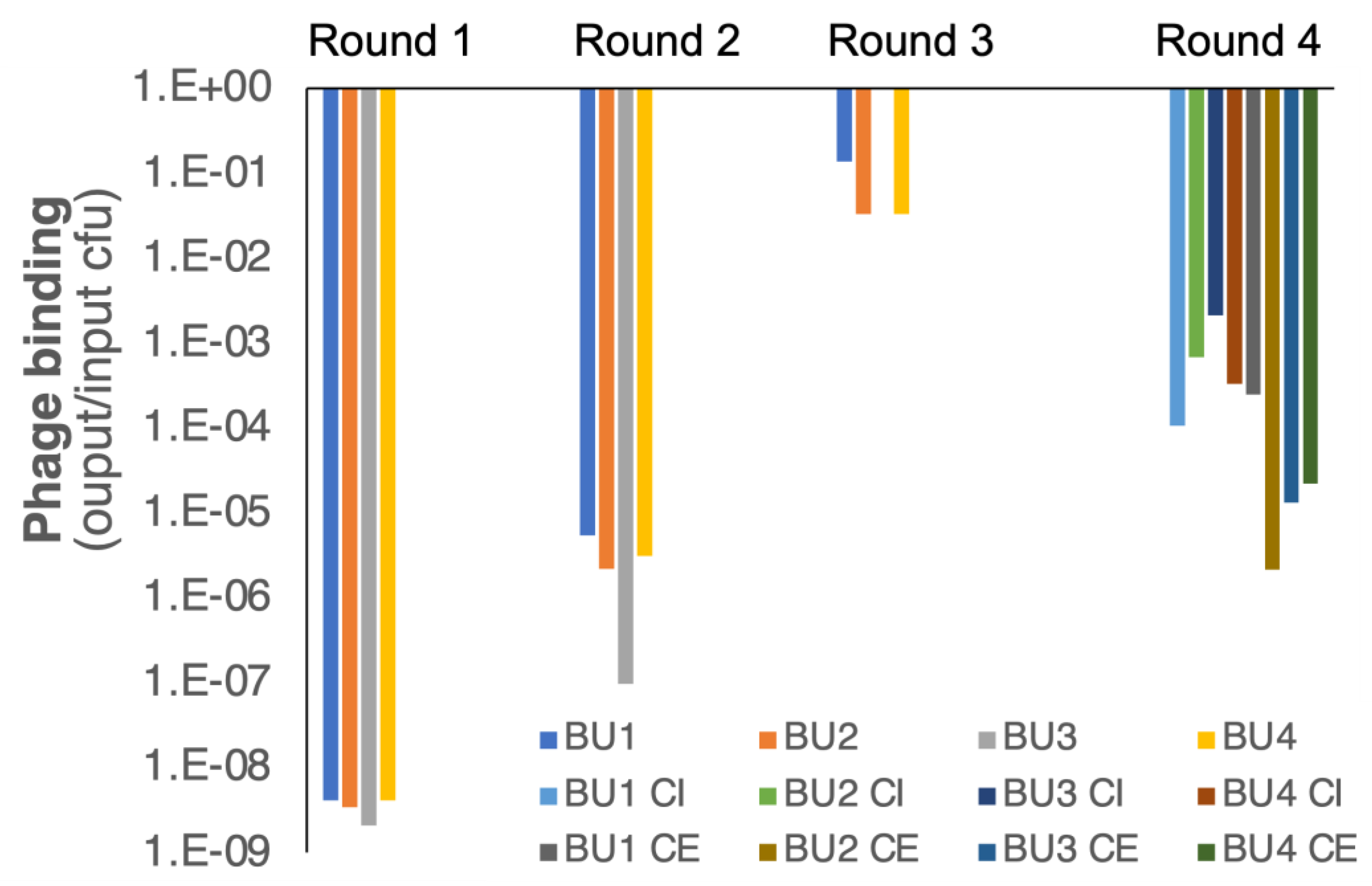
3.2. Antibody Selections
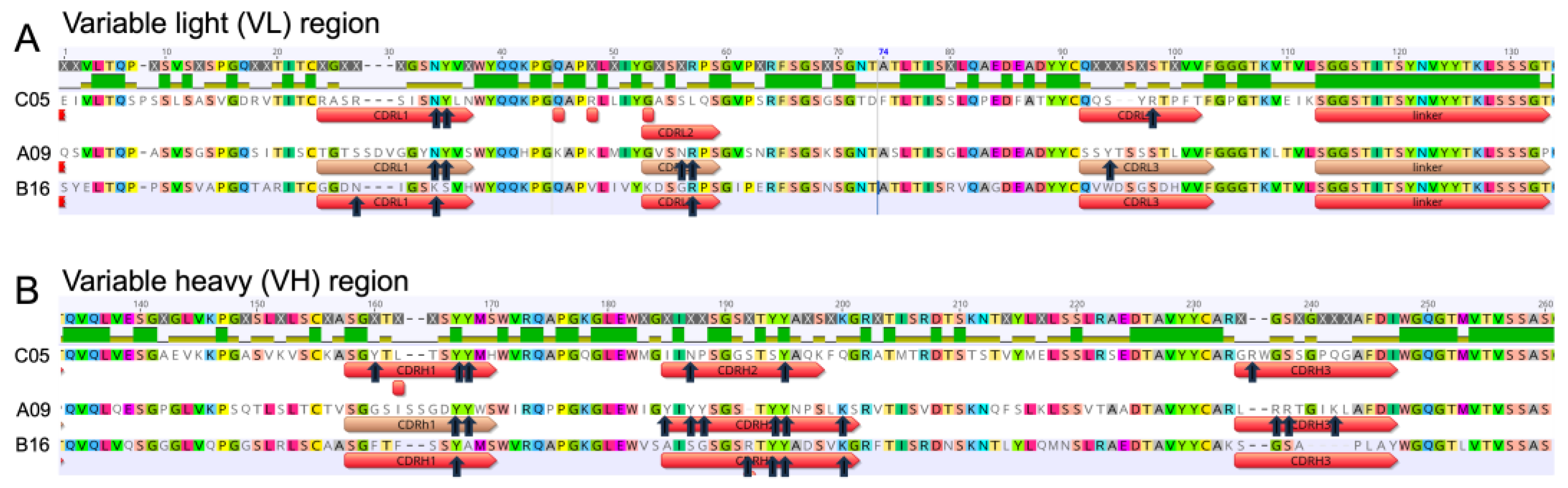
3.3. Single Yeast Clone Characterization
3.4. Characterization of Antibodies in Soluble Form
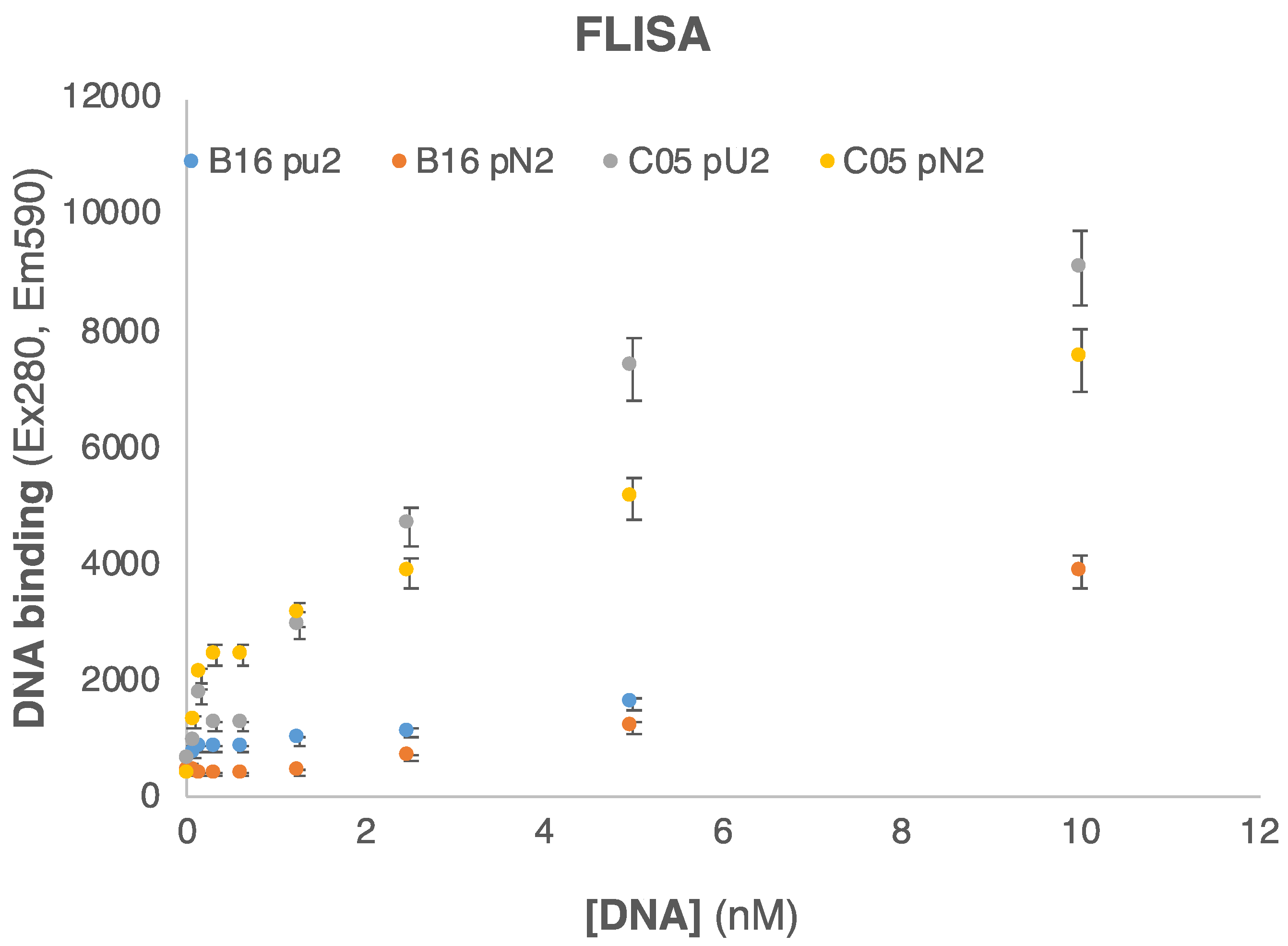
3.5. Production of Real Sample Surrogates
3.6. Immunoassays for Detection of Real Samples Surrogates
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BSA | bovine serum albumin |
| BU | biotinylated U |
| BN | biotinylated N |
| pU | plasmid containing U |
| CI | competition at the incubation step |
| CE | competition at the elution step |
| CDR | complementarity determining regions |
| ELISA | enzyme-linked immunosorbent assay |
| EMSA | electrophoretic mobility assay |
| FLISA | fluorescence-linked immunosorbent assay |
| HRP | horse radish peroxidase |
| IgG | Immunoglobulin G |
| N | all-natural DNA oligo |
| PAGE | polyacrylamide gel electrophoresis |
| PBS | phosphate-buffered saline |
| PCR | polymerase chain reaction |
| pN | plasmid containing N |
| scFv | single chain fragment crystallizable |
| SSO | semisynthetic organism |
| TBE | tris/borate/EDTA |
| UBPs | unnatural base pairs |
| U | unnatural base pair-containing DNA oligo |
References
- Dien, V.T.; Morris, S.E.; Karadeema, R.J.; Romesberg, F.E. Expansion of the genetic code via expansion of the genetic alphabet. Curr. Opin. Chem. Biol. 2018, 46, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Hoshika, S.; Leal, N.A.; Kim, M.-J.; Kim, M.-S.; Karalkar, N.B.; Kim, H.-J.; Bates, A.M.; Watkins, N.E., Jr.; SantaLucia, H.A.; Meyer, A.J. Hachimoji DNA and RNA: A genetic system with eight building blocks. Science 2019, 363, 884–887. [Google Scholar] [CrossRef]
- Kimoto, M.; Yamashige, R.; Matsunaga, K.-i.; Yokoyama, S.; Hirao, I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat. Biotechnol. 2013, 31, 453–457. [Google Scholar] [CrossRef]
- Sefah, K.; Yang, Z.; Bradley, K.M.; Hoshika, S.; Jiménez, E.; Zhang, L.; Zhu, G.; Shanker, S.; Yu, F.; Turek, D. In vitro selection with artificial expanded genetic information systems. Proc. Natl. Acad. Sci. USA 2014, 111, 1449–1454. [Google Scholar] [CrossRef] [PubMed]
- Akram, F.; Ali, H.; Laghari, A.T. Trends to store digital data in DNA: An overview. Mol. Biol. Rep. 2018, 45, 1479–1490. [Google Scholar] [CrossRef] [PubMed]
- Ceze, L.; Nivala, J.; Strauss, K. Molecular digital data storage using DNA. Nat. Rev. Genet. 2019, 20, 456–466. [Google Scholar] [CrossRef] [PubMed]
- de la Torre, D.; Chin, J.W. Reprogramming the genetic code. Nat. Rev. Genet. 2021, 22, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Romesberg, F.E. Creation, Optimization, and Use of Semi-Synthetic Organisms that Store and Retrieve Increased Genetic Information. J. Mol. Biol. 2021, 434, 167331. [Google Scholar] [CrossRef] [PubMed]
- Romesberg, F.E. Discovery, implications and initial use of semi-synthetic organisms with an expanded genetic alphabet/code. Philos. Trans. R. Soc. B 2023, 378, 20220030. [Google Scholar] [CrossRef]
- Synthorx Inc., a.S.c. A Study Evaluating Safety and Therapeutic Activity of THOR-707 in Adult Subjects with Advanced or Metastatic Solid Tumors (THOR-707-101); Synthorx Inc.: La Jolla, CA, USA, 2019. [Google Scholar]
- Craig, J.M.; Laszlo, A.H.; Derrington, I.M.; Ross, B.C.; Brinkerhoff, H.; Nova, I.C.; Doering, K.; Tickman, B.I.; Svet, M.T.; Gundlach, J.H. Direct detection of unnatural DNA nucleotides dNaM and d5SICS using the MspA nanopore. PLoS ONE 2015, 10, e0143253. [Google Scholar] [CrossRef] [PubMed]
- Ledbetter, M.P.; Craig, J.M.; Karadeema, R.J.; Noakes, M.T.; Kim, H.C.; Abell, S.J.; Huang, J.R.; Anderson, B.A.; Krishnamurthy, R.; Gundlach, J.H. Nanopore sequencing of an expanded genetic alphabet reveals high-fidelity replication of a predominantly hydrophobic unnatural base pair. J. Am. Chem. Soc. 2020, 142, 2110–2114. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Fang, L.; Yu, G.; Wang, D.; Xiao, C.-L.; Wang, K. Detection of DNA base modifications by deep recurrent neural network on Oxford Nanopore sequencing data. Nat. Commun. 2019, 10, 2449. [Google Scholar] [CrossRef]
- Stollar, B. The origin and pathogenic role of anti-DNA autoantibodies. Curr. Opin. Immunol. 1990, 2, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Stollar, B.D. Immunochemistry of DNA. Int. Rev. Immunol. 1989, 5, 1–22. [Google Scholar] [CrossRef]
- Reynaud, C.; Bruno, C.; Boullanger, P.; Grange, J.; Barbesti, S.; Niveleau, A. Monitoring of urinary excretion of modified nucleosides in cancer patients using a set of six monoclonal antibodies. Cancer Lett. 1992, 61, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Levine, L.; Plescia, O. Nucleic Acids as Antigens. In Progress in Immunology; Elsevier: Amsterdam, The Netherlands, 1971; pp. 1211–1213. [Google Scholar]
- Erlanger, B.F.; Beiser, S.M. Antibodies specific for ribonucleosides and ribonucleotides and their reaction with DNA. Proc. Natl. Acad. Sci. USA 1964, 52, 68–74. [Google Scholar] [CrossRef]
- Feederle, R.; Schepers, A. Antibodies specific for nucleic acid modifications. RNA Biol. 2017, 14, 1089–1098. [Google Scholar] [CrossRef]
- Scott, J.K.; Smith, G.P. Searching for Peptide Ligands with an Epitope Library. Science 1990, 249, 386–390. [Google Scholar] [CrossRef]
- Bradbury, A.R.M.; Marks, J.D. Antibodies from phage antibody libraries. J. Immunol. Methods 2004, 290, 29–49. [Google Scholar] [CrossRef]
- Boder, E.T.; Wittrup, K.D. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 1997, 15, 553–557. [Google Scholar] [CrossRef]
- Ferrara, F.; Soluri, M.F.; Sblattero, D. Recombinant antibody selections by combining phage and yeast display. In Human Monoclonal Antibodies; Springer: Berlin/Heidelberg, Germany, 2019; pp. 339–352. [Google Scholar]
- Smith, G.P.; Petrenko, V.A. Phage display. Chem. Rev. 1997, 97, 391–410. [Google Scholar] [CrossRef] [PubMed]
- Boder, E.T.; Wittrup, K.D. Yeast surface display for directed evolution of protein expression, affinity, and stability. Methods Enzymol. 2000, 328, 430–444. [Google Scholar]
- Velappan, N.; Micheva-Viteva, S.; Adikari, S.H.; Waldo, G.S.; Lillo, A.M.; Bradbury, A.R. Selection and verification of antibodies against the cytoplasmic domain of M2 of influenza, a transmembrane protein. Proc. Mabs 2020, 12, 1843754. [Google Scholar] [CrossRef]
- Lillo, A.M.; Ayriss, J.E.; Shou, Y.; Graves, S.W.; Bradbury, A.R. Development of phage-based single chain Fv antibody reagents for detection of Yersinia pestis. PLoS ONE 2011, 6, e27756. [Google Scholar] [CrossRef] [PubMed]
- Lillo, A.M.; Velappan, N.; Kelliher, J.M.; Watts, A.J.; Merriman, S.P.; Vuyisich, G.; Lilley, L.M.; Coombs, K.E.; Mastren, T.; Teshima, M. Development of Anti-Yersinia pestis Human Antibodies with Features Required for Diagnostic and Therapeutic Applications. ImmunoTargets Ther. 2020, 9, 299. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lillo, A.M.; Steiniger, S.C.; Liu, Y.; Ballatore, C.; Anichini, A.; Mortarini, R.; Kaufmann, G.F.; Zhou, B.; Felding-Habermann, B. Targeting heat shock proteins on cancer cells: Selection, characterization, and cell-penetrating properties of a peptidic GRP78 ligand. Biochemistry 2006, 45, 9434–9444. [Google Scholar] [CrossRef]
- Lillo, A.M.; Sun, C.; Gao, C.; Ditzel, H.; Parrish, J.; Gauss, C.-M.; Moss, J.; Felding-Habermann, B.; Wirsching, P.; Boger, D.L. A human single-chain antibody specific for integrin α3β1 capable of cell internalization and delivery of antitumor agents. Chem. Biol. 2004, 11, 897–906. [Google Scholar] [CrossRef][Green Version]
- Velappan, N.; Mahajan, A.; Naranjo, L.; Velappan, P.; Andrews, N.; Tiee, N.; Chakraborti, S.; Hemez, C.; Gaiotto, T.; Wilson, B. Selection and characterization of FcεRI phospho-ITAM specific antibodies. Proc. MAbs 2019, 11, 1206–1218. [Google Scholar] [CrossRef] [PubMed]
- Velappan, N.L.; Nguyen, H.B.; Micheva-Viteva, S.; Bedinger, D.; Ye, C.; Mangadu, B.; Watts, A.J.; Meagher, R.; Bradfute, S.; Hu, B.; et al. Healthy humans can be a source of antibodies countering COVID-19. Bioengineered 2022, in press. [CrossRef]
- Shusta, E.V.; Kieke, M.C.; Parke, E.; Kranz, D.M.; Wittrup, K.D. Yeast polypeptide fusion surface display levels predict thermal stability and soluble secretion efficiency. J. Mol. Biol. 1999, 292, 949–956. [Google Scholar] [CrossRef]
- Li, B.; Fouts, A.E.; Stengel, K.; Luan, P.; Dillon, M.; Liang, W.-C.; Feierbach, B.; Kelley, R.F.; Hötzel, I. In vitro affinity maturation of a natural human antibody overcomes a barrier to in vivo affinity maturation. Proc. MAbs 2014, 6, 437–445. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hirao, I.; Mitsui, T.; Kimoto, M.; Yokoyama, S. An efficient unnatural base pair for PCR amplification. J. Am. Chem. Soc. 2007, 129, 15549–15555. [Google Scholar] [CrossRef] [PubMed]
- Sblattero, D.; Bradbury, A. Exploiting recombination in single bacteria to make large phage antibody libraries. Nat. Biotechnol. 2000, 18, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Feldhaus, M.J.; Siegel, R.W.; Opresko, L.K.; Coleman, J.R.; Feldhaus, J.M.W.; Yeung, Y.A.; Cochran, J.R.; Heinzelman, P.; Colby, D.; Swers, J. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat. Biotechnol. 2003, 21, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Wentz, A.E.; Shusta, E.V. A novel high-throughput screen reveals yeast genes that increase secretion of heterologous proteins. Appl. Environ. Microbiol. 2007, 73, 1189–1198. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xia, Y. Anti-double Stranded DNA Antibodies: Origin, Pathogenicity, and Targeted Therapies. Front. Immunol. 2019, 10, 1667. [Google Scholar] [CrossRef]
- Richardson, C.; Chida, A.S.; Adlowitz, D.; Silver, L.; Fox, E.; Jenks, S.A.; Palmer, E.; Wang, Y.; Heimburg-Molinaro, J.; Li, Q.-Z.; et al. Molecular Basis of 9G4 B Cell Autoreactivity in Human Systemic Lupus Erythematosus. J. Immunol. 2013, 191, 4926–4939. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, K.; Herrmann, M.; Winkler, T.H. The role of somatic hypermutation in the generation of pathogenic antibodies in SLE. Autoimmunity 2013, 46, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Akbar, R.; Robert, P.A.; Pavlovic, M.; Jeliazkov, J.R.; Snapkov, I.; Slabodkin, A.; Weber, C.R.; Scheffer, L.; Miho, E.; Haff, I.H.; et al. A compact vocabulary of paratope-epitope interactions enables predictability of antibody-antigen binding. Cell Rep. 2021, 34, 108856. [Google Scholar] [CrossRef]
- Wan, Z.K.; Lee, J.; Xu, W.; Erbe, D.V.; Joseph-McCarthy, D.; Follows, B.C.; Zhang, Y.L. Monocyclic thiophenes as protein tyrosine phosphatase 1B inhibitors: Capturing interactions with Asp48. Bioorganic Med. Chem. Lett. 2006, 16, 4941–4945. [Google Scholar] [CrossRef]
- Anan, Y.; Itakura, M.; Shimoda, T.; Yamaguchi, K.; Lu, P.; Nagata, K.; Dong, J.; Ueda, H.; Uchida, K. Molecular and structural basis of anti-DNA antibody specificity for pyrrolated proteins. Commun. Biol. 2024, 7, 149. [Google Scholar] [CrossRef] [PubMed]
- Koide, S.; Sidhu, S.S. The importance of being tyrosine: Lessons in molecular recognition from minimalist synthetic binding proteins. ACS Chem. Biol. 2009, 4, 325–334. [Google Scholar] [CrossRef] [PubMed]


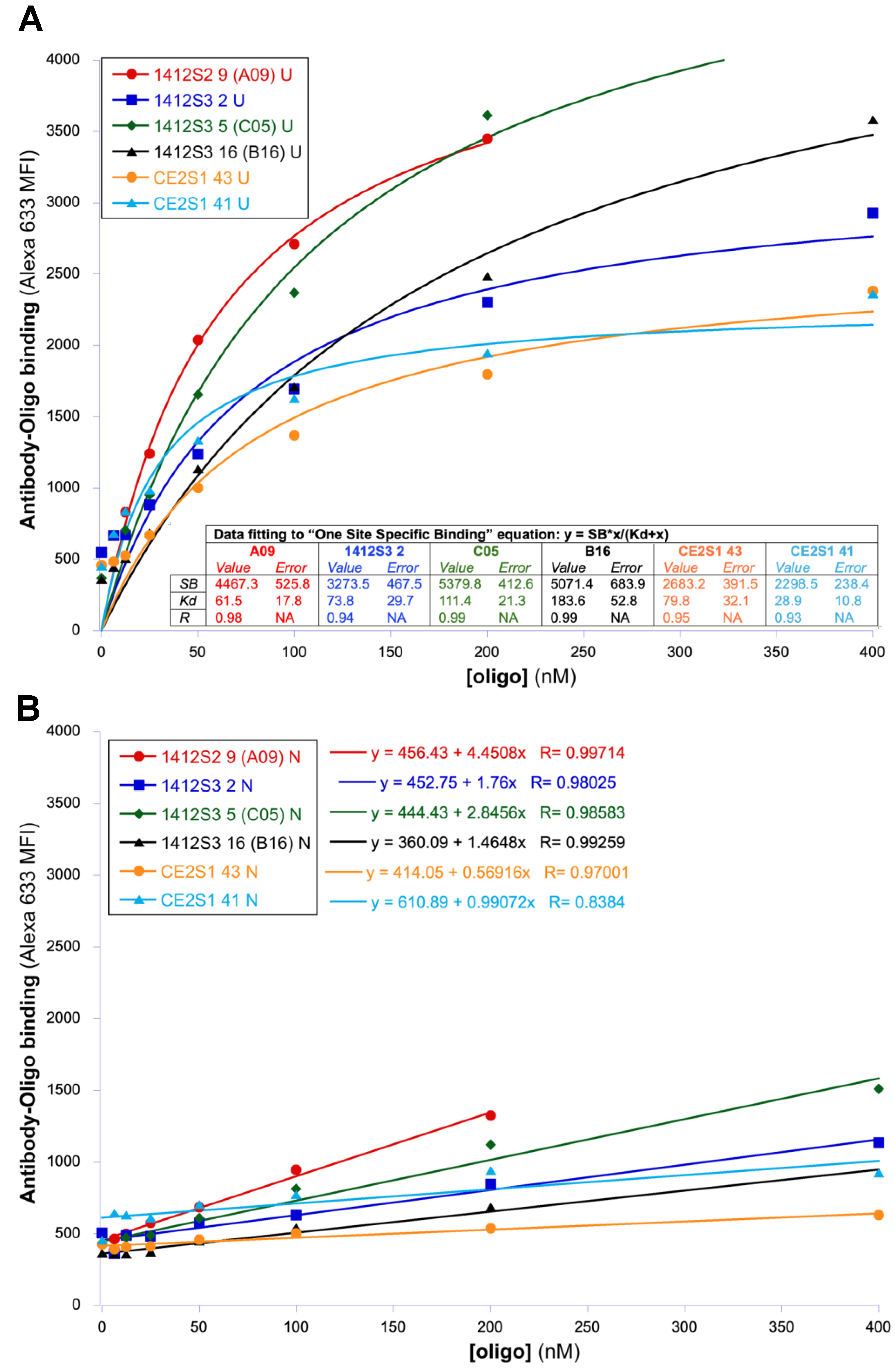
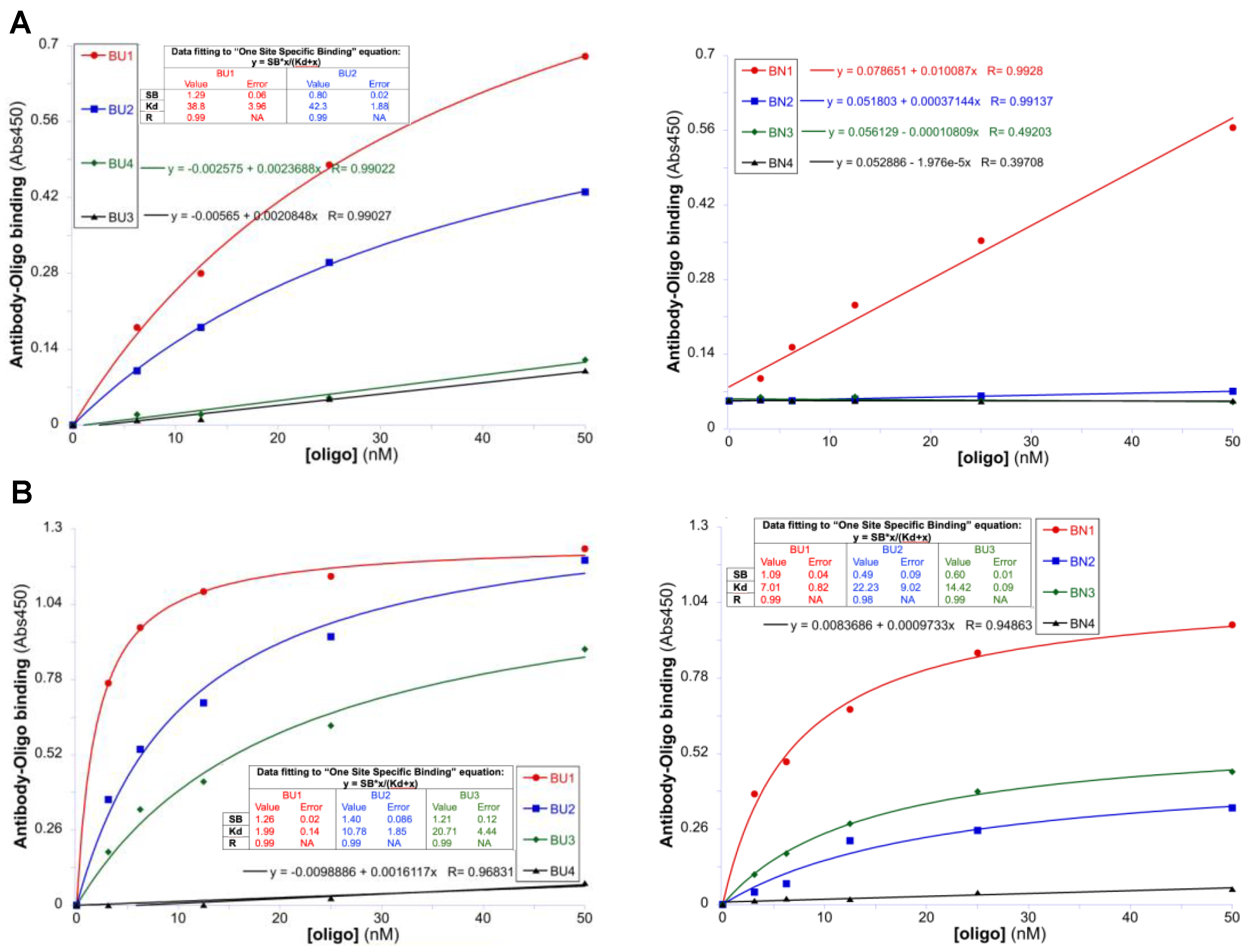
| Antibody Name | Oligo | Interaction with BUs and BNs | Antibody Format | |||
|---|---|---|---|---|---|---|
| Affinity (Kd, nM) a | Saturation Binding b | |||||
| BU | BN | BU | BN | |||
| A09 | 1 | 61.5 ± 17.8 | L/B c | 4467.3 ± 528.8 | N/A | Yeast displayed scFv |
| 1 | 38.8 ± 3.96 | L/B c | 1.3 ± 0.06 | 1.0 ± 0.2 | IgG | |
| 2 | 42.3 ± 1.88 | L/B c | 0.8 ± 0.02 | N/A e | ||
| 3/4 | L/B | N/B d | N/A f | N/A e | ||
| 1 | 29.6 ± 3.3 | L/B c | 0.7 ± 0.04 | N/A e | minibody | |
| 2/3/4 | N/B | N/B d | N/A f | N/A e | ||
| B16 | 1 | 183.6 ± 52.8 | L/B c | 5071.4 ± 683.9 | N/A e | Yeast displayed scFv |
| 1/2/3/4 | N/A f | N/A f | N/A f | N/A f | IgG | |
| 1 | 48.7 ± 11.8 | L/B c | 1.1 ± 0.1 | N/A e | minibody | |
| 2 | 24.4 ± 4.0 | L/B c | 1.1 ± 0.06 | N/A e | ||
| 3 | 77.5 ± 31.5 | L/B c | 0.8 ± 0.2 | N/A e | ||
| 4 | L/B | N/B d | N/A e | N/A e | ||
| C05 | 1 | 111.4 ± 21.3 | L/B c | 5379.8 ± 412.6 | N/A e | Yeast displayed scFv |
| 1 | 2.0 ± 0.1 | 7.0 ± 0.8 | 1.3 ± 0.02 | 1.1 ± 0.04 | IgG | |
| 2 | 10.8 ± 1.8 | 22.2 ± 9.0 | 1.4 ± 0.09 | 0.5 ± 0.09 | ||
| 3 | 20.7 ± 4.4 | 14.4 ± 0.9 | 1.2 ± 0.1 | 0.6 ± 0.01 | ||
| 4 | L/B c | L/B c | N/A e | N/A e | ||
| 1 | 39.3 ± 8.6 | L/B c | 3.1 ± 0.4 | N/A e | minibody | |
| 2 | 41.3 ± 8.6 | L/B c | 1.4 ± 0.2 | N/A e | ||
| 3 | 42.8 ± 8.6 | L/B c | 1.5 ± 0.3 | N/A e | ||
| 4 | 97.2 ± 31.3 | N/B d | N/A e | N/A e | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lillo, A.M.; Velappan, N.; Wu, R.; Bolding, M.R. Selection and Characterization of Antibodies Recognizing Unnatural Base Pairs. Biologics 2024, 4, 423-443. https://doi.org/10.3390/biologics4040026
Lillo AM, Velappan N, Wu R, Bolding MR. Selection and Characterization of Antibodies Recognizing Unnatural Base Pairs. Biologics. 2024; 4(4):423-443. https://doi.org/10.3390/biologics4040026
Chicago/Turabian StyleLillo, Antonietta M., Nileena Velappan, Ruilian Wu, and Madeline R. Bolding. 2024. "Selection and Characterization of Antibodies Recognizing Unnatural Base Pairs" Biologics 4, no. 4: 423-443. https://doi.org/10.3390/biologics4040026
APA StyleLillo, A. M., Velappan, N., Wu, R., & Bolding, M. R. (2024). Selection and Characterization of Antibodies Recognizing Unnatural Base Pairs. Biologics, 4(4), 423-443. https://doi.org/10.3390/biologics4040026






