Unveiling Novel Insights in Helminth Proteomics: Advancements, Applications, and Implications for Parasitology and Beyond
Abstract
1. Introduction
2. Methods
3. Proteomics in Nematodes
3.1. Heligmosomoides polygyrus
3.2. Ascaris suum
3.3. Toxocara canis
3.4. Toxocara cati
3.5. Nippostrongylus brasiliensis
3.6. Ancylostoma caninum
3.7. Haemonchus contortus
3.8. Trichostrongylus colubriformis
3.9. Trichinella spiralis
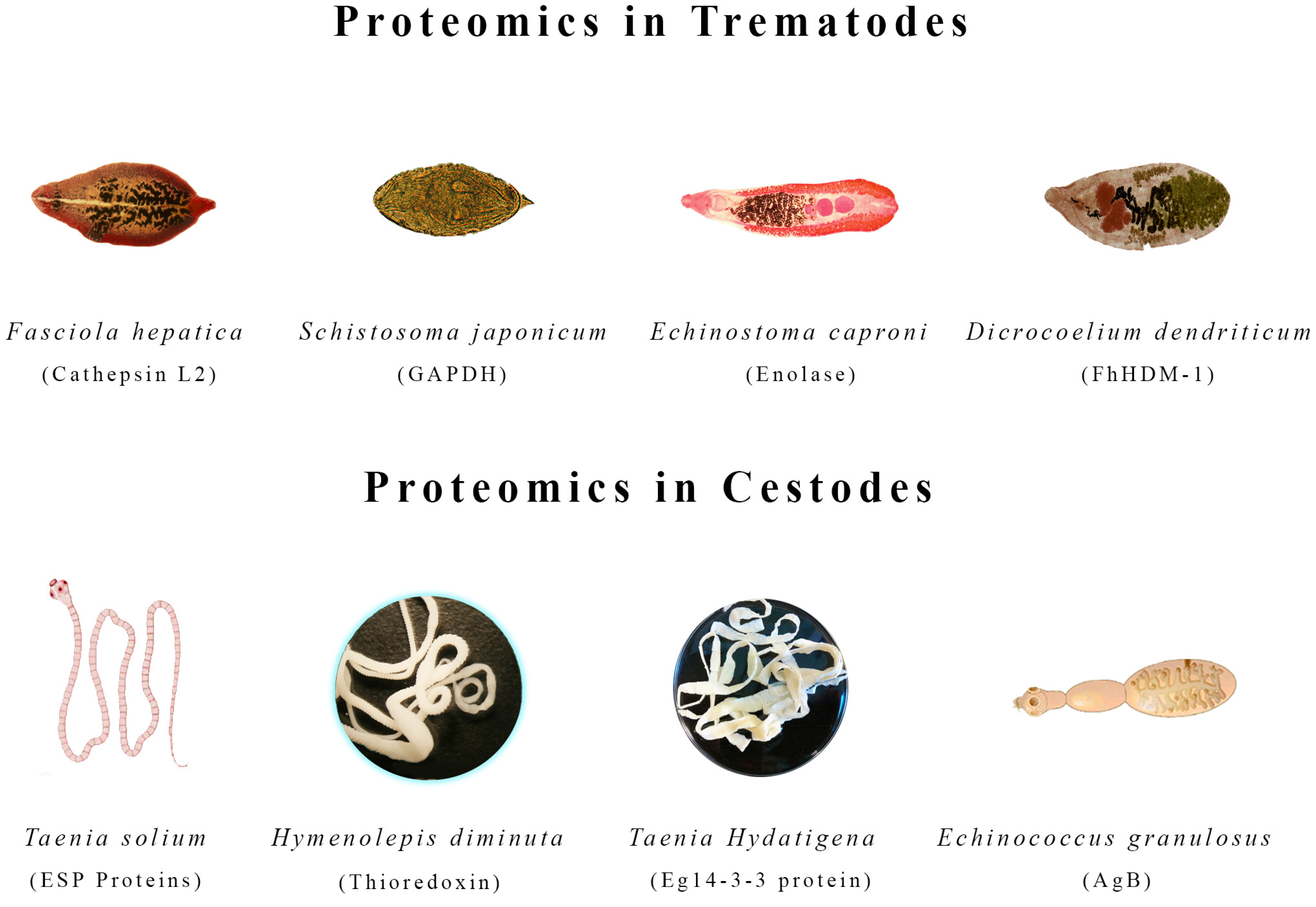
4. Proteomics in Trematodes
4.1. Fasciola hepatica
4.2. Dicrocoelium dendriticum
4.3. Schistosoma japonicum
4.4. Echinostoma caproni
5. Proteomics in Cestodes
5.1. Echinococcus granulosus
5.1.1. Protoscolex Proteins
5.1.2. Germinal Layer and Hydatid Cyst Fluid Proteins
5.2. Taenia solium
5.3. Taenia hydatigena
5.4. Hymenolepis diminuta
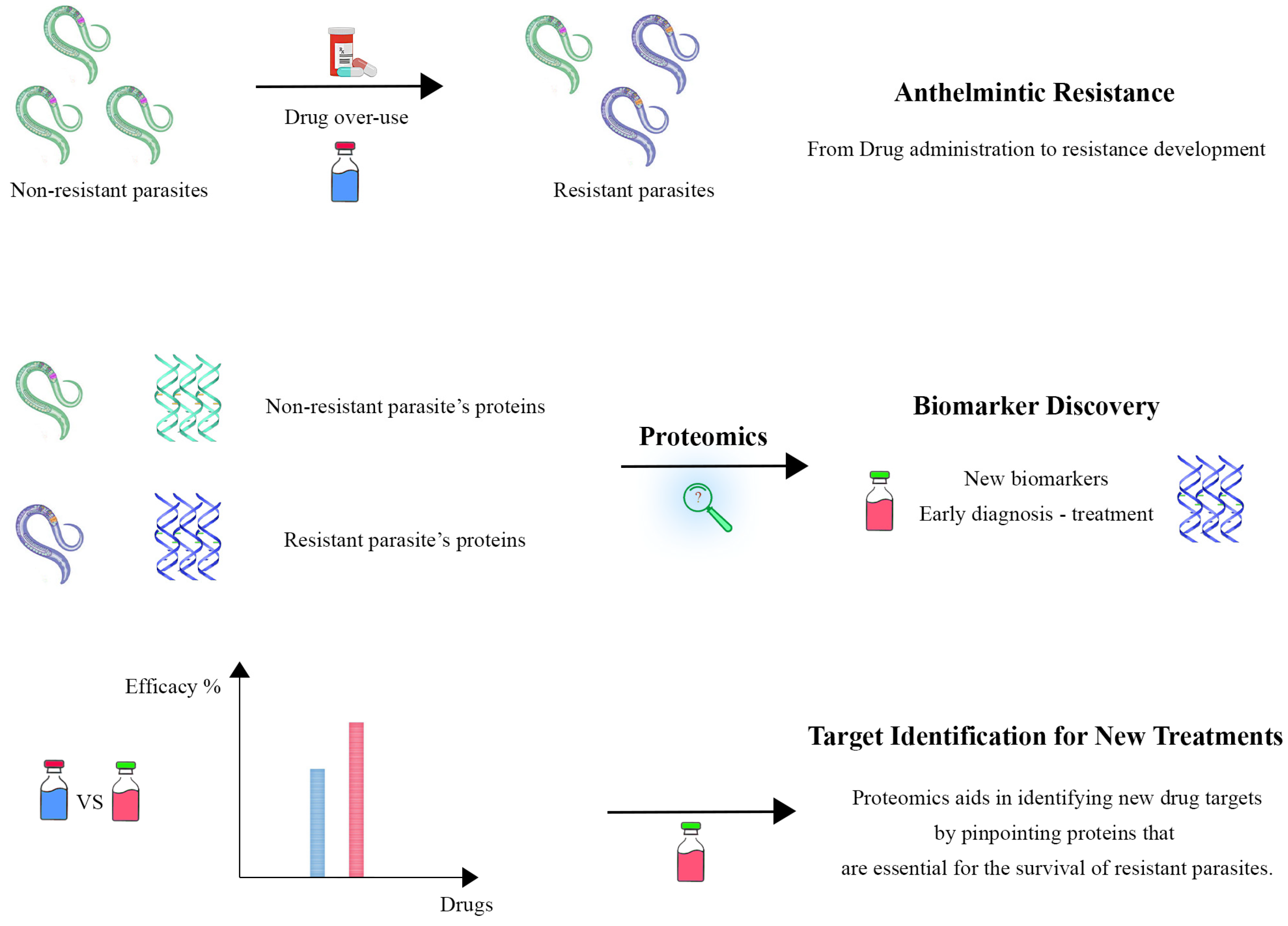
6. Anthelmintic Resistance
7. Vaccine Production
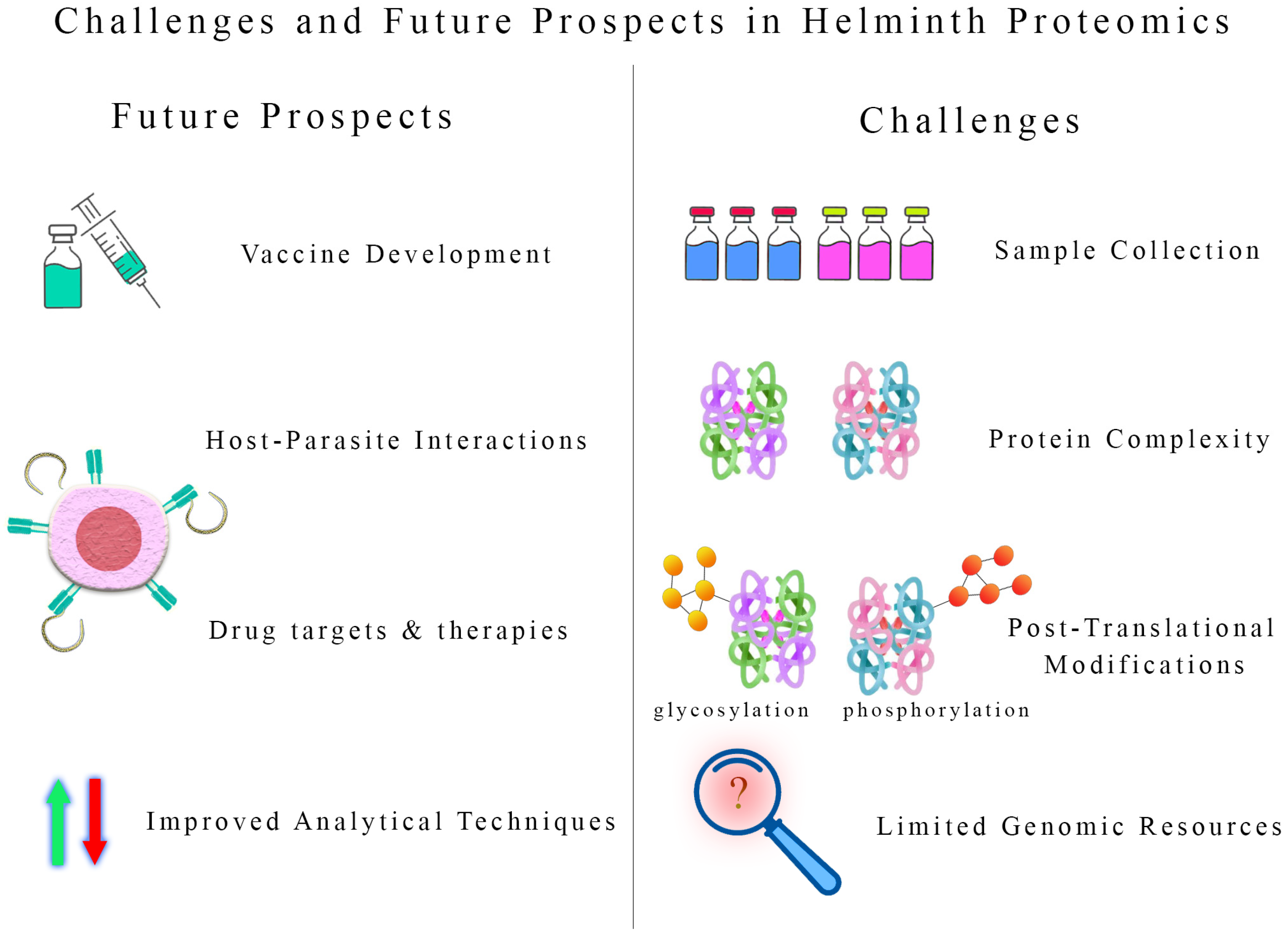
8. Challenges and Future Prospects
9. Discussion
10. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Serin, Y.; Acar Tek, N. Effect of circadian rhythm on metabolic processes and the regulation of energy balance. Ann. Nutr. Metab. 2019, 74, 322–330. [Google Scholar] [CrossRef] [PubMed]
- Nechushtai, R.; Karmi, O.; Zuo, K.; Marjault, H.-B.; Darash-Yahana, M.; Sohn, Y.-S.; King, S.D.; Zandalinas, S.I.; Carloni, P.; Mittler, R. The balancing act of NEET proteins: Iron, ROS, calcium and metabolism. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118805. [Google Scholar] [CrossRef] [PubMed]
- Kong, P.; Cui, Z.-Y.; Huang, X.-F.; Zhang, D.-D.; Guo, R.-J.; Han, M. Inflammation and atherosclerosis: Signaling pathways and therapeutic intervention. Signal Transduct. Target. Ther. 2022, 7, 131. [Google Scholar] [CrossRef] [PubMed]
- Dupree, E.J.; Jayathirtha, M.; Yorkey, H.; Mihasan, M.; Petre, B.A.; Darie, C.C. A critical review of bottom-up proteomics: The good, the bad, and the future of this field. Proteomes 2020, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Mergner, J.; Kuster, B. Plant proteome dynamics. Annu. Rev. Plant Biol. 2022, 73, 67–92. [Google Scholar] [CrossRef]
- Gajahin Gamage, N.T.; Miyashita, R.; Takahashi, K.; Asakawa, S.; Senevirathna, J.D.M. Proteomic applications in aquatic environment studies. Proteomes 2022, 10, 32. [Google Scholar] [CrossRef]
- Hood, L.E.; Omenn, G.S.; Moritz, R.L.; Aebersold, R.; Yamamoto, K.R.; Amos, M.; Hunter-Cevera, J.; Locascio, L.; Participants, W. New and improved proteomics technologies for understanding complex biological systems: Addressing a grand challenge in the life sciences. Proteomics 2012, 12, 2773–2783. [Google Scholar] [CrossRef]
- Zhou, H.; Ning, Z.; Starr, A.E.; Abu-Farha, M.; Figeys, D. Advancements in top-down proteomics. Anal. Chem. 2012, 84, 720–734. [Google Scholar] [CrossRef]
- O’Donnell, S.T.; Ross, R.P.; Stanton, C. The progress of multi-omics technologies: Determining function in lactic acid bacteria using a systems level approach. Front. Microbiol. 2020, 10, 3084. [Google Scholar] [CrossRef]
- Öztürk, M.; Freiwald, A.; Cartano, J.; Schmitt, R.; Dejung, M.; Luck, K.; Al-Sady, B.; Braun, S.; Levin, M.; Butter, F. Proteome effects of genome-wide single gene perturbations. Nat. Commun. 2022, 13, 6153. [Google Scholar] [CrossRef]
- Reitz, C.J.; Kuzmanov, U.; Gramolini, A.O. Multi-omic analyses and network biology in cardiovascular disease. Proteomics 2023, 23, 2200289. [Google Scholar] [CrossRef] [PubMed]
- Hotez, P.J.; Brindley, P.J.; Bethony, J.M.; King, C.H.; Pearce, E.J.; Jacobson, J. Helminth infections: The great neglected tropical diseases. J. Clin. Investig. 2008, 118, 1311–1321. [Google Scholar] [CrossRef] [PubMed]
- Ianiro, G.; Iorio, A.; Porcari, S.; Masucci, L.; Sanguinetti, M.; Perno, C.F.; Gasbarrini, A.; Putignani, L.; Cammarota, G. How the gut parasitome affects human health. Therap. Adv. Gastroenterol. 2022, 15, 17562848221091524. [Google Scholar] [CrossRef]
- Braseth, A.L.; Elliott, D.E.; Ince, M.N. Parasitic infections of the gastrointestinal track and liver. Gastroenterol. Clin. 2021, 50, 361–381. [Google Scholar] [CrossRef]
- Yeshi, K.; Ruscher, R.; Loukas, A.; Wangchuk, P. Immunomodulatory and biological properties of helminth-derived small molecules: Potential applications in diagnostics and therapeutics. Front. Parasitol. 2022, 1, 984152. [Google Scholar] [CrossRef]
- Stijlemans, B.; Caljon, G.; Van Den Abbeele, J.; Van Ginderachter, J.A.; Magez, S.; De Trez, C. Immune evasion strategies of Trypanosoma brucei within the mammalian host: Progression to pathogenicity. Front. Immunol. 2016, 7, 233. [Google Scholar] [CrossRef]
- Gómez-Arreaza, A.; Acosta, H.; Quiñones, W.; Concepción, J.L.; Michels, P.A.; Avilán, L. Extracellular functions of glycolytic enzymes of parasites: Unpredicted use of ancient proteins. Mol. Biochem. Parasitol. 2014, 193, 75–81. [Google Scholar] [CrossRef]
- Jackson, J.A.; Friberg, I.M.; Little, S.; Bradley, J.E. Review series on helminths, immune modulation and the hygiene hypothesis: Immunity against helminths and immunological phenomena in modern human populations: Coevolutionary legacies? Immunology 2009, 126, 18–27. [Google Scholar] [CrossRef] [PubMed]
- McSorley, H.J.; Maizels, R.M. Helminth infections and host immune regulation. Clin. Microbiol. Rev. 2012, 25, 585–608. [Google Scholar] [CrossRef]
- Sotillo, J.; Toledo, R.; Mulvenna, J.; Loukas, A. Exploiting helminth–host interactomes through big data. Trends Parasitol. 2017, 33, 875–888. [Google Scholar] [CrossRef]
- Gazzinelli-Guimarães, A.C.; Gazzinelli-Guimarães, P.; Weatherhead, J.E. A historical and systematic overview of Ascaris vaccine development. Parasitology 2021, 148, 1795–1805. [Google Scholar] [CrossRef] [PubMed]
- You, H.; Jones, M.K.; Gordon, C.A.; Arganda, A.E.; Cai, P.; Al-Wassiti, H.; Pouton, C.W.; McManus, D.P. The mRNA vaccine technology era and the future control of Parasitic infections. Clin. Microbiol. Rev. 2023, 36, e00241-21. [Google Scholar] [CrossRef] [PubMed]
- Okakpu, O.K.; Dillman, A.R. Review of the role of parasitic nematode excretory/secretory proteins in host immunomodulation. J. Parasitol. 2022, 108, 199–208. [Google Scholar] [CrossRef]
- Joshi, P.; Mishra, P.K.K. Functional diversity of the excretory/secretory proteins of nematode parasites. Acta Parasitol. 2022, 67, 619–627. [Google Scholar] [CrossRef]
- Sadr, S.; Ghiassi, S.; Lotfalizadeh, N.; Simab, P.A.; Hajjafari, A.; Borji, H. Antitumor mechanisms of molecules secreted by Trypanosoma cruzi in colon and breast cancer: A review. Anti-Cancer Agents Med. Chem. 2023, 23, 1710–1721. [Google Scholar] [CrossRef] [PubMed]
- Sadr, S.; Borji, H. Echinococcus granulosus as a promising therapeutic agent against triple-negative breast cancer. Curr. Cancer Ther. Rev. 2023, 19, 292–297. [Google Scholar] [CrossRef]
- Lotfalizadeh, N.; Sadr, S.; Morovati, S.; Lotfalizadeh, M.; Hajjafari, A.; Borji, H. A potential cure for tumor-associated immunosuppression by Toxoplasma gondii. Cancer Rep. 2024, 7, e1963. [Google Scholar] [CrossRef]
- Marcilla, A.; Trelis, M.; Cortés, A.; Sotillo, J.; Cantalapiedra, F.; Minguez, M.T.; Valero, M.L.; Sánchez del Pino, M.M.; Muñoz-Antoli, C.; Toledo, R. Extracellular vesicles from parasitic helminths contain specific excretory/secretory proteins and are internalized in intestinal host cells. PLoS ONE 2012, 7, e45974. [Google Scholar] [CrossRef]
- Sadr, S.; Yousefsani, Z.; Simab, P.A.; Alizadeh, A.J.R.; Lotfalizadeh, N.; Borji, H. Trichinella spiralis as a potential antitumor agent: An update. World’s Vet. J. 2023, 13, 65–74. [Google Scholar] [CrossRef]
- Wangchuk, P.; Kouremenos, K.; Eichenberger, R.M.; Pearson, M.; Susianto, A.; Wishart, D.S.; McConville, M.J.; Loukas, A. Metabolomic profiling of the excretory–secretory products of hookworm and whipworm. Metabolomics 2019, 15, 101. [Google Scholar] [CrossRef]
- Gomez-Fuentes, S.; Morales-Ruiz, V.; López-Recinos, D.; Guevara-Salinas, A.; Arce-Sillas, A.; Rodríguez, J.; Parada-Colin, C.; Adalid-Peralta, L. Biological role of excretory–secretory proteins in endemic parasites of Latin America and the Caribbean. J. Helminthol. 2020, 94, e53. [Google Scholar] [CrossRef]
- Wu, F.; Chen, X.; Du, Z.; Chen, Y.; Tong, D.; Zhang, J.; Yang, Y.; Ma, G.; Du, A. Proteomic differences between extracellular vesicles and extracellular vesicle-depleted excretory/secretory products of barber’s pole worm. Parasit. Vectors 2024, 17, 17. [Google Scholar] [CrossRef]
- Vanhamme, L.; Souopgui, J.; Ghogomu, S.; Ngale Njume, F. The functional parasitic worm secretome: Mapping the place of Onchocerca volvulus excretory secretory products. Pathogens 2020, 9, 975. [Google Scholar] [CrossRef]
- Hotterbeekx, A.; Perneel, J.; Vieri, M.K.; Colebunders, R.; Kumar-Singh, S. The secretome of filarial nematodes and its role in host-parasite interactions and pathogenicity in onchocerciasis-associated epilepsy. Front. Cell. Infect. Microbiol. 2021, 11, 662766. [Google Scholar] [CrossRef]
- Pan, W.; Shen, Y.; Han, X.; Wang, Y.; Liu, H.; Jiang, Y.; Zhang, Y.; Wang, Y.; Xu, Y.; Cao, J. Transcriptome profiles of the protoscoleces of Echinococcus granulosus reveal that excretory-secretory products are essential to metabolic adaptation. PLoS Negl. Trop. Dis. 2014, 8, e3392. [Google Scholar] [CrossRef][Green Version]
- Huang, S.-Y.; Yue, D.-M.; Hou, J.-L.; Zhang, X.-X.; Zhang, F.-k.; Wang, C.-R.; Zhu, X.-Q. Proteomic analysis of Fasciola gigantica excretory and secretory products (FgESPs) interacting with buffalo serum of different infection periods by shotgun LC-MS/MS. Parasitol. Res. 2019, 118, 453–460. [Google Scholar] [CrossRef]
- Sánchez-López, C.M.; Trelis, M.; Bernal, D.; Marcilla, A. Overview of the interaction of helminth extracellular vesicles with the host and their potential functions and biological applications. Mol. Immunol. 2021, 134, 228–235. [Google Scholar] [CrossRef]
- White, R.R.; Artavanis-Tsakonas, K. How helminths use excretory secretory fractions to modulate dendritic cells. Virulence 2012, 3, 668–677. [Google Scholar] [CrossRef] [PubMed]
- van der Ree, A.M.; Mutapi, F. The helminth parasite proteome at the host–parasite interface–informing diagnosis and control. Exp. Parasitol. 2015, 157, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Marzano, V.; Pane, S.; Foglietta, G.; Levi Mortera, S.; Vernocchi, P.; Onetti Muda, A.; Putignani, L. Mass spectrometry based-proteomic analysis of Anisakis spp.: A preliminary study towards a new diagnostic tool. Genes 2020, 11, 693. [Google Scholar] [CrossRef] [PubMed]
- Fissiha, W.; Kinde, M.Z. Anthelmintic resistance and its mechanism: A review. Infect. Drug Resist. 2021, 14, 5403–5410. [Google Scholar] [CrossRef] [PubMed]
- Shalaby, H.A. Anthelmintics resistance; how to overcome it? Iran. J. Parasitol. 2013, 8, 18–32. [Google Scholar]
- Sadr, S.; Ahmadi Simab, P.; Niazi, M.; Yousefsani, Z.; Lotfalizadeh, N.; Hajjafari, A.; Borji, H. Anti-inflammatory and immunomodulatory effects of mesenchymal stem cell therapy on parasitic drug resistance. Expert Rev. Anti-Infect. Ther. 2024, 22, 435–451. [Google Scholar] [CrossRef] [PubMed]
- Zaiss, M.M.; Harris, N.L. Interactions between the intestinal microbiome and helminth parasites. Parasite Immunol. 2016, 38, 5–11. [Google Scholar] [CrossRef]
- Moreno, Y.; Gros, P.-P.; Tam, M.; Segura, M.; Valanparambil, R.; Geary, T.G.; Stevenson, M.M. Proteomic analysis of excretory-secretory products of Heligmosomoides polygyrus assessed with next-generation sequencing transcriptomic information. PLOS. Negl. Trop. Dis. 2011, 5, e1370. [Google Scholar] [CrossRef]
- Maruszewska-Cheruiyot, M.; Szewczak, L.; Krawczak-Wójcik, K.; Głaczyńska, M.; Donskow-Łysoniewska, K. The production of excretory-secretory molecules from Heligmosomoides polygyrus bakeri fourth stage larvae varies between mixed and single sex cultures. Parasit. Vectors 2021, 14, 106. [Google Scholar] [CrossRef]
- Zhou, C.; Chen, J.; Niu, H.; Ouyang, S.; Wu, X. Study on the population evolution of Ascaris lumbricoides and Ascaris suum based on whole genome resequencing. Vet. Parasitol. 2020, 279, 109062. [Google Scholar] [CrossRef]
- Eamsobhana, P.; Yong, H.-S.; Boonyong, S.; Wanachiwanawin, D.; Tungtrongchitr, A. Genetic diversity and identity of Ascaris worms from human and pig hosts in Thailand. Vet. Parasitol. Reg. Stud. Rep. 2022, 33, 100752. [Google Scholar] [CrossRef]
- Leung, A.K.; Leung, A.A.; Wong, A.H.; Hon, K.L. Human ascariasis: An updated review. Recent. Pat. Inflamm. Allergy. Drug. Discov. 2020, 14, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Fauziah, N.; Aviani, J.K.; Agrianfanny, Y.N.; Fatimah, S.N. Intestinal parasitic infection and nutritional status in children under five years old: A systematic review. Trop. Med. Infect. Dis. 2022, 7, 371. [Google Scholar] [CrossRef]
- Roesel, K.; Dohoo, I.; Baumann, M.; Dione, M.; Grace, D.; Clausen, P.-H. Prevalence and risk factors for gastrointestinal parasites in small-scale pig enterprises in Central and Eastern Uganda. J. Parasitol. Res. 2017, 116, 335–345. [Google Scholar] [CrossRef]
- Vismarra, A.; Lenti, A.; Genchi, M.; Kramer, L.; Geldhof, P. Seroprevalence of Ascaris suum compared to milk spot prevalence at slaughter in Italian fattening pigs. Vet. Parasitol. Reg. Stud. Rep. 2023, 37, 100828. [Google Scholar] [CrossRef]
- Urban Jr, J.F.; Douvres, F.W. In vitro development of Ascaris suum from third-to fourth-stage larvae and detection of metabolic antigens in multi-well culture systems. J. Parasitol. 1981, 67, 800–806. [Google Scholar] [CrossRef]
- Wang, T.; Van Steendam, K.; Dhaenens, M.; Vlaminck, J.; Deforce, D.; Jex, A.R.; Gasser, R.B.; Geldhof, P. Proteomic analysis of the excretory-secretory products from larval stages of Ascaris suum reveals high abundance of glycosyl hydrolases. PLOS. Negl. Trop. Dis. 2013, 7, e2467. [Google Scholar] [CrossRef]
- Okulewicz, A.; Perec-Matysiak, A.; Buńkowska, K.; Hildebrand, J. Toxocara canis Toxocara cati and Toxascaris leonina in wild and domestic carnivores. Helminthologia 2012, 49, 3–10. [Google Scholar] [CrossRef]
- Schwartz, R.; Bidaisee, S.; Fields, P.J.; Macpherson, M.L.; Macpherson, C.N. The epidemiology and control of Toxocara canis in puppies. Parasite Epidemiol. Control 2022, 8, e00232. [Google Scholar] [CrossRef]
- Choi, D.; Lim, J.H.; Choi, D.-C.; Lee, K.S.; Paik, S.W.; Kim, S.-H.; Choi, Y.-H.; Huh, S. Transmission of Toxocara canis via ingestion of raw cow liver: A cross-sectional study in healthy adults. Korean. J. Parasitol. 2012, 50, 23–27. [Google Scholar] [CrossRef]
- Omonijo, A.; Kalinda, C.; Mukaratirwa, S. A systematic review and meta-analysis of canine, feline and human Toxocara infections in sub-Saharan Africa. J. Helminthol. 2020, 94, e96. [Google Scholar] [CrossRef]
- da Silva, M.B.; Oviedo, Y.; Cooper, P.J.; Pacheco, L.G.; Pinheiro, C.S.; Ferreira, F.; Briza, P.; Alcantara-Neves, N.M. The somatic proteins of Toxocara canis larvae and excretory-secretory products revealed by proteomics. Vet. Parasitol. 2018, 259, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Sperotto, R.L.; Kremer, F.S.; Berne, M.E.A.; de Avila, L.F.C.; da Silva Pinto, L.; Monteiro, K.M.; Caumo, K.S.; Ferreira, H.B.; Berne, N.; Borsuk, S. Proteomic analysis of Toxocara canis excretory and secretory (TES) proteins. Mol. Biochem. Parasitol. 2017, 211, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Raulf, M.-K.; Lepenies, B.; Strube, C. Toxocara canis and Toxocara cati somatic and excretory-secretory antigens are recognised by C-type lectin receptors. Pathogens 2021, 10, 321. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.-B.; Zou, Y.; Zhu, X.-Q.; Liu, G.-H. Toxocara “omics” and the promises it holds for medicine and veterinary medicine. J. Adv. Parasitol. 2020, 109, 89–108. [Google Scholar] [CrossRef]
- Torkan, S.; Ghandehari-Alavijeh, M.; Khamesipour, F. Survey of the prevalence of Toxocara cati in stray cats in Isfahan city, Iran by PCR method. Trop. Biomed. 2017, 34, 550–555. [Google Scholar] [PubMed]
- Zanzani, S.A.; Gazzonis, A.L.; Scarpa, P.; Berrilli, F.; Manfredi, M.T. Intestinal parasites of owned dogs and cats from metropolitan and micropolitan areas: Prevalence, zoonotic risks, and pet owner awareness in northern Italy. Biomed. Res. Int. 2014, 2014, 696508. [Google Scholar] [CrossRef]
- Maciag, L.; Morgan, E.R.; Holland, C. Toxocara: Time to let cati ‘out of the bag’. Trends. Parasitol. 2022, 38, 280–289. [Google Scholar] [CrossRef]
- Wu, T.; Bowman, D.D. Visceral larval migrans of Toxocara canis and Toxocara cati in non-canid and non-felid hosts. Adv. Parasitol. 2020, 109, 63–88. [Google Scholar] [CrossRef]
- Abbas, I.; Al-Araby, M.; Elmishmishy, B.; El-Alfy, E.-S. Gastrointestinal parasites of cats in Egypt: High prevalence high zoonotic risk. BMC. Vet. Res. 2022, 18, 420. [Google Scholar] [CrossRef]
- Soleyman, N.M.; Darnhofer, B.; Gruenberger, R.B.; Abnous, K.; Borji, H. Proteomic analysis of soluble protein extract of adult Toxocara cati. Comp. Immunol. Microbiol. Infect. Dis. 2020, 73, 101528. [Google Scholar] [CrossRef]
- Soleymani, N.; Grunberger, R.B.; Abnous, K.; Borji, H.; Vahdati, F. Identification and immunological characterization of somatic proteins from adults of Toxocara cati by proteomics technique. Iran. J. Parasitol. 2021, 16, 23. [Google Scholar] [CrossRef]
- Yeshi, K.; Creek, D.J.; Anderson, D.; Ritmejerytė, E.; Becker, L.; Loukas, A.; Wangchuk, P. Metabolomes and lipidomes of the infective stages of the gastrointestinal nematodes, Nippostrongylus brasiliensis and Trichuris muris. Metabolites 2020, 10, 446. [Google Scholar] [CrossRef]
- Thuma, N.; Döhler, D.; Mielenz, D.; Sticht, H.; Radtke, D.; Reimann, L.; Warscheid, B.; Voehringer, D. A newly identified secreted larval antigen elicits basophil-dependent protective immunity against N. brasiliensis infection. Front. Immunol. 2022, 13, 979491. [Google Scholar] [CrossRef] [PubMed]
- Loukas, A.; Hotez, P.J.; Diemert, D.; Yazdanbakhsh, M.; McCarthy, J.S.; Correa-Oliveira, R.; Croese, J.; Bethony, J.M. Hookworm infection. Nat. Rev. Dis. Primers 2016, 2, 16088. [Google Scholar] [CrossRef] [PubMed]
- Sotillo, J.; Sanchez-Flores, A.; Cantacessi, C.; Harcus, Y.; Pickering, D.; Bouchery, T.; Camberis, M.; Tang, S.-C.; Giacomin, P.; Mulvenna, J. Secreted proteomes of different developmental stages of the gastrointestinal nematode Nippostrongylus brasiliensis. Mol. Cell. Proteom. 2014, 13, 2736–2751. [Google Scholar] [CrossRef] [PubMed]
- Hawdon, J.M.; Wise, K.A. Ancylostoma caninum and other canine hookworms. In Dog Parasites Endangering Human Health; Springer: Berlin/Heidelberg, Germany, 2021; pp. 147–193. [Google Scholar] [CrossRef]
- Traub, R.J. Ancylostoma ceylanicum, a re-emerging but neglected parasitic zoonosis. Int. J. Parasitol. 2013, 43, 1009–1015. [Google Scholar] [CrossRef]
- Mulvenna, J.; Hamilton, B.; Nagaraj, S.H.; Smyth, D.; Loukas, A.; Gorman, J.J. Proteomics analysis of the excretory/secretory component of the blood-feeding stage of the hookworm, Ancylostoma caninum. Mol. Cell. Proteom. 2009, 8, 109–121. [Google Scholar] [CrossRef]
- Ju, C.; Feng, Z.; Brindley, P.J.; McManus, D.P.; Han, Z.; Peng, J.-x.; Hu, W. Our wormy world: Genomics, proteomics and transcriptomics in East and Southeast Asia. Adv. Parasitol. 2010, 73, 327–371. [Google Scholar] [CrossRef]
- Crilly, J.P.; Evans, M.; Tähepõld, K.; Sargison, N. Haemonchosis: Dealing with the increasing threat of the barber’s pole worm. Livestock 2020, 25, 237–246. [Google Scholar] [CrossRef]
- Adduci, I.; Sajovitz, F.; Hinney, B.; Lichtmannsperger, K.; Joachim, A.; Wittek, T.; Yan, S. Haemonchosis in sheep and goats, control strategies and development of vaccines against Haemonchus contortus. Animals 2022, 12, 2339. [Google Scholar] [CrossRef]
- Besier, R.; Kahn, L.; Sargison, N.; Van Wyk, J. Diagnosis, treatment and management of Haemonchus contortus in small ruminants. Adv. Parasitol. 2016, 93, 181–238. [Google Scholar] [CrossRef]
- Zheng, Y.; Young, N.D.; Song, J.; Gasser, R.B. Genome-Wide Analysis of Haemonchus contortus Proteases and Protease Inhibitors Using Advanced Informatics Provides Insights into Parasite Biology and Host–Parasite Interactions. Int. J. Mol. Sci. 2023, 24, 12320. [Google Scholar] [CrossRef]
- De Vries, E.; Bakker, N.; Krijgsveld, J.; Knox, D.P.; Heck, A.J.; Yatsuda, A.P. An AC-5 cathepsin B-like protease purified from Haemonchus contortus excretory secretory products shows protective antigen potential for lambs. Vet. Res. 2009, 40, 41. [Google Scholar] [CrossRef] [PubMed]
- Yatsuda, A.P.; Bakker, N.; Krijgsveld, J.; Knox, D.P.; Heck, A.J.; de Vries, E. Identification of secreted cysteine proteases from the parasitic nematode Haemonchus contortus detected by biotinylated inhibitors. Infect. Immun. 2006, 74, 1989–1993. [Google Scholar] [CrossRef] [PubMed]
- Jasmer, D.P.; Mitreva, M.D.; McCarter, J.P. mRNA sequences for Haemonchus contortus intestinal cathepsin B-like cysteine proteases display an extreme in abundance and diversity compared with other adult mammalian parasitic nematodes. Mol. Biochem. Parasitol. 2004, 137, 297–305. [Google Scholar] [CrossRef]
- Hosseinnezhad, H.; Sharifdini, M.; Ashrafi, K.; Atrkar Roushan, Z.; Mirjalali, H.; Rahmati, B. Trichostrongyloid nematodes in ruminants of northern Iran: Prevalence and molecular analysis. BMC. Vet. Res. 2021, 17, 371. [Google Scholar] [CrossRef]
- Tafere, A.; Terefe, G.; Mamo, G.; Kaba, T.; Shiferaw, J. A Comparative Study on Pathological Changes in the Small Intestine of Sheep and Goat Experimentally Infected with Trichostrongylus colubriformis. Vet. Med. 2022, 13, 213–233. [Google Scholar] [CrossRef]
- Kaba, T.; Terefe, G.; Waktole, H. Sheep and goat response to Trichostrongylus colubriformis infection based on egg output and worm burden. Trop. Anim. Health. Prod. 2023, 55, 177. [Google Scholar] [CrossRef]
- Kiel, M.; Josh, P.; Jones, A.; Windon, R.; Hunt, P.; Kongsuwan, K. Identification of immuno-reactive proteins from a sheep gastrointestinal nematode, Trichostrongylus colubriformis, using two-dimensional electrophoresis and mass spectrometry. Int. J. Parasitol. 2007, 37, 1419–1429. [Google Scholar] [CrossRef]
- Ortega-Pierres, G.; Vaquero-Vera, A.; Fonseca-Linan, R.; Bermudez-Cruz, R.; Argüello-García, R. Induction of protection in murine experimental models against Trichinella spiralis: An up-to-date review. J. Helminthol. 2015, 89, 526–539. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Yang, Y.; Liu, X.; Xu, F.; Wang, Y.; Liu, L.; Yang, Y.; Liu, M.; Bai, X. Extracellular vesicles from Trichinella spiralis: Proteomic analysis and protective immunity. PLoS Negl. Trop. Dis. 2022, 16, e0010528. [Google Scholar] [CrossRef]
- Xu, J.; Pang, Z.; Zhang, J.; Xia, S.; Wang, R.; Zhang, Y.; Zhen, J.; Song, X.; Lin, L.; Sun, F. Regulatory effects of Trichinella spiralis and a serine protease inhibitor on the endoplasmic reticulum stress response of intestinal epithelial cells. Vet. Res. 2022, 53, 18. [Google Scholar] [CrossRef]
- Thawornkuno, C.; Nogrado, K.; Adisakwattana, P.; Thiangtrongjit, T.; Reamtong, O. Identification and profiling of Trichinella spiralis circulating antigens and proteins in sera of mice with trichinellosis. PLoS ONE 2022, 17, e0265013. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yang, Y.; Wang, Y.; Zhan, B.; Gu, Y.; Cheng, Y.; Zhu, X. Excretory/secretory products from Trichinella spiralis adult worms ameliorate DSS-induced colitis in mice. PLoS ONE 2014, 9, e96454. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.D.; Cui, J.; Liu, X.L.; Jiang, P.; Sun, G.G.; Zhang, X.; Long, S.R.; Wang, L.; Wang, Z.Q. Comparative proteomic analysis of surface proteins of Trichinella spiralis muscle larvae and intestinal infective larvae. Acta Trop. 2015, 150, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, L.; Wei, X.; Wang, Y.; Zhang, H.; Shi, A.; Liu, T.; Yang, X.; Fang, Q. Proteomic analysis of the response of Trichinella spiralis muscle larvae to exogenous nitric oxide. PLoS ONE 2018, 13, e0198205. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, N.; Li, W.; Li, L.; Yan, H.; Qu, Z.; Li, T.; Cui, J.; Yang, Y.; Jia, W. Proteomic analysis of differentially expressed proteins in the three developmental stages of Trichinella spiralis. Vet. Parasitol. 2016, 231, 32–38. [Google Scholar] [CrossRef]
- Ren, H.N.; Liu, R.D.; Song, Y.Y.; Zhuo, T.X.; Guo, K.X.; Zhang, Y.; Jiang, P.; Wang, Z.Q.; Cui, J. Label-free quantitative proteomic analysis of molting-related proteins of Trichinella spiralis intestinal infective larvae. Vet. Res. 2019, 50, 70. [Google Scholar] [CrossRef]
- Cui, J.; Liu, R.D.; Wang, L.; Zhang, X.; Jiang, P.; Liu, M.Y.; Wang, Z.Q. Proteomic analysis of surface proteins of Trichinella spiralis muscle larvae by two-dimensional gel electrophoresis and mass spectrometry. Parasites Vectors 2013, 6, 355. [Google Scholar] [CrossRef]
- Ren, H.N.; Zhuo, T.X.; Bai, S.J.; Bai, Y.; Sun, X.Y.; Liu, R.D.; Long, S.R.; Cui, J.; Wang, Z.Q. Proteomic analysis of hydrolytic proteases in excretory/secretory proteins from Trichinella spiralis intestinal infective larvae using zymography combined with shotgun LC-MS/MS approach. Acta Trop. 2021, 216, 105825. [Google Scholar] [CrossRef]
- Jiang, H.-Y.; Zhao, N.; Zhang, Q.-L.; Gao, J.-M.; Liu, L.-L.; Wu, T.-F.; Wang, Y.; Huang, Q.-H.; Gou, Q.; Chen, W. Intestinal microbes influence the survival, reproduction and protein profile of Trichinella spiralis in vitro. Int. J. Parasitol. 2016, 46, 51–58. [Google Scholar] [CrossRef]
- Wang, Z.Q.; Wang, L.; Cui, J. Proteomic analysis of Trichinella spiralis proteins in intestinal epithelial cells after culture with their larvae by shotgun LC–MS/MS approach. J. Proteom. 2012, 75, 2375–2383. [Google Scholar] [CrossRef]
- Janwan, P.; Intapan, P.M.; Laummaunwai, P.; Rodpai, R.; Wongkham, C.; Insawang, T.; Thanchomnang, T.; Sanpool, O.; Maleewong, W. Proteomic analysis identification of antigenic proteins in Gnathostoma spinigerum larvae. Exp. Parasitol. 2015, 159, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Soblik, H.; Younis, A.E.; Mitreva, M.; Renard, B.Y.; Kirchner, M.; Geisinger, F.; Steen, H.; Brattig, N.W. Life cycle stage-resolved proteomic analysis of the excretome/secretome from Strongyloides ratti—Identification of stage-specific proteases. Mol. Cell. Proteom. 2011, 10, M111.010157. [Google Scholar] [CrossRef] [PubMed]
- Vo, D.; Le, K.; Nguyen, T.; Nguyen, H. Identification of excretory/secretory antigens produced by L2 stage larvae of Toxocara canis involving in induction of IgG response in mice by proteomics approach. In 6th International Conference on the Development of Biomedical Engineering in Vietnam (BME6); Vo Van, T., Nguyen Le, T., Nguyen Duc, T., Eds.; BME 2017. IFMBE Proceedings; Springer: Singapore, 2017; Volume 63, pp. 633–636. [Google Scholar] [CrossRef]
- Dea-Ayuela, M.; Bolás-Fernández, F. Two-dimensional electrophoresis and mass spectrometry for the identification of species-specific Trichinella antigens. Vet. Parasitol. 2005, 132, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.W.; Connolly, B. Proteomic analysis of the excretory-secretory proteins of the Trichinella spiralis L1 larva, a nematode parasite of skeletal muscle. Proteomics 2005, 5, 4525–4532. [Google Scholar] [CrossRef] [PubMed]
- Else, K.J.; Keiser, J.; Holland, C.V.; Grencis, R.K.; Sattelle, D.B.; Fujiwara, R.T.; Bueno, L.L.; Asaolu, S.O.; Sowemimo, O.A.; Cooper, P.J. Whipworm and roundworm infections. Nat. Rev. Dis. Primers 2020, 6, 44. [Google Scholar] [CrossRef]
- Bennett, A.P.; Robinson, M.W. Trematode proteomics: Recent advances and future directions. Pathogens 2021, 10, 348. [Google Scholar] [CrossRef]
- McManus, D.P.; Dalton, J.P. Vaccines against the zoonotic trematodes Schistosoma japonicum, Fasciola hepatica and Fasciola gigantica. Parasitology 2006, 133, 43–61. [Google Scholar]
- Toledo, R.; Bernal, M.D.; Marcilla, A. Proteomics of foodborne trematodes. J. Proteom. 2011, 74, 1485–1503. [Google Scholar] [CrossRef]
- Sripa, B.; Kaewkes, S.; Intapan, P.M.; Maleewong, W.; Brindley, P.J. Food-borne trematodiases in Southeast Asia: Epidemiology, pathology, clinical manifestation and control. Adv. Parasitol. 2010, 72, 305–350. [Google Scholar] [CrossRef]
- Moazeni, M.; Ahmadi, A. Controversial aspects of the life cycle of Fasciola hepatica. Exp. Parasitol. 2016, 169, 81–89. [Google Scholar] [CrossRef]
- Lalor, R.; Cwiklinski, K.; Calvani, N.E.D.; Dorey, A.; Hamon, S.; Corrales, J.L.; Dalton, J.P.; De Marco Verissimo, C. Pathogenicity and virulence of the liver flukes Fasciola hepatica and Fasciola gigantica that cause the zoonosis Fasciolosis. Virulence 2021, 12, 2839–2867. [Google Scholar] [CrossRef] [PubMed]
- Cwiklinski, K.; Jewhurst, H.; McVeigh, P.; Barbour, T.; Maule, A.G.; Tort, J.; O’Neill, S.M.; Robinson, M.W.; Donnelly, S.; Dalton, J.P. Infection by the helminth parasite Fasciola hepatica requires rapid regulation of metabolic, virulence, and invasive factors to adjust to its mammalian host. Mol. Cell. Proteom. 2018, 17, 792–809. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.W.; Menon, R.; Donnelly, S.M.; Dalton, J.P.; Ranganathan, S. An integrated transcriptomics and proteomics analysis of the secretome of the helminth pathogen Fasciola hepatica: Proteins associated with invasion and infection of the mammalian host. Mol. Cell. Proteom. 2009, 8, 1891–1907. [Google Scholar] [CrossRef]
- Cwiklinski, K.; Robinson, M.W.; Donnelly, S.; Dalton, J.P. Complementary transcriptomic and proteomic analyses reveal the cellular and molecular processes that drive growth and development of Fasciola hepatica in the host liver. BMC Genom. 2021, 22, 46. [Google Scholar] [CrossRef]
- Robinson, M.W.; Tort, J.F.; Lowther, J.; Donnelly, S.M.; Wong, E.; Xu, W.; Stack, C.M.; Padula, M.; Herbert, B.; Dalton, J.P. Proteomics and phylogenetic analysis of the cathepsin L protease family of the helminth pathogen Fasciola hepatica: Expansion of a repertoire of virulence-associated factors. Mol. Cell. Proteom. 2008, 7, 1111–1123. [Google Scholar] [CrossRef]
- Stack, C.; Dalton, J.P.; Robinson, M.W. The phylogeny, structure and function of trematode cysteine proteases, with particular emphasis on the Fasciola hepatica cathepsin L family. Cysteine proteases of pathogenic organisms. In Cysteine Proteases of Pathogenic Organisms; Springer: Berlin/Heidelberg, Germany, 2011; pp. 116–135. [Google Scholar] [CrossRef]
- McVeigh, P.; Maule, A.G.; Dalton, J.P.; Robinson, M.W. Fasciola hepatica virulence-associated cysteine peptidases: A systems biology perspective. Microbes Infect. 2012, 14, 301–310. [Google Scholar] [CrossRef]
- Cwiklinski, K.; Donnelly, S.; Drysdale, O.; Jewhurst, H.; Smith, D.; Verissimo, C.D.M.; Pritsch, I.C.; O’Neill, S.; Dalton, J.P.; Robinson, M.W. The cathepsin-like cysteine peptidases of trematodes of the genus Fasciola. Adv. Parasitol. 2019, 104, 113–164. [Google Scholar] [CrossRef] [PubMed]
- Barbour, T.; Cwiklinski, K.; Lalor, R.; Dalton, J.P.; De Marco Verissimo, C. The zoonotic helminth parasite Fasciola hepatica: Virulence-associated cathepsin B and cathepsin L cysteine peptidases secreted by infective newly excysted juveniles (NEJ). Animals 2021, 11, 3495. [Google Scholar] [CrossRef]
- Corrales, J.L.; Cwiklinski, K.; Verissimo, C.D.M.; Dorey, A.; Lalor, R.; Jewhurst, H.; McEvoy, A.; Diskin, M.; Duffy, C.; Cosby, S.L. Diagnosis of sheep fasciolosis caused by Fasciola hepatica using cathepsin L enzyme-linked immunosorbent assays (ELISA). Vet. Parasitol. 2021, 298, 109517. [Google Scholar] [CrossRef]
- Collett, C.F.; Phillips, H.C.; Fisher, M.; Smith, S.; Fenn, C.; Goodwin, P.; Morphew, R.M.; Brophy, P.M. Fasciola hepatica cathepsin L zymogens: Immuno-proteomic evidence for highly immunogenic zymogen-specific conformational epitopes to support diagnostics development. J. Proteome Res. 2022, 21, 1997–2010. [Google Scholar] [CrossRef]
- De Marco Verissimo, C.; Jewhurst, H.L.; Tikhonova, I.G.; Urbanus, R.T.; Maule, A.G.; Dalton, J.P.; Cwiklinski, K. Fasciola hepatica serine protease inhibitor family (serpins): Purposely crafted for regulating host proteases. PLoS Negl. Trop. Dis. 2020, 14, e0008510. [Google Scholar] [CrossRef] [PubMed]
- Lucius, R. Dicrocoelium dendriticum. Trends Parasitol. 2022, 38, 1089–1090. [Google Scholar] [CrossRef] [PubMed]
- Kahl, A.; von Samson-Himmelstjerna, G.; Krücken, J.; Ganter, M. Chronic wasting due to liver and rumen flukes in sheep. Animals 2021, 11, 549. [Google Scholar] [CrossRef] [PubMed]
- Cézilly, F.; Thomas, F.; Médoc, V.; Perrot-Minnot, M.-J. Host-manipulation by parasites with complex life cycles: Adaptive or not? Trends. Parasitol. 2010, 26, 311–317. [Google Scholar] [CrossRef]
- Martínez-Ibeas, A.; González-Lanza, C.; Manga-González, M.Y. Proteomic analysis of the tegument and excretory–secretory products of Dicrocoelium dendriticum (Digenea) adult worms. Exp. Parasitol. 2013, 133, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Montaner, S.; Galiano, A.; Trelis, M.; Martin-Jaular, L.; Del Portillo, H.A.; Bernal, D.; Marcilla, A. The role of extracellular vesicles in modulating the host immune response during parasitic infections. Front. Immunol. 2014, 5, 433. [Google Scholar] [CrossRef]
- Bernal, D.; Trelis, M.; Montaner, S.; Cantalapiedra, F.; Galiano, A.; Hackenberg, M.; Marcilla, A. Surface analysis of Dicrocoelium dendriticum. The molecular characterization of exosomes reveals the presence of miRNAs. J. Proteom. 2014, 105, 232–241. [Google Scholar] [CrossRef]
- Rojo-Vázquez, F.A.; Meana, A.; Valcárcel, F.; Martínez-Valladares, M. Update on trematode infections in sheep. Vet. Parasitol. 2012, 189, 15–38. [Google Scholar] [CrossRef]
- Di Maggio, L.S.; Tirloni, L.; Pinto, A.F.; Diedrich, J.K.; Yates III, J.R.; Benavides, U.; Carmona, C.; da Silva Vaz Jr, I.; Berasain, P. Across intra-mammalian stages of the liver fluke Fasciola hepatica: A proteomic study. Sci. Rep. 2016, 6, 32796. [Google Scholar] [CrossRef]
- Lo, N.C.; Bezerra, F.S.M.; Colley, D.G.; Fleming, F.M.; Homeida, M.; Kabatereine, N.; Kabole, F.M.; King, C.H.; Mafe, M.A.; Midzi, N.; et al. Review of 2022 WHO guidelines on the control and elimination of schistosomiasis. Lancet. Infec. Dis 2022, 22, E327–E335. [Google Scholar] [CrossRef]
- Xue, Q.; Deng, Y.; Liu, Y.; Wang, Y.; Hu, W.; Huang, Y.; Yang, K. A retrospective analysis of schistosomiasis related literature from 2011-2020: Focusing on the next decade. Acta Trop. 2023, 238, 106750. [Google Scholar] [CrossRef] [PubMed]
- Lima, R.R.; Lima, J.V.; Ribeiro, J.F.; Nascimento, J.B.; Oliveira, W.F.; Cabral Filho, P.E.; Fontes, A. Emerging biomedical tools for biomarkers detection and diagnostics in schistosomiasis. Talanta 2023, 265, 124900. [Google Scholar] [CrossRef] [PubMed]
- Lowe, C.; Ahmadabadi, Z.; Gray, D.; Kelly, M.; McManus, D.P.; Williams, G. Systematic review of applied mathematical models for the control of Schistosoma japonicum. Acta Trop. 2023, 241, 106873. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Lu, J.; Hu, W.; Wang, S.-Y.; Cui, S.-J.; Chi, M.; Yan, Q.; Wang, X.-R.; Song, H.-D.; Xu, X.-N. New perspectives on host-parasite interplay by comparative transcriptomic and proteomic analyses of Schistosoma japonicum. PLoS Pathog. 2006, 2, e29. [Google Scholar] [CrossRef]
- Tritten, L.; Geary, T.G. Helminth extracellular vesicles in host–parasite interactions. Curr. Opin. Microbiol. 2018, 46, 73–79. [Google Scholar] [CrossRef]
- Liao, Q.; Yuan, X.; Xiao, H.; Liu, C.; Lv, Z.; Zhao, Y.; Wu, Z. Identifying Schistosoma japonicum excretory/secretory proteins and their interactions with host immune system. PLoS ONE 2011, 6, e23786. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.-l.; Lv, Z.-y.; Hu, S.-m.; He, S.-j.; Li, Z.-y.; Zhang, S.-m.; Zheng, H.-q.; Li, M.-t.; Yu, X.-b.; Fung, M.-C. Schistosoma japonicum: Proteomics analysis of differentially expressed proteins from ultraviolet-attenuated cercariae compared to normal cercariae. Parasitol. Res. 2009, 105, 237–248. [Google Scholar] [CrossRef]
- Cao, X.; Fu, Z.; Zhang, M.; Han, Y.; Han, Q.; Lu, K.; Li, H.; Zhu, C.; Hong, Y.; Lin, J. Excretory/secretory proteome of 14-day schistosomula, Schistosoma japonicum. J. Proteom. 2016, 130, 221–230. [Google Scholar] [CrossRef]
- Han, Z.-G.; Brindley, P.J.; Wang, S.-Y.; Chen, Z. Schistosoma genomics: New perspectives on schistosome biology and host-parasite interaction. Annu. Rev. Genomics. Hum. Genet. 2009, 10, 211–240. [Google Scholar] [CrossRef]
- Cortés, A.; Muñoz-Antoli, C.; Sotillo, J.; Fried, B.; Esteban, J.; Toledo, R. Echinostoma caproni (T rematoda): Differential in vivo mucin expression and glycosylation in high-and low-compatible hosts. Parasit. Immunol. 2015, 37, 32–42. [Google Scholar] [CrossRef]
- Trelis, M.; Sotillo, J.; Monteagudo, C.; Fried, B.; Marcilla, A.; Esteban, J.G.; Toledo, R. Echinostoma caproni (Trematoda): Differential in vivo cytokine responses in high and low compatible hosts. Exp. Parasitol. 2011, 127, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Eichenberger, R.M.; Sotillo, J.; Loukas, A. Immunobiology of parasitic worm extracellular vesicles. Immunol. Cell Biol. 2018, 96, 704–713. [Google Scholar] [CrossRef] [PubMed]
- Cortés, A.; Sotillo, J.; Muñoz-Antolí, C.; Trelis, M.; Esteban, J.G.; Toledo, R. Definitive host influences the proteomic profile of excretory/secretory products of the trematode Echinostoma caproni. Parasites Vectors 2016, 9, 185. [Google Scholar] [CrossRef]
- Toledo, R.; Espert, A.; Carpena, I.; Muñoz-Antoli, C.; Fried, B.; Esteban, J.G. Immunological characterization of somatic and excretory–secretory antigens of Echinostoma caproni (Trematoda: Echinostomatidae) in experimentally infected rats. Comp. Parasitol. 2004, 71, 42–48. [Google Scholar] [CrossRef]
- Sotillo, J.; Valero, L.; Sanchez Del Pino, M.; Fried, B.; Esteban, J.; Marcilla, A.; Toledo, R. Identification of antigenic proteins from Echinostoma caproni (Trematoda) recognized by mouse immunoglobulins M, A and G using an immunoproteomic approach. Parasite Immunol. 2008, 30, 271–279. [Google Scholar] [CrossRef]
- Higón, M.; Monteagudo, C.; Fried, B.; Esteban, J.; Toledo, R.; Marcilla, A. Molecular cloning and characterization of Echinostoma caproni heat shock protein-70 and differential expression in the parasite derived from low-and high-compatible hosts. Parasitology 2008, 135, 1469–1477. [Google Scholar] [CrossRef]
- Sotillo, J.; Valero, M.L.; Sanchez Del Pino, M.M.; Fried, B.; Esteban, J.G.; Marcilla, A.; Toledo, R. Excretory/secretory proteome of the adult stage of Echinostoma caproni. Parasitol. Res. 2010, 107, 691–697. [Google Scholar] [CrossRef]
- Toledo, R.; Esteban, J.G.; Fried, B. Chapter 3 recent advances in the biology of echinostomes. Adv. Parasitol. 2009, 69, 147–204. [Google Scholar] [CrossRef] [PubMed]
- Casulli, A.; Massolo, A.; Saarma, U.; Umhang, G.; Santolamazza, F.; Santoro, A. Species and genotypes belonging to Echinococcus granulosus sensu lato complex causing human cystic echinococcosis in Europe (2000–2021): A systematic review. Parasites Vectors 2022, 15, 109. [Google Scholar] [CrossRef]
- Tamarozzi, F.; Legnardi, M.; Fittipaldo, A.; Drigo, M.; Cassini, R. Epidemiological distribution of Echinococcus granulosus s.l. infection in human and domestic animal hosts in European Mediterranean and Balkan countries: A systematic review. PLoS Neg. Trop. Dis. 2020, 14, e0008519. [Google Scholar] [CrossRef]
- Karshima, S.N.; Ahmed, M.I.; Adamu, N.B.; Magaji, A.A.; Zakariah, M.; Mohammed, K. Africa-wide meta-analysis on the prevalence and distribution of human cystic echinococcosis and canine Echinococcus granulosus infections. Parasites Vectors 2022, 15, 357. [Google Scholar] [CrossRef] [PubMed]
- Pal, M.; Alemu, H.H.; Marami, L.M.; Garedo, D.R.; Bodena, E.B. Cystic Echincoccoosis: A comprehensive review on life cycle, epidemiology, pathogenesis, clinical spectrum, diagnosis, public health and economic implications, treatment, and control. Int. J. Clin. Exp. Med. Res. 2022, 6, 131–141. [Google Scholar] [CrossRef]
- Alshoabi, S.A.; Alkalady, A.H.; Almas, K.M.; Magram, A.O.; Algaberi, A.K.; Alareqi, A.A.; Hamid, A.M.; Alhazmi, F.H.; Qurashi, A.A.; Abdulaal, O.M. Hydatid disease: A radiological pictorial review of a great neoplasms mimicker. Diagnostics 2023, 13, 1127. [Google Scholar] [CrossRef] [PubMed]
- Gessese, A.T. Review on epidemiology and public health significance of hydatidosis. Vet. Med. Int. 2020, 2020, 8859116. [Google Scholar] [CrossRef]
- Tamarozzi, F.; Deplazes, P.; Casulli, A. Reinventing the wheel of Echinococcus granulosus sensu lato transmission to humans. Trends. Parasitol. 2020, 36, 427–434. [Google Scholar] [CrossRef]
- Woolsey, I.D.; Miller, A.L. Echinococcus granulosus sensu lato and Echinococcus multilocularis: A review. Res. Vet. Sci. 2021, 135, 517–522. [Google Scholar] [CrossRef]
- Ohiolei, J.A.; Yan, H.-B.; Odeniran, P.O.; Li, L.; Shumuye, N.A.; Qurishi, S.A.; Isaac, C.; Fu, B.-Q.; Jia, W.-Z. Echinococcus granulosus sensu lato in animal intermediate hosts: What is with the organ location? Vet. Parasitol. 2022, 304, 109695. [Google Scholar] [CrossRef]
- Serra, E.; Masu, G.; Chisu, V.; Cappai, S.; Masala, G.; Loi, F.; Piseddu, T. Environmental contamination by Echinococcus spp. eggs as a risk for human health in educational farms of Sardinia, Italy. Vet. Sci. 2022, 9, 143. [Google Scholar] [CrossRef]
- Zhang, X.; Gong, W.; Cao, S.; Yin, J.; Zhang, J.; Cao, J.; Shen, Y. Comprehensive analysis of non-coding RNA profiles of exosome-like vesicles from the protoscoleces and hydatid cyst fluid of Echinococcus granulosus. Front. Cell. Infect. Microbiol. 2020, 10, 316. [Google Scholar] [CrossRef]
- Wen, H.; Vuitton, L.; Tuxun, T.; Li, J.; Vuitton, D.A.; Zhang, W.; McManus, D.P. Echinococcosis: Advances in the 21st century. Clin. Microbiol. Rev. 2019, 32, 10–1128. [Google Scholar] [CrossRef]
- Debarba, J.A.; Monteiro, K.M.; Moura, H.; Barr, J.R.; Ferreira, H.B.; Zaha, A. Identification of newly synthesized proteins by Echinococcus granulosus protoscoleces upon induction of strobilation. PLoS Negl. Trop. Dis. 2015, 9, e0004085. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, K.M.; de Carvalho, M.O.; Zaha, A.; Ferreira, H.B. Proteomic analysis of the Echinococcus granulosus metacestode during infection of its intermediate host. Proteomics 2010, 10, 1985–1999. [Google Scholar] [CrossRef] [PubMed]
- Diaz, A.; Casaravilla, C.; Barrios, A.A.; Ferreira, A.M. Parasite molecules and host responses in cystic echinococcosis. Parasite Immunol. 2016, 38, 193–205. [Google Scholar] [CrossRef]
- Cancela, M.; Paes, J.A.; Moura, H.; Barr, J.R.; Zaha, A.; Ferreira, H.B. Unraveling oxidative stress response in the cestode parasite Echinococcus granulosus. Sci. Rep. 2019, 9, 15876. [Google Scholar] [CrossRef]
- Wang, Y.; Cheng, Z.; Lu, X.; Tang, C. Echinococcus multilocularis: Proteomic analysis of the protoscoleces by two-dimensional electrophoresis and mass spectrometry. Exp. Parasitol. 2009, 123, 162–167. [Google Scholar] [CrossRef]
- Siracusano, A.; Delunardo, F.; Teggi, A.; Ortona, E. Host-parasite relationship in cystic echinococcosis: An evolving story. J. Immunol. Res. 2012, 639362. [Google Scholar] [CrossRef] [PubMed]
- Hewitson, J.P.; Maizels, R.M. Vaccination against helminth parasite infections. Expert Rev. Vaccines 2014, 13, 473–487. [Google Scholar] [CrossRef]
- Wu, Z.; Wang, L.; Li, J.; Wang, L.; Wu, Z.; Sun, X. Extracellular vesicle-mediated communication within host-parasite interactions. Front. Immunol. 2019, 9, 3066. [Google Scholar] [CrossRef]
- Zeghir-Bouteldja, R.; Touil-Boukoffa, C. Identification of proteins of laminated layer of Echinococcus granulosus: Interface among host and parasite. Veterinaria 2022, 71, 53–60. [Google Scholar] [CrossRef]
- Wang, Y.; Xiao, D.; Shen, Y.; Han, X.; Zhao, F.; Li, X.; Wu, W.; Zhou, H.; Zhang, J.; Cao, J. Proteomic analysis of the excretory/secretory products and antigenic proteins of Echinococcus granulosus adult worms from infected dogs. BMC Vet. Res. 2015, 11, 1–7. [Google Scholar] [CrossRef]
- Drurey, C.; Coakley, G.; Maizels, R.M. Extracellular vesicles: New targets for vaccines against helminth parasites. Int. J. Parasitol. 2020, 50, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Makepeace, B.L.; Martin, C.; Turner, J.D.; Specht, S. Granulocytes in helminth infection-who is calling the shots? Curr. Med. Chem. 2012, 19, 1567–1586. [Google Scholar] [CrossRef]
- Schmidt, V.; O’Hara, M.-C.; Ngowi, B.; Herbinger, K.-H.; Noh, J.; Wilkins, P.P.; Richter, V.; Kositz, C.; Matuja, W.; Winkler, A.S. Taenia solium cysticercosis and taeniasis in urban settings: Epidemiological evidence from a health-center based study among people with epilepsy in Dar es Salaam, Tanzania. PLoS Negl. Trop. Dis. 2019, 13, e0007751. [Google Scholar] [CrossRef]
- Acosta Soto, L.; Parker, L.A.; Irisarri-Gutiérrez, M.J.; Bustos, J.A.; Castillo, Y.; Perez, E.; Muñoz-Antoli, C.; Esteban, J.G.; García, H.H.; Bornay-Llinares, F.J. Evidence for transmission of Taenia solium taeniasis/cysticercosis in a rural area of Northern Rwanda. Front. Vet. Sci. 2021, 8, 645076. [Google Scholar] [CrossRef]
- Sciutto, E.; Fragoso, G.; Fleury, A.; Laclette, J.P.; Sotelo, J.; Aluja, A.; Vargas, L.; Larralde, C. Taenia solium disease in humans and pigs: An ancient parasitosis disease rooted in developing countries and emerging as a major health problem of global dimensions. Microbes Infect. 2000, 2, 1875–1890. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.-W.; Ito, A.; Ai, L.; Zhou, X.-N.; Acosta, L.P.; Willingham III, A.L. Cysticercosis/taeniasis endemicity in Southeast Asia: Current status and control measures. Acta Trop. 2017, 165, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Yanagida, T.; Nakao, M. Recent advances and perspectives in molecular epidemiology of Taenia solium cysticercosis. Infect. Genet. Evol. 2016, 40, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Victor, B.; Kanobana, K.; Gabriël, S.; Polman, K.; Deckers, N.; Dorny, P.; Deelder, A.M.; Palmblad, M. Proteomic analysis of Taenia solium metacestode excretion-secretion proteins. Proteomics 2012, 12, 1860–1869. [Google Scholar] [CrossRef]
- Santivanez, S.; Hernandez-Gonzalez, A.; Chile, N.; Oleaga, A.; Arana, Y.; Palma, S.; Verastegui, M.; Gonzalez, A.; Gilman, R.; Garcia, H. Proteomic study of activated Taenia solium oncospheres. Mol. Biochem. Parasitol. 2010, 171, 32–39. [Google Scholar] [CrossRef]
- Kaur, R.; Arora, N.; Jamakhani, M.A.; Malik, S.; Kumar, P.; Anjum, F.; Tripathi, S.; Mishra, A.; Prasad, A. Development of multi-epitope chimeric vaccine against Taenia solium by exploring its proteome: An in silico approach. Expert Rev. Vaccines 2020, 19, 105–114. [Google Scholar] [CrossRef]
- Li, L.; He, W.; Fan, X.; Liu, M.; Luo, B.; Yang, F.; Jiang, N.; Wang, L.; Zhou, B. Proteomic analysis of Taenia solium cysticercus and adult stages. Front. Vet. Sci. 2023, 9, 934197. [Google Scholar] [CrossRef]
- Navarrete-Perea, J.; Isasa, M.; Paulo, J.A.; Corral-Corral, R.; Flores-Bautista, J.; Hernández-Téllez, B.; Bobes, R.J.; Fragoso, G.; Sciutto, E.; Soberón, X. Quantitative multiplexed proteomics of Taenia solium cysts obtained from the skeletal muscle and central nervous system of pigs. PLoS Negl. Trop. Dis. 2017, 11, e0005962. [Google Scholar] [CrossRef]
- Arora, N.; Prasad, A. Taenia solium proteins: A beautiful kaleidoscope of pro and anti-inflammatory antigens. Expert Rev. Proteom. 2020, 17, 609–622. [Google Scholar] [CrossRef]
- Lee, E.G.; Kim, S.H.; Bae, Y.A.; Chung, J.Y.; Suh, M.; Na, B.K.; Kim, T.S.; Kang, I.; Ma, L.; Kong, Y. A hydrophobic ligand-binding protein of the Taenia solium metacestode mediates uptake of the host lipid: Implication for the maintenance of parasitic cellular homeostasis. Proteomics 2007, 7, 4016–4030. [Google Scholar] [CrossRef]
- Kim, S.-H.; Bae, Y.-A.; Yang, Y.; Hong, S.-T.; Kong, Y. Paralogous proteins comprising the 150 kDa hydrophobic-ligand-binding-protein complex of the Taenia solium metacestode have evolved non-overlapped binding affinities toward fatty acid analogs. Int. J. Parasitol. 2011, 41, 1207–1215. [Google Scholar] [CrossRef]
- Kim, S.-H.; Bae, Y.-A.; Yang, H.-J.; Shin, J.-H.; Diaz-Camacho, S.P.; Nawa, Y.; Kang, I.; Kong, Y. Structural and binding properties of two paralogous fatty acid binding proteins of Taenia solium metacestode. PLoS Negl. Trop. Dis. 2012, 6, e1868. [Google Scholar] [CrossRef]
- Kaur, R.; Arora, N.; Rawat, S.S.; Keshri, A.K.; Singh, G.; Kumar, R.; Prasad, A. Recognition of immune reactive proteins as a potential multiepitope vaccine candidate of Taenia solium cysticerci through proteomic approach. J. Cell. Biochem. 2023, 124, 1587–1602. [Google Scholar] [CrossRef]
- Kaur, R.; Arora, N.; Rawat, S.S.; Keshri, A.K.; Sharma, S.R.; Mishra, A.; Singh, G.; Prasad, A. Vaccine for a neglected tropical disease Taenia solium cysticercosis: Fight for eradication against all odds. Expert Rev. Vaccines 2021, 20, 1447–1458. [Google Scholar] [CrossRef]
- Jarošová, J.; Antolová, D.; Iglodyová, A.; Königová, A.; Dolinská, M.U.; Víchová, B. Molecular identification of Taenia hydatigena from domestic and free-living animals in Slovakia, Central Europe. Parasitol. Res. 2022, 121, 1345–1354. [Google Scholar] [CrossRef]
- Ulziijargal, G.; Yeruult, C.; Khulan, J.; Gantsetseg, C.; Wandra, T.; Yamasaki, H.; Narankhajid, M. Molecular identification of Taenia hydatigena and Mesocestoides species based on copro-DNA analysis of wild carnivores in Mongolia. Int. J. Parasitol. Parasit. Wild Life 2020, 11, 72–82. [Google Scholar] [CrossRef]
- Velusamy, R.; Annamalai, L.; Vijayasarathi, M.K. Parasites in the gastrointestinal system of dogs and cats. In Organ-Specific Parasitic Diseases of Dogs and Cats; Elsevier: Amsterdam, The Netherlands, 2023; pp. 205–238. [Google Scholar] [CrossRef]
- Wang, S.; Liu, X.; Liu, Z.; Wang, Y.; Guo, A.; Huang, W.; Wang, Q.; Zhang, S.; Zhu, G.; Luo, X. The genome of the thin-necked bladder worm Taenia hydatigena reveals evolutionary strategies for helminth survival. Commun. Biol. 2021, 4, 1004. [Google Scholar] [CrossRef]
- Cai, M.; Li, Y.; He, G.; Guo, X.; Zhang, S.; Yan, L.; Zhang, J.; Ding, J. Comparative Proteomic Analysis of Different Parts of Taenia Hydatigena. Front. Vet. Sci. 2021, 8, 626579. [Google Scholar] [CrossRef]
- Zheng, Y. Proteomic analysis of Taenia hydatigena cyst fluid reveals unique internal microenvironment. Acta Trop. 2017, 176, 224–227. [Google Scholar] [CrossRef]
- Guo, X. Proteomics analysis of Hydatigera taeniaeformis metacestode stage. Front. Vet. Sci. 2020, 7, 474. [Google Scholar] [CrossRef]
- Knox, D.P. Development of vaccines against gastrointestinal nematodes. Parasitology 2000, 120, 43–61. [Google Scholar] [CrossRef]
- Panti-May, J.A.; Rodríguez-Vivas, R.I.; García-Prieto, L.; Servián, A.; Costa, F. Worldwide overview of human infections with Hymenolepis diminuta. Parasitol. Res. 2020, 119, 1997–2004. [Google Scholar] [CrossRef]
- Thompson, R. Neglected zoonotic helminths: Hymenolepis nana, Echinococcus canadensis and Ancylostoma ceylanicum. Clin. Microbiol. Infect. 2015, 21, 426–432. [Google Scholar] [CrossRef]
- Sulima-Celińska, A.; Kalinowska, A.; Młocicki, D. The Tapeworm Hymenolepis diminuta as an important model organism in the experimental parasitology of the 21st century. Pathogens 2022, 11, 1439. [Google Scholar] [CrossRef]
- Galoș, F.; Anghel, M.; Ioan, A.; Ieșanu, M.-I.; Boboc, C.; Boboc, A.A. Hymenolepis diminuta infection in a Romanian Child from an urban area. Pathogens 2022, 11, 322. [Google Scholar] [CrossRef]
- Zawistowska-Deniziak, A.; Basałaj, K.; Strojny, B.; Młocicki, D. New data on human macrophages polarization by Hymenolepis diminuta tapeworm-an in-vitro study. Front. Immunol. 2017, 8, 148. [Google Scholar] [CrossRef]
- Sulima, A.; Savijoki, K.; Bień, J.; Näreaho, A.; Sałamatin, R.; Conn, D.B.; Młocicki, D. Comparative proteomic analysis of Hymenolepis diminuta cysticercoid and adult stages. Front. Microbiol. 2018, 8, 2672. [Google Scholar] [CrossRef] [PubMed]
- Mazanec, H.; Koník, P.; Gardian, Z.; Kuchta, R. Extracellular vesicles secreted by model tapeworm Hymenolepis diminuta: Biogenesis, ultrastructure and protein composition. Int. J. Parasitol. 2021, 51, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Sulima, A.; Bień, J.; Savijoki, K.; Näreaho, A.; Sałamatin, R.; Conn, D.B.; Młocicki, D. Identification of immunogenic proteins of the cysticercoid of Hymenolepis diminuta. Parasites Vectors 2017, 10, 577. [Google Scholar] [CrossRef] [PubMed]
- Bień, J.; Sałamatin, R.; Sulima, A.; Savijoki, K.; Conn, D.B.; Näreaho, A.; Młocicki, D. Mass spectrometry analysis of the excretory-secretory (ES) products of the model cestode Hymenolepis diminuta reveals their immunogenic properties and the presence of new ES proteins in cestodes. Acta Parasitol. 2016, 61, 429–442. [Google Scholar] [CrossRef]
- Młocicki, D.; Sulima, A.; Bień, J.; Näreaho, A.; Zawistowska-Deniziak, A.; Basałaj, K.; Sałamatin, R.; Conn, D.B.; Savijoki, K. Immunoproteomics and surfaceomics of the adult tapeworm Hymenolepis diminuta. Front. Immunol. 2018, 9, 2487. [Google Scholar] [CrossRef]
- Chemale, G.; van Rossum, A.J.; Jefferies, J.R.; Barrett, J.; Brophy, P.M.; Ferreira, H.B.; Zaha, A. Proteomic analysis of the larval stage of the parasite Echinococcus granulosus: Causative agent of cystic hydatid disease. PROTEOMICS Int. Ed. 2003, 3, 1633–1636. [Google Scholar] [CrossRef]
- Monteiro, K.M.; Lorenzatto, K.R.; de Lima, J.C.; Dos Santos, G.B.; Förster, S.; Paludo, G.P.; Carvalho, P.C.; Brehm, K.; Ferreira, H.B. Comparative proteomics of hydatid fluids from two Echinococcus multilocularis isolates. J. Proteom. 2017, 162, 40–51. [Google Scholar] [CrossRef]
- Blanco, M.G.; De Rosa, M.J.; Rayes, D. Anthelmintic drug discovery: Current situation and future perspectives. Front. Clin. Drug Res. Anti-Infect. 2021, 8, 209. [Google Scholar] [CrossRef]
- Riaz, A.; Bano, F.; Marescotti, M.; Saba, E.; Manzoor, Z. Anthelmintic Resistance. In Antiparasitic Drug Resistance in Veterinary Practice; CAB International 2024; Rizwan, H.M., Naeem, M.A., Younus, M., Sajid, M.S., Chen, X., Eds.; CAB International: Willingford, UK, 2024; pp. 41–57. [Google Scholar] [CrossRef]
- McVeigh, P. Post-genomic progress in helminth parasitology. Parasitology 2020, 147, 835–840. [Google Scholar] [CrossRef]
- Kim, E.; Park, S.; Park, H.; Choi, J.; Yoon, H.J.; Kim, J.-H. Determination of anthelmintic and antiprotozoal drug residues in fish using liquid chromatography-tandem mass spectrometry. Molecules 2021, 26, 2575. [Google Scholar] [CrossRef]
- Morales-Montor, J.; Del Río-Araiza, V.H.; Hernandéz-Bello, R. Parasitic Helminths and Zoonoses: From Basic to Applied Research; BoD–Books on Demand: Norderstedt, Germany, 2022. [Google Scholar]
- Malik, M.A.; Sajid, M.S.; Abbas, R.Z.; Aleem, M.T.; Anjum, F.R.; Khan, A.; Farhab, M.; Maqbool, M.; Zeeshan, M.; Hussain, K. Anthelmintic drug resistance in livestock: Current understanding and future trends. In Parasitic Helminths and Zoonoses-From Basic to Applied Research; IntechOpen: London, UK, 2022. [Google Scholar]
- Khodadadi, E.; Zeinalzadeh, E.; Taghizadeh, S.; Mehramouz, B.; Kamounah, F.S.; Khodadadi, E.; Ganbarov, K.; Yousefi, B.; Bastami, M.; Kafil, H.S. Proteomic applications in antimicrobial resistance and clinical microbiology studies. Infect. Drug Resist. 2020, 13, 1785–1806. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, S.; Rastogi, P.; Mendiratta, N. Bioinformatics: Methods and Applications-Genomics, Proteomics and Drug Discovery; PHI Learning Pvt. Ltd.: New Delhi, India, 2022. [Google Scholar]
- Chagas, A.C.S.; Ribeiro, D.M.; Osório, H.; Abreu, A.A.; Okino, C.H.; Niciura, S.C.; Amarante, A.F.; Bello, H.J.; Melito, G.R.; Esteves, S.N. Molecular signatures of Haemonchus contortus infection in sheep: A comparative serum proteomic study on susceptible and resistant sheep breeds. Vet. Parasitol. 2024, 331, 110280. [Google Scholar] [CrossRef] [PubMed]
- Ibarra-Meneses, A.V.; Corbeil, A.; Wagner, V.; Beaudry, F.; do Monte-Neto, R.L.; Fernandez-Prada, C. Exploring direct and indirect targets of current antileishmanial drugs using a novel thermal proteomics profiling approach. Front. Cell. Infect. Microbiol. 2022, 12, 954144. [Google Scholar] [CrossRef]
- Ahire, D.; Kruger, L.; Sharma, S.; Mettu, V.S.; Basit, A.; Prasad, B. Quantitative proteomics in translational absorption, distribution, metabolism, and excretion and precision medicine. Pharmacol. Rev. 2022, 74, 771–798. [Google Scholar] [CrossRef]
- Seebacher, N.A.; Krchniakova, M.; Stacy, A.E.; Skoda, J.; Jansson, P.J. Tumour microenvironment stress promotes the development of drug resistance. Antioxidants 2021, 10, 1801. [Google Scholar] [CrossRef]
- Saracino, M.P.; Vila, C.C.; Baldi, P.C.; Gonzalez Maglio, D.H. Searching for the one (s): Using probiotics as anthelmintic treatments. Front. Pharmacol. 2021, 12, 714198. [Google Scholar] [CrossRef]
- Rahime, Ş.; Aqsa, F.; Salah-Ud-Din, K.; Shahanavaj, K. Parasitic infections: Immune responses and therapeutics. Chapter 1, Back to the future‒solutions for parasitic problems. In Parasitic Infections: Immune Responses and Therapeutics; Wiley: Hoboken, NJ, USA, 2023. [Google Scholar] [CrossRef]
- Zhang, C.; Liang, C.; Zhang, C.; Ma, Y. Symbiosis evolution model and behavior of multiple resource agents in the smart elderly care service ecosystem. Symmetry 2021, 13, 570. [Google Scholar] [CrossRef]
- McIntyre, J.R. Genetic Markers of Anthelmintic Resistance in Gastrointestinal Parasites of Ruminants. Ph.D. Thesis, University of Glasgow, Glasgow, UK, 2020. [Google Scholar]
- Liu, Y.; Wang, X.; Luo, X.; Wang, R.; Zhai, B.; Wang, P.; Li, J.; Yang, X. Transcriptomics and proteomics of Haemonchus contortus in response to Ivermectin treatment. Animals 2023, 13, 919. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, C.; Alonso, P.; Ringwald, P. Current and emerging strategies to combat antimalarial resistance. Expert Rev. Anti-infect. Ther. 2022, 20, 353–372. [Google Scholar] [CrossRef]
- Wang, T.; Gasser, R.B. Prospects of using high-throughput proteomics to underpin the discovery of animal host–nematode interactions. Pathogens 2021, 10, 825. [Google Scholar] [CrossRef]
- Vashisht, V.; Vashisht, A.; Mondal, A.K.; Farmaha, J.; Alptekin, A.; Singh, H.; Ahluwalia, P.; Srinivas, A.; Kolhe, R. Genomics for emerging pathogen identification and monitoring: Prospects and obstacles. BioMedInformatics 2023, 3, 1145–1177. [Google Scholar] [CrossRef]
- Geddes-McAlister, J.; Roux-Dalvai, F.; Droit, A. Proteomics, bioinformatics, and infectious diseases. In Genetics and Evolution of Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2024; pp. 465–492. [Google Scholar] [CrossRef]
- Wijnant, G.-J.; Dumetz, F.; Dirkx, L.; Bulté, D.; Cuypers, B.; Van Bocxlaer, K.; Hendrickx, S. Tackling drug resistance and other causes of treatment failure in leishmaniasis. Front. Trop. Dis. 2022, 3, 837460. [Google Scholar] [CrossRef]
- Peraman, R.; Sure, S.K.; Dusthackeer, V.A.; Chilamakuru, N.B.; Yiragamreddy, P.R.; Pokuri, C.; Kutagulla, V.K.; Chinni, S. Insights on recent approaches in drug discovery strategies and untapped drug targets against drug resistance. Futur. J. Pharm. Sci. 2021, 7, 56. [Google Scholar] [CrossRef] [PubMed]
- Chulanetra, M.; Chaicumpa, W. Revisiting the mechanisms of immune evasion employed by human parasites. Front. Cell. Infect. Microbiol. 2021, 11, 702125. [Google Scholar] [CrossRef]
- Van den Kerkhof, M.; Sterckx, Y.G.-J.; Leprohon, P.; Maes, L.; Caljon, G. Experimental strategies to explore drug action and resistance in kinetoplastid parasites. Microorganisms 2020, 8, 950. [Google Scholar] [CrossRef] [PubMed]
- Sukumaran, A.; Woroszchuk, E.; Ross, T.; Geddes-McAlister, J. Proteomics of host–bacterial interactions: New insights from dual perspectives. Can. J. Microbiol. 2021, 67, 213–225. [Google Scholar] [CrossRef]
- Vasala, A.; Hytönen, V.P.; Laitinen, O.H. Modern tools for rapid diagnostics of antimicrobial resistance. Front. Cell. Infect. Microbiol. 2020, 10, 308. [Google Scholar] [CrossRef]
- Aboshady, H.M.; Stear, M.J.; Johansson, A.; Jonas, E.; Bambou, J.-C. Immunoglobulins as biomarkers for gastrointestinal nematodes resistance in small ruminants: A systematic review. Sci. Rep. 2020, 10, 7765. [Google Scholar] [CrossRef]
- Bhanot, P. Introduction to Parasites. In Practical Handbook of Microbiology; CRC Press: Boca Raton, FL, USA, 2021; pp. 665–682. [Google Scholar] [CrossRef]
- Azad, A.K.; Hakim, A.; Sohag, M.M.H.; Rahman, M. Metabolomics in clinical diagnosis, prognosis, and treatment of infectious diseases. In Metabolomics; Elsevier: Amsterdam, The Netherlands, 2023; pp. 71–119. [Google Scholar] [CrossRef]
- Rivara-Espasandín, M.; Palumbo, M.C.; Sosa, E.J.; Radío, S.; Turjanski, A.G.; Sotelo-Silveira, J.; Fernandez Do Porto, D.; Smircich, P. Omics data integration facilitates target selection for new antiparasitic drugs against TriTryp infections. Front. Pharmacol. 2023, 14, 1136321. [Google Scholar] [CrossRef]
- Sotillo, J.; Pearson, M.; Becker, L.; Mulvenna, J.; Loukas, A. A quantitative proteomic analysis of the tegumental proteins from Schistosoma mansoni schistosomula reveals novel potential therapeutic targets. Int. J. Parasitol. 2015, 45, 505–516. [Google Scholar] [CrossRef]
- Shao, G.; Hua, R.; Song, H.; Chen, Y.; Zhu, X.; Hou, W.; Li, S.; Yang, A.; Yang, G. Protective efficacy of six recombinant proteins as vaccine candidates against Echinococcus granulosus in dogs. PLoS Negl. Trop. Dis. 2023, 17, e0011709. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Z.-A.; Wu, C.-L.; Wang, D.-Y.; Zhong, S.-H.; Yang, X.; Rao, G.-S.; Peng, H.; Feng, S.-W.; Li, J.; Huang, W.-Y. Proteomic analysis of exosome-like vesicles from Fasciola gigantica adult worm provides support for new vaccine targets against fascioliasis. Parasites Vectors 2023, 16, 62. [Google Scholar] [CrossRef] [PubMed]
- Dicker, A.J.; Inglis, N.F.; Manson, E.D.; Subhadra, S.; Illangopathy, M.; Muthusamy, R.; Knox, D.P. Proteomic analysis of Mecistocirrus digitatus and Haemonchus contortus intestinal protein extracts and subsequent efficacy testing in a vaccine trial. PLoS Negl. Trop. Dis. 2014, 8, e2909. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Sharan, U.; Kaur, R.; Rawat, S.S.; Keshri, A.K.; Prasad, A. An Inclusive Approach to Design a Multi-Epitope Chimeric Vaccine for Taenia Infections by Integrating Proteomics and Reverse Vaccinology. Front. Trop. Dis. 2024, 5, 1393570. [Google Scholar] [CrossRef]
- Castro-Borges, W.; Dowle, A.; Curwen, R.S.; Thomas-Oates, J.; Wilson, R.A. Enzymatic shaving of the tegument surface of live schistosomes for proteomic analysis: A rational approach to select vaccine candidates. PLoS Negl. Trop. Dis. 2011, 5, e993. [Google Scholar] [CrossRef]
- Sotillo, J.; Doolan, D.; Loukas, A. Recent advances in proteomic applications for schistosomiasis research: Potential clinical impact. Expert Rev. Proteom. 2017, 14, 171–183. [Google Scholar] [CrossRef]
- Miles, S.; Portela, M.; Cyrklaff, M.; Ancarola, M.E.; Frischknecht, F.; Durán, R.; Dematteis, S.; Mourglia-Ettlin, G. Combining proteomics and bioinformatics to explore novel tegumental antigens as vaccine candidates against Echinococcus granulosus infection. J. Cell. Biochem. 2019, 120, 15320–15336. [Google Scholar] [CrossRef]
- Hewitson, J.P.; Ivens, A.C.; Harcus, Y.; Filbey, K.J.; McSorley, H.J.; Murray, J.; Bridgett, S.; Ashford, D.; Dowle, A.A.; Maizels, R.M. Secretion of protective antigens by tissue-stage nematode larvae revealed by proteomic analysis and vaccination-induced sterile immunity. PLoS Pathog. 2013, 9, e1003492. [Google Scholar] [CrossRef]
- Newton, S.E.; Meeusen, E.N.T. Progress and new technologies for developing vaccines against gastrointestinal nematode parasites of sheep. Parasit. Immunol. 2003, 25, 283–296. [Google Scholar] [CrossRef]
- Rehman, A.; Ahmad, S.; Shahid, F.; Albutti, A.; Alwashmi, A.S.; Aljasir, M.A.; Alhumeed, N.; Qasim, M.; Ashfaq, U.A.; Tahir ul Qamar, M. Integrated core proteomics, subtractive proteomics, and immunoinformatics investigation to unveil a potential multi-epitope vaccine against schistosomiasis. Vaccines 2021, 9, 658. [Google Scholar] [CrossRef]
- Mutapi, F. Helminth parasite proteomics: From experimental models to human infections. Parasitology 2012, 139, 1195–1204. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wilson, R.A.; Curwen, R.S.; Braschi, S.; Hall, S.L.; Coulson, P.S.; Ashton, P.D. From genomes to vaccines via the proteome. Mem. Inst. Oswaldo Cruz 2004, 99, 45–50. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soleymani, N.; Sadr, S.; Santucciu, C.; Dianaty, S.; Lotfalizadeh, N.; Hajjafari, A.; Heshmati, F.; Borji, H. Unveiling Novel Insights in Helminth Proteomics: Advancements, Applications, and Implications for Parasitology and Beyond. Biologics 2024, 4, 314-344. https://doi.org/10.3390/biologics4030020
Soleymani N, Sadr S, Santucciu C, Dianaty S, Lotfalizadeh N, Hajjafari A, Heshmati F, Borji H. Unveiling Novel Insights in Helminth Proteomics: Advancements, Applications, and Implications for Parasitology and Beyond. Biologics. 2024; 4(3):314-344. https://doi.org/10.3390/biologics4030020
Chicago/Turabian StyleSoleymani, Nooshinmehr, Soheil Sadr, Cinzia Santucciu, Shiva Dianaty, Narges Lotfalizadeh, Ashkan Hajjafari, Fatemeh Heshmati, and Hassan Borji. 2024. "Unveiling Novel Insights in Helminth Proteomics: Advancements, Applications, and Implications for Parasitology and Beyond" Biologics 4, no. 3: 314-344. https://doi.org/10.3390/biologics4030020
APA StyleSoleymani, N., Sadr, S., Santucciu, C., Dianaty, S., Lotfalizadeh, N., Hajjafari, A., Heshmati, F., & Borji, H. (2024). Unveiling Novel Insights in Helminth Proteomics: Advancements, Applications, and Implications for Parasitology and Beyond. Biologics, 4(3), 314-344. https://doi.org/10.3390/biologics4030020





