Abstract
Extracellular vesicles (EVs) encompass a diverse array of cell-derived vesicles, originating either from the endosomal compartment (exosomes) or generated through shedding from the cell membrane. These lipid bilayer nanovesicles carry a diverse cargo consisting of nucleic acids, various macromolecules, and growth factors, capable of being assimilated by nearby or distant cells through biofluids, thereby triggering a wide range of cellular responses. Given their distinctive biological characteristics and crucial roles in intercellular communication, EVs have garnered significant attention, especially concerning potential clinical applications. Inheriting cargo from their parent cells, EVs present promising resources for diverse disease biomarkers. Research elucidating the specific impacts of cargo on target cells has sparked enthusiasm for their therapeutic potential. Compelling evidence indicates that RNA cargo housed within EVs can modulate gene expression and influence cellular functions in recipient cells. However, despite significant progress, numerous aspects of EV biology remain obscure, encompassing selective cargo-loading mechanisms that yield distinct compositions from source cells, variability in size and content, and undisclosed pathways governing uptake and cargo fate in recipient cells. A thorough understanding of core EV mechanisms—such as generation, trafficking, and payload delivery—is essential for their effective clinical utilization. This review explores the current understanding of RNA loading and transportation within EVs, shedding light on the advancements made toward clinical applications.
1. Introduction
Extracellular vesicles (EVs), lipid bilayer packages secreted by all living cells, serve as messengers transporting proteins, lipids, RNA, and DNA between cells, thereby influencing their behavior and function. EVs carry a distinct RNA cargo, enriched with small non-coding RNAs (ncRNAs) like microRNAs, alongside fragments and intact mRNAs, rRNAs, and lncRNAs [1,2,3,4]. These RNAs profoundly affect recipient cells, influencing processes like wound healing, neurodegeneration, and even cancer progression. However, deciphering the exact mechanism of RNA communication within EVs remains a topic of interest for scientists. The intricate interplay between various biomolecules carried by EVs and the incomplete understanding of many ncRNA functions adds further complexity. Additionally, the journey of these exosomal RNAs, including their packaging into specific EV types, uptake by target cells, and subsequent fate, is still under investigation.
Presently, four primary categories of EV subtypes are acknowledged: “exosomes” (50–150 nanometers), “microvesicles” (100–1000 nanometers), “large oncosomes” (1000–10,000 nanometers), and “apoptotic bodies” (100–5000 nanometers) [5,6,7]. Distinguishing these from other particles, such as lipoproteins and protein aggregates, remains a hurdle, prompting ongoing efforts to standardize EV characterization to enhance research reproducibility. The small size of EVs limits their cargo capacity, making them invisible with standard microscopy and sorting techniques. The overlapping size and properties among subclasses, combined with the absence of unique markers, further complicate cargo identification, including RNA. Furthermore, technical variations in both EV isolation and RNA analysis can significantly impact the interpretation of data.
Despite these challenges, researchers are developing sophisticated approaches to tackle them, such as culturing cells in serum-free media to prevent contamination and employing size-based separation techniques to isolate specific EV subtypes [8,9]. These efforts are pivotal in understanding the biology of EV RNAs. The presence of these RNAs in easily accessible biofluids enables the non-invasive monitoring of cellular and tissue health. Additionally, EVs possess the capability to shield their cargo from degradation and deliver it precisely to target cells without eliciting an immune response. These characteristics make them ideal carriers for RNA therapies like siRNAs, miRNAs, and mRNAs. This review delves into the types of EV RNAs, packaging mechanisms, selective loading, and clinical applications. It emphasizes the key challenges encountered when working with EVs. By overcoming these hurdles, a deeper understanding of EV-mediated RNA delivery can be achieved, thereby paving the way for the diagnosis and treatment of various diseases.
2. Isolation and Characterization of Extracellular Vesicles
Extracellular vesicles (EVs) are membrane-bound nanoparticles released by cells into the extracellular environment, playing essential roles in intercellular communication and disease pathogenesis. To study EVs effectively, researchers employ various enrichment techniques to isolate and concentrate these vesicles from complex biological samples. Herein is an elaboration on each of the EV enrichment techniques.
2.1. Ultracentrifugation
Ultracentrifugation is a classical method for EV enrichment, involving the centrifugation of samples at high speeds (typically exceeding 100,000× g) to separate EVs from other components based on their size and density. Differential ultracentrifugation and gradient ultracentrifugation are two common approaches that offer varying degrees of purity and yield.
2.2. Co-Precipitation
Co-precipitation methods exploit the differential solubility of EVs and contaminants to selectively precipitate EVs from a solution. Common co-precipitants include polyethylene glycol (PEG) and protamine sulfate, which induce EV aggregation and subsequent precipitation. While simple and cost-effective, co-precipitation may lead to the co-isolation of non-EV contaminants.
2.3. Size-Exclusion Chromatography (SEC)
SEC separates EVs based on their size by passing the sample through a porous matrix that selectively retains larger particles while allowing smaller EVs to elute first. This technique offers gentle and scalable EV enrichment, with minimal sample manipulation and no requirement for specialized equipment.
2.4. Field-Flow Fractionation (FFF)
FFF is a chromatographic technique that separates particles based on their size and shape as they flow through a thin channel under the influence of a perpendicular field. FFF can effectively enrich EVs while removing contaminants based on their hydrodynamic properties. This method offers high resolution and flexibility but requires sophisticated instrumentation.
2.5. Microfluidic Filtering
Microfluidic devices leverage precise control over fluid flow and particle manipulation at the microscale, making them well-suited for EV enrichment. These devices employ nanoporous membranes or channels to selectively trap EVs while allowing smaller molecules to pass through, enabling rapid and efficient EV isolation with minimal sample loss.
2.6. Contact-Free Cell Sorting
Contact-free cell sorting, also known as acoustic-based sorting, utilizes acoustic waves to manipulate EVs within a sample, enabling gentle and label-free separation based on size and density. This technique offers the advantage of preserving EV integrity and functionality while enabling high-throughput sorting with minimal sample handling.
2.7. Immunoaffinity Enrichment
Immunoaffinity enrichment involves capturing EVs using antibodies specific to surface markers expressed on EVs. This method enables highly specific isolation of EV subpopulations based on their molecular profiles. Immunoaffinity enrichment is particularly useful for studying EV heterogeneity and identifying EV biomarkers.
Physical characterization of extracellular vesicles (EVs) plays a crucial role in comprehending their structure, size distribution, and heterogeneity. Numerous techniques are utilized for EV characterization, encompassing microscopy-based methods and particle-sizing techniques. These methods enable the visualization of EVs with nanometer-scale precision, facilitating the observation of structural attributes like shape, size, and surface characteristics. Below is a list of the following techniques employed for EV characterization.
2.8. Transmission Electron Microscopy (TEM)
TEM enables the visualization of EVs at higher magnifications, providing detailed information about their internal structures and ultrastructural features. This method offers superior resolution, allowing researchers to observe EV contents, membrane morphology, and potential cargo.
2.9. Cryo-Electron Microscopy (Cryo-EM)
Cryo-EM is a powerful technique for imaging EVs in their native hydrated state without the need for fixation or staining. This method preserves EV structure and integrity, offering detailed information about their three-dimensional architecture and organization.
2.10. Atomic Force Microscopy (AFM)
AFM enables the imaging of EVs at the nanoscale by scanning a sharp probe across the sample surface. This technique provides topographical information about EVs, including their size, shape, and surface roughness, with high resolution and minimal sample preparation requirements.
2.11. Dynamic Light Scattering (DLS)
DLS measures the fluctuation in light scattering caused by the Brownian motion of EVs in solution. By analyzing the intensity and correlation of scattered light, DLS provides information about EV size distribution and hydrodynamic radius. It is a rapid and non-destructive technique suitable for analyzing EVs in solution.
2.12. Nanoparticle Tracking Analysis (NTA)
NTA tracks individual EVs in solution using light scattering and Brownian motion analysis. This technique provides real-time visualization and quantification of the EV size distribution and concentration, offering valuable insights into EV heterogeneity and dynamics.
2.13. Tunable Resistive Pulse Sensing (TRPS)
TRPS measures the changes in electrical resistance as EVs pass through a nanopore, allowing for the precise sizing and counting of individual EVs. This technique offers high resolution and sensitivity, enabling the accurate characterization of the EV size distribution and concentration.
2.14. Single-EV Analysis (SEA) Methods
SEA methods, such as microfluidic-based approaches and single-particle interferometric reflectance imaging sensing (SP-IRIS), enable the analysis of individual EVs. These techniques offer a single-vesicle resolution, allowing for the characterization of the EV size, concentration, and cargo composition at the single-particle level.
2.15. Flow Cytometry
Flow cytometry utilizes fluorescently labeled antibodies to analyze EVs based on their size, concentration, and surface marker expression. It enables high-throughput analysis of EVs and is particularly useful for quantifying specific EV subpopulations.
3. Types of RNA in Extracellular Vesicle Cargo
Extracellular vesicles’ RNA profile displays distinct features, with small non-coding RNAs (ncRNAs/miRNAs) predominating the cargo, alongside full-length or degraded mRNAs, ribosomal RNAs (rRNAs), long non-coding RNAs (lncRNAs), and DNA fragments (Table 1). This unique composition highlights the selective packaging process that governs RNA inclusion into EVs, ensuring targeted delivery of specific messages to recipient cells. EVs encapsulate a diverse range of RNA species, including the following.
3.1. MicroRNAs (miRNAs)
These non-coding RNAs are approximately 22 nucleotides long and regulate gene expression post-transcriptionally. They achieve this by interacting with the three prime untranslated region (3′ UTR) of target mRNAs, leading to mRNA degradation and translational repression [10,11]. Among RNA species found in EVs, miRNAs stand as the most abundant and extensively studied, implicated in various biological processes such as development, cell differentiation, immune response, and disease progression [12,13,14]. For instance, in the nervous system, EVs transporting miR-193b exhibit a capability to reduce amyloid precursor protein levels within neurons, potentially mitigating the pathology associated with Alzheimer’s disease [15]. Interestingly, the abundance of this particular miRNA enclosed in EVs holds promise as a potential diagnostic biomarker for the disease. Similarly, miR-124a-loaded neuronal EVs play a crucial role in regulating glutamate transporters within astrocytes, thereby impacting synaptic transmission [16,17]. In brain cancer scenarios, astrocyte-derived EVs carrying miR-19 inhibit PTEN, a critical tumor suppressor, thereby promoting the growth of brain metastases [18,19]. Moving beyond neural contexts, EVs derived from mesenchymal stem cells, enriched with miR-375, demonstrate the ability to induce bone marrow stem cells to participate in bone regeneration processes [20]. Furthermore, dendritic cell-derived EVs facilitate the transfer of miR-155 and miR-146a in vivo, where the latter suppresses while the former stimulates endotoxin-induced inflammation [21].
3.2. Long Non-Coding RNAs (lncRNAs)
These non-coding RNAs, typically around 200 nucleotides in length, possess the ability to regulate gene expression across multiple levels, encompassing transcription, post-transcriptional processing, and chromatin modification [22,23]. These RNAs have demonstrated the potential to regulate cell proliferation, migration, invasion, and drug resistance in cancer cells [24,25]. Studies indicate that hypoxic cardiomyocytes release extracellular vesicles enriched with lncRNA NEAT1, which, upon uptake by fibroblasts, upregulate a gene expression profile linked to promoting fibrosis [26]. Another study showed that hypoxia-inducible factor 1α-stabilizing lncRNA was found to be integrated into extracellular vesicles released by tumor-associated macrophages. These vesicles, upon uptake by breast cancer cells, supported their viability, indicating a potential role in tumor progression [27]. Decoding the precise mechanisms through which these lncRNAs enclosed within extracellular vesicles exert their actions remains a challenge.
3.3. Messenger RNAs (mRNAs)
These coding RNAs function by conveying genetic information from DNA to ribosomes, enabling the synthesis of proteins. While the existence of intact mRNAs within extracellular vesicles (EVs) remains a subject of debate, fragmented forms of these messenger RNAs (mRNAs) have been identified, suggesting their involvement in cell-to-cell communication and the pathogenesis of various diseases [28]. Typically, most mRNAs detected within EVs measure as smaller than 1 kb [29]. This finding hints at the potential of these mRNA molecules to serve as a reservoir for generating novel proteins within the cells that receive them. However, releasing mRNAs from the endosome represents a challenge in ensuring functional mRNA delivery.
Recent studies have provided evidence of the active translation of EV-transported mRNAs upon uptake by recipient cells. For instance, the expression of reporter proteins resulting from transferred mRNA enclosed in EVs has been observed between mast cells [2] and from glioblastoma to endothelial cells [30]. Co-culture experiments with glioblastoma and HEK293T cells demonstrated translated mRNAs carried by EVs, eliciting phenotypic responses, while evidence also suggests that EV-borne mRNA can induce the production of functional proteins. Additionally, the transfer of Cre recombinase mRNA via EVs resulted in recombination events in the brains of floxed reporter mice [2,30]. However, while detecting Cre mRNA in EVs was achieved through reverse transcription–PCR, the corresponding protein detection by Western blot was unsuccessful. This detection sensitivity discrepancy poses challenges requiring further investigation.
Studies have shown the transfer of mRNAs between human pulmonary artery smooth muscle cells and arterial endothelial cells through EVs, which carry various mRNAs, including those involved in regulating the actin cytoskeleton and extracellular matrix remodeling. This process potentially influences the pathology of pulmonary arterial hypertension, particularly involving transforming growth factor-β [31]. Moreover, extracellular vesicles isolated from human blood were electroporated with Cas9 mRNA and were shown to facilitate CRISPR editing in recipient cells [32]. These findings support the notion that mRNA carried by EVs can indeed undergo translation within recipient cells.
3.4. Small Nucleolar RNAs (snoRNAs)
These non-coding RNAs are involved in various essential cellular processes, primarily related to ribosomal RNA (rRNA) processing and modification. Their presence within extracellular vesicles (EVs) has opened a new avenue for research, raising intriguing questions about their potential functions and roles in intercellular communication and disease progression. Studies show that the snoRNA content within extracellular vesicles derived from immune cells, such as dendritic cells, is loaded with immunomodulatory factors [33,34,35]. EVs from activated macrophages are loaded with altered snoRNA profiles, which, upon uptake by recipient cells, exhibit increased levels of 2′-O-methylation of RNA—a modification typically directed by the released snoRNAs onto their canonical rRNA targets [36]. Consequently, snoRNAs carried within extracellular vesicles have the potential to serve as modulators of inflammation.
3.5. Transfer RNAs (tRNAs)
These are small RNAs (approximately 75–90 nucleotides long) that play a crucial role in protein translation by delivering amino acids to the ribosome. However, fragmented tRNAs (tRFs) have also been identified in EVs, with potential regulatory roles in gene expression and epigenetic processes, particularly in contexts such as immunomodulation [37] and cancer progression [38]. The exact functional role of tRNAs and their fragments in recipient cells is unclear. Further research is needed to ascertain their specific effects and mechanisms of action in the recipient cells.
Ribosomal RNAs (rRNAs): These are the major structural components of ribosomes, which are essential for protein translation. While intact rRNAs are not commonly found in EVs, rRNA fragments have been identified, and their functional significance in extracellular communication remains an area of ongoing investigation.

Table 1.
Types of RNA found inside extracellular vesicles.
Table 1.
Types of RNA found inside extracellular vesicles.
| RNA Type | Size (nt) | Function | Implication in EVs | References |
|---|---|---|---|---|
| miRNAs | 22 | Post-transcriptional gene regulation | Regulate various biological processes in recipient cells | [39,40] |
| lncRNAs | >200 | Diverse regulatory roles | Impact various biological functions, including cell proliferation, migration, invasion, and drug resistance | [23,41] |
| tRNAs | 75–90 | Protein translation | Fragmented tRNAs (tRFs) regulate gene expression, cell signaling, and stress response | [42,43] |
| rRNAs | Variable | Ribosome structure | rRNA fragments are linked to cell proliferation, migration, and inflammation | [29] |
| mRNAs | Variable | Protein synthesis | Fragmented mRNAs may play a role in cell communication and disease pathogenesis | [44,45] |
| piRNAs | 26–31 | Transposon silencing and germline development | Potential role in regulating gene expression and cell communication | [46,47,48] |
| snoRNAs | 60–300 | rRNA processing and modification | Function in EVs remains unknown | [49] |
| Y RNAs | 85–100 | Regulatory role | Potential role in RNA processing and modification | [49,50,51] |
4. Mechanisms for RNA Packaging Inside Extracellular Vesicles
The RNA cargo of EVs plays a crucial role in intercellular communication, influencing various cellular processes in recipient cells. Understanding the mechanisms of RNA packaging into EVs is essential for deciphering their function and developing therapeutic strategies. The precise mechanisms of RNA packaging into EVs remain under investigation, but several key pathways are emerging.
4.1. Direct Sorting
RNA-binding proteins (RBPs) play a significant role in trafficking, packaging, and stabilizing RNA cargo within these vesicles (Figure 1A). RBPs are a diverse group of proteins that interact with RNA molecules, influencing their processing, localization, stability, and function [29,52,53]. In the context of EV-mediated RNA transport, RBPs significantly contribute to the selective packaging of specific RNA species into these vesicles. The incorporation of RNAs into EVs is a selective process, where RBPs recognize and bind to specific motifs or sequences within the RNA molecules, facilitating their encapsulation into EVs. Various RNA-binding proteins (RBPs) have been identified as playing a part in packaging distinct RNA populations into EVs (Table 2). Among these, heterogeneous nuclear ribonucleoproteins (hnRNPs) have been implicated in recognizing specific RNA sequences or secondary structures to aid in the encapsulation of mRNA and non-coding RNAs into EVs. Additionally, RBPs like Ago2 (Argonaute 2) and GW182 have been linked to loading microRNAs (miRNAs) into EVs, thus regulating miRNA stability and sorting. These findings underscore the diverse roles of RBPs in orchestrating RNA packaging within EVs [29,52,53,54].
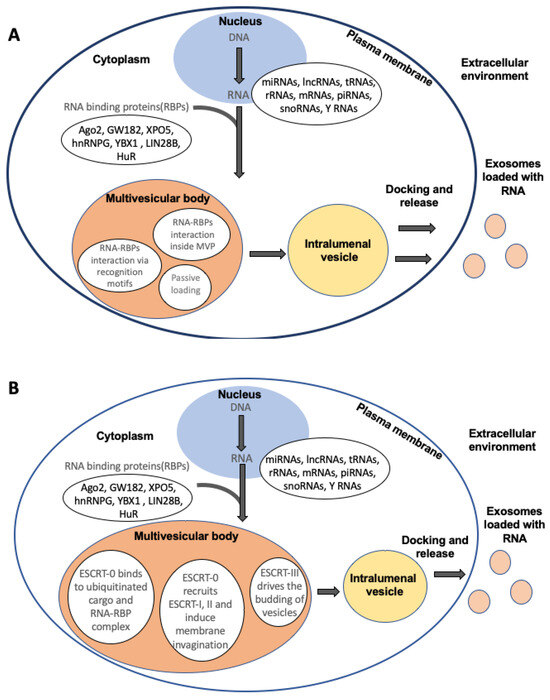
Figure 1.
Schematic of RNA packaging inside extracellular vesicles. (A) RNA and RNA-binding proteins (RBPs) interact in the cytoplasm, where RBPs bind to RNA via specific sequence recognition motifs or secondary structures. The RNA-RBP complex then interacts with certain proteins in the multivesicular body (MVB). In cases of higher abundance of RNA in the cytosol, there might be passive diffusion of RNA moieties inside the extracellular vesicles: (B) RNA-RBP complexes bind to ESCRT-0 inside the MVB, which recruits ESCRT-1 and -II, initiating membrane invaginations. Subsequently, ESCRT-III drives the budding of the vesicles inside the MVB. Cargo is transported from the MVB to intraluminal vesicles and then released from the cell via exocytosis.
The interaction between RBPs and RNA cargo takes place within endosomal compartments during the formation of multivesicular bodies and intraluminal vesicles (ILVs). Subsequently, these vesicles are transported to the plasma membrane for fusion and the release of exosomes [29,55,56]. Additionally, RBPs not only facilitate the sorting of specific RNA molecules but also contribute to the protection of encapsulated RNAs from degradation. By binding to and shielding RNA cargo, RBPs enhance the stability of these molecules within EVs, preserving their integrity and functionality during transport and delivery to recipient cells [57,58,59]. The functional implications of RNA packaging into EVs using RBPs extend beyond intercellular communication. EV-mediated transfer of RNA cargo packaged by RBPs can influence various cellular processes in recipient cells, including gene expression, signaling pathways, and modulation of the immune response. These transferred RNAs can regulate gene expression by affecting mRNA translation or stability, modulating cellular phenotypes, and potentially contributing to pathological conditions or physiological responses.
4.2. ESCRT-Mediated Sorting
The ESCRT machinery comprises four main protein complexes: ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III, alongside associated proteins such as ALIX (ALG-2-interacting protein X). These complexes operate sequentially to facilitate the inward budding of the endosomal membrane, leading to the formation of intraluminal vesicles (ILVs) within multivesicular bodies (MVBs) (Figure 1B). These ILVs encapsulate cytoplasmic contents, including RNA molecules, and are subsequently released as exosomes upon fusion of the MVBs with the plasma membrane [60,61,62]. The mechanisms of ESCRT-mediated RNA packaging involve the following steps.
4.2.1. Recognition and Sequestration of RNA Cargo
Within the cytoplasm, specific RNA molecules are recognized and bound by RNA-binding proteins (RBPs) or other sorting machinery. These RBPs may interact with the ESCRT components or be involved in ESCRT recruitment for RNA cargo sequestration.
4.2.2. ESCRT Complex Assembly and Cargo Selection
The ESCRT-0 complex, including proteins like HRS and STAM, identifies and binds ubiquitinated cargo proteins and RBP-RNA complexes present on the endosomal membrane, initiating the recruitment and stepwise assembly of ESCRT-I and ESCRT-II complexes. Alongside the protein cargo, RNA molecules associated with these cargo proteins or RNA-binding proteins (RBPs) are selectively incorporated into the developing intraluminal vesicles (ILVs).
4.2.3. ESCRT-III-Associated Membrane Budding
The ESCRT-III complex, composed of multiple proteins including CHMPs (charged multivesicular body proteins), orchestrates the final steps of ILV formation by mediating membrane scission. ESCRT-III components constrict the endosomal membrane, resulting in the formation of EVs inside MVBs and ILVs containing the selected RNA cargo.
4.2.4. Exosome Formation and Release
These MVBs and ILVs either merge with lysosomes for degradation or fuse with the cell membrane, resulting in the exocytosis of exosomes near the cell periphery. These EVs carry the packaged RNA cargo, protected by the lipid bilayer, and are then available for intercellular communication and functional transfer to recipient cells.
The ESCRT machinery’s involvement in RNA sorting and packaging into exosomes highlights the intricate coordination between cellular components in regulating the selective incorporation of RNA molecules. This selective sorting mechanism ensures the packaging of specific RNA species into exosomes, influencing the diversity and functionality of exosomal RNA content.
4.3. Passive Inclusion
In this process, the RNA in EVs lacks the specificity seen in selective packaging mechanisms. Instead, it relies on the abundance and availability of RNA molecules within the cytoplasmic milieu during the formation of ILVs. As the ILVs are formed, they can encapsulate a wide range of cytoplasmic components, including RNAs, present in the vicinity, without distinct sorting signals or specific interactions with sorting machinery. This non-discriminatory process results in the encapsulation of various RNA species including coding and non-coding RNA inside the EVs. The encapsulated RNA cargo may represent a snapshot of the cellular RNA landscape at the time of EV biogenesis, capturing a diverse repertoire of RNA molecules present within the producing cell.

Table 2.
RNA-binding proteins and their implication in RNA packaging inside exosomes.
Table 2.
RNA-binding proteins and their implication in RNA packaging inside exosomes.
| RNA-Binding Protein | Implication in RNA Packaging | Cargo Type | References |
|---|---|---|---|
| Heterogeneous nuclear ribonucleoproteins (hnRNPs) | Recognition of specific RNA sequences and secondary structures | mRNA and non-coding RNAs | [63,64] |
| Argonaute 2 (Ago2) | Loading of microRNAs (miRNAs) into EVs | miRNAs | [29,65] |
| GW182 | Co-packaging with Ago2, regulating miRNA stability and sorting | miRNAs | [66,67,68] |
| Exportin 5 (XPO5) | Nuclear export of specific RNAs; potential involvement in EV loading | diverse | [69,70] |
| hnRNPG | Involved in exosome biogenesis and RNA loading | diverse | [53,71] |
| YBX1 | Recognition of specific RNA sequences, promoting exosome release | diverse | [72] |
| LIN28B | Binds and stabilizes specific RNAs, potentially influencing EV packaging | diverse | [53,73] |
| HuR | Binds and stabilizes specific miRNAs, promoting their inclusion in EVs | miRNAs | [40,74] |
5. Selective Loading of RNA Cargo in Extracellular Vesicles
There are two main approaches for loading cargo into extracellular vesicles (EVs). The first approach involves modifying donor cells to promote the incorporation of the desired cargo into EVs. This is called the cell-based method. Alternatively, the cargo can be directly loaded into isolated EVs using techniques such as electroporation, sonication, incubation, or transfection. This is known as the non-cell-based method [29,75].
The cell-based loading approach involves the overexpression of RNA in the selected cell line, typically achieved through transfection or lentiviral transduction. This typically leads to higher RNA levels in EVs and potentially elevated expression of any encoded protein [75]. The second strategy relies on donor cells expressing proteins with specific RNA-binding motifs. These motifs, acting like molecular locks, seek out and bind to the cargo RNA equipped with the right sequence [29,53,76]. This binding guides the RNA to its destination: either the plasma membrane for budding or multivesicular bodies (MVBs) for subsequent incorporation and release as EVs. Expressing fusion proteins that combine an MVB-enriched protein with an RNA-binding motif offers an alternative and effective strategy for targeted RNA loading in EVs [75,77,78].
One method for selectively enriching RNA cargo in extracellular vesicles (EVs) involves leveraging arrestin-domain-containing protein 1 (ARRDC1)-mediated microvesicles (ARMMs), a type of EV that buds directly from the plasma membrane. This approach involves engineering ARRDC1 to recognize and bind the RNA cargo. This is achieved by incorporating the HIV Tat peptide into ARRDC1 and equipping the RNA with the corresponding transactivating response (TAR) element. This interaction enables efficient RNA loading into ARMMs. This strategy has successfully transferred p53 mRNA, which was translated into recipient cells after uptake [29,79].
In addition, other approaches involve “zipcode” sequences and exploiting the unique lipid composition of extracellular vesicles (EVs) to facilitate targeted RNA loading. These sequences, typically incorporated into the 3′ untranslated region of the cargo RNA, act as recognition signals for specific RNA-binding proteins (RBPs) associated with membrane-targeted or MVB-targeted proteins [80,81]. Additionally, there are reports which suggest that engineered RBPs with tailored peptides bind more efficiently to specific lipids on the EV membrane. For example, annexin A5 targets the negatively curved lipid phosphatidylserine, while the myristoylated alanine-rich C-kinase substrate (MARCKS) protein binds the positively curved lipid phosphatidylinositol 4,5-bisphosphate on plasma membrane-derived vesicles (residues 151–175 of MARCKS are particularly effective) [82,83]. This targeted binding facilitates the integration of RNA–RBP complexes into EVs. Additional options involve integrating binding domains from proteins with an affinity for specific intracellular membranes, such as phosphatidylinositol 4,5-bisphosphate or phosphatidylinositol 3,4,5-trisphosphate, thereby expanding the toolkit for targeted RNA loading in EVs [82,83].
Non-cell-based methods, also known as exogenous or direct loading, involve loading cargo into isolated EVs. Various substances, including siRNA, miRNA, proteins, CRISPR/Cas9, natural products derived from hydrophobic compounds, and anticancer drugs, can be incorporated into EVs using techniques such as sonication, electroporation, transfection reagents, and specific buffer agents [75,84]. Notably, some compounds effortlessly integrate with EVs through simple room-temperature mixing. These loading methods are categorized as either passive (via diffusion) or active (by disrupting the membrane through electroporation or sonication), and they can be combined to optimize cargo uptake. Following loading, the disrupted membrane naturally restores itself through incubation at room temperature or 37 °C, ensuring the stability and functionality of the EVs for further research or therapeutic applications [29,75]. This non-cell-based approach provides enhanced control over cargo incorporation while minimizing potential alterations to the intrinsic properties of EVs. When exploring various methods for loading RNA into isolated extracellular vesicles, it is crucial to consider the vesicles’ integrity and the possible inclusion of antigenic or toxic components during the loading process. These factors could limit the feasibility of subsequent administrations of externally loaded vesicles.
6. Applications of Extracellular Vesicles
While in this review, our primary focus lies on the RNA content in extracellular vesicles for their potential as biomarkers and therapeutic delivery agents, it is important to acknowledge that their rich repertoire of proteins and other macromolecules may also hold a wealth of information about the physiological state of their source cells or tissues. The RNA cargo inside EVs reflects the physiological state of the cells or tissues from which they originate, rendering them highly valuable for biomarker discovery. These vesicles are present in diverse bodily fluids, including blood serum, urine, saliva, cerebrospinal fluid, and breast milk. An advantage of utilizing RNA from EVs as biomarkers lies in their protection against degradation by RNases, ensuring stability. Furthermore, these vesicles display surface markers and RNA profiles that correspond to the cells they originated from, facilitating the enrichment of vesicles from specific tissues [85,86].
Novel technologies, such as droplet digital reverse transcription–PCR, enhance the detection of specific RNA types in extracellular vesicles. This method reveals various alterations and anomalies in the RNA cargo, which might be linked to disease and could serve as a biomarker for a particular condition. For example, the detection of tumor-specific mRNA in the EVs isolated from serum samples of glioblastoma patients [87,88]. Increased levels of certain miRNAs have been discovered in EVs isolated from the cerebrospinal fluid of patients with neurological disorders and cancer [89,90,91]. Changes in mRNA splice variants have been identified in urine from patients with myotonic dystrophy, while long non-coding RNA profiles in blood samples correlate with heart functions in patients with type 2 diabetes [23,92,93]. The utilization of extracellular vesicles as biomarkers has already undergone significant progress in clinical settings. Pathogenic mRNAs and various other macromolecules found within these vesicles are now utilized as biomarkers for diagnosing blood disorders and prognosticating prostate, lung, and other solid tumors. For instance, tests like the ExoDx prostate test from Bio-Techne utilize these biomarkers for diagnosis and prognostic evaluation [94,95,96,97].
There is considerable enthusiasm surrounding the utilization of extracellular vesicles for delivering RNA therapies. These vesicles offer diverse sourcing options, including patients’ derived vesicles, which evade immune responses, shield the enclosed cargo, display surface proteins, and accommodate multiple therapeutic substances alongside RNA [98,99,100]. However, their application in therapies is somewhat constrained due to their inherent diversity, preventing the attainment of entirely pure vesicle groups. The preparations used in experiments, termed ‘secretomes’, constitute all kinds of vesicles along with EVs. Despite these challenges, clinical research has initiated the use of extracellular vesicles for RNA treatments. Animal studies demonstrate the ability of these vesicles derived from cells like mesenchymal stem cells to traverse the body and aid in various diseases affecting vital organs and systems.
Researchers are exploring diverse avenues to harvest abundant vesicles from varied sources, including easily cultivable cells, high-volume biofluids, and even plant materials [101], which come under natural substances. Primary approaches under exploration for utilizing these vesicles in treatments encompass strategies like using vesicle-releasing cells on-site, injecting purified or modified vesicles, and implanting modified cells that produce tailored vesicles within the body [102,103,104]. Examples include investigating mesenchymal stem cell-derived vesicles for treating conditions like cerebellar ataxia, bone repair, and eye issues [105,106]. Additionally, there has been extensive research into loading isolated vesicles with specific substances or modifying their surfaces for enhanced delivery, targeting ailments such as stroke and pancreatic cancer [99,100,107]. Furthermore, there are protocols to modify vesicles to address specific targets, like bone diseases or muscular dystrophy [103]. Some studies also contemplate using modified cells to produce tailored vesicles targeting conditions like brain tumors [108,109]. EVs show lower immunogenicity and toxicity, a better ability to traverse barriers within cells and tissues, and the potential for more efficient RNA delivery while incorporating targeting molecules [98]. They allow the loading of both natural and synthetic RNA cargo, presenting potential in combined therapies [75,110]. Clinical trials involve using extracellular vesicles for tumor-targeted vaccines, enhancing the immune response by delivering tumor-specific antigens [111,112]. Additionally, these vesicles can potentially deliver pathogenic protein mRNA to dendritic cells, aiding vaccination without generating infectious agents [113,114,115]. Please refer to Table 3 to find the list of exosomal microRNA-based clinical trials conducted by the National Institute of Health (NIH).

Table 3.
Extracellular vesicle-derived microRNAs in clinical trials.
Table 3.
Extracellular vesicle-derived microRNAs in clinical trials.
| Clinical Trial Name | Clinical Phase | Clinical Trial Identifier |
|---|---|---|
| Treatment of Cerebellar Ataxia With Mesenchymal Stem Cells-Derived Exosomes | Phase 1/2 | NCT01649687 |
| Allogenic Mesenchymal Stem Cell-Derived Exosome in Patients With Acute Ischemic Stroke | Phase 1/2 | NCT03384433 |
| Mesenchymal Stem Cells-Derived Exosomes for Promoting Healing of Large and Refractory Macular Holes (MHs) | Early phase 1 | NCT03437759 |
| iExosomes in Treating Participants With Metastatic Pancreas Cancer With KrasG12D Mutation | Phase 1 | NCT03608631 |
| Role of the Serum Exosomal miRNA in Diabetic Retinopathy | Observational | NCT03264976 |
| Circulating Exosomal miRNA Expression on Patients With Heart Transplantation | Observational | NCT04921774 |
| Evaluation of Safety and Efficiency of Method of Exosome Inhalation in SARS-CoV-2-Associated Two-Sided Pneumonia | Phase 1/2 | NCT04491240 |
| Serum Exosomal miRNA Predicting the Therapeutic Efficiency in Lung Squamous Carcinoma | Observational | NCT05854030 |
| Screening of Serum Exosomal miRNA as a Biomarker for Ocular Muscle Myasthenia Gravis | Observational | NCT05888558 |
| Exosomal microRNA in Predicting the Aggressiveness of Prostate Cancer in Chinese Patients | Observational | NCT03911999 |
| U01-Biomarkers for Noninvasive and Early Detection of Pancreatic Cancer | Observational | NCT03886571 |
| MicroRNAs to Predict Response to Androgen Deprivation Therapy | Observational | NCT02366494 |
| Early Detection of Lung Cancer by Combining Exosomal Analysis of Hypoxia With Standard of Care Imaging (LungExoDETECT) | Observational | NCT04629079 |
| Research on the Early and Prognosis Diagnosis of Vascular Dementia | Observational | NCT03152630 |
7. Conclusions and Future Perspectives
Despite the promising therapeutic and diagnostic potential of extracellular vesicles (EVs) in human diseases, several challenges persist. Technical barriers, such as isolation techniques, characterization, and the establishment of standardized procedures for clinically viable EV preparations, continue to be major hurdles. Current isolation methods, such as ultracentrifugation, have limitations in scalability. However, emerging techniques like tangential flow filtration hold promise for larger-scale EV production. Nonetheless, scaling up cell cultures for clinical application remains challenging, exacerbated by issues related to bioreactor usage and media supplements like fetal bovine serum, which contains EVs, necessitating rigorous characterization during scaling.
Effective dosing and understanding the mechanism of action, particularly with stem cell-derived EVs, require further investigation. The heterogeneity of EVs poses challenges in refining isolation methods to enrich functional vesicles. Developing potency assays to measure efficacy is critical for regulatory approval. Concerns arise regarding the use of EVs as drug carriers, as exogenous cargo may interfere with endogenous content, potentially causing off-target effects, especially in complex conditions like cancer. Identifying the safest cell sources for EV isolation and therapeutic use is vital to mitigate immunogenicity and unwanted cargo delivery. Improving EV targeting to specific sites necessitates a deeper understanding of delivery mechanisms while minimizing off-target effects. Although engineering EVs for specific tropism shows promise, scaling production remains an obstacle.
The development of EV-based biomarkers for diseases such as cancer demands technological advancements, particularly in high-throughput assays and standardized methodologies for clinical translation. Emerging trends and technologies offer potential solutions to these challenges. For instance, advancements in isolation techniques, such as microfluidic-based methods, may overcome scalability issues and enhance EV purity. Integration of omics technologies, including genomics, proteomics, and lipidomics, can provide comprehensive insights into EV cargo and function, facilitating the development of potent therapeutic EVs. Furthermore, the application of artificial intelligence and machine learning algorithms to analyze EV data can accelerate biomarker discovery and therapeutic optimization. Utilizing three-dimensional cell culture systems and organ-on-a-chip platforms can better mimic physiological conditions, improving our understanding of EV biology and therapeutic efficacy. Moreover, the development of standardized protocols and quality control measures for EV production and characterization is essential for reproducibility and clinical translation.
Areas for future investigation include elucidating the role of EVs in intercellular communication and disease pathogenesis, exploring novel EV-based therapeutic strategies, and optimizing EV delivery systems for targeted drug delivery. Addressing these challenges and leveraging emerging technologies will drive the translation of EV-based therapies and diagnostics from bench to bedside, ultimately improving patient outcomes in various disease settings.
Funding
HaystackAnalytics Pvt. Limited funded this work.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The author Alakesh Das was employed by the company HaystackAnalytics Pvt. Ltd. The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Narang, P.; Shah, M.; Beljanski, V. Exosomal RNAs in diagnosis and therapies. Noncoding RNA Res. 2022, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Wang, J.; Yue, B.L.; Huang, Y.Z.; Lan, X.Y.; Liu, W.J.; Chen, H. Exosomal RNAs: Novel Potential Biomarkers for Diseases—A Review. Int. J. Mol. Sci. 2022, 23, 2461. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Jia, Y.; Yu, L.; Ma, T.; Xu, W.; Qian, H.; Sun, Y.; Shi, H. Small extracellular vesicles isolation and separation: Current techniques, pending questions and clinical applications. Theranostics 2022, 12, 6548. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef] [PubMed]
- Liangsupree, T.; Multia, E.; Riekkola, M.L. Modern isolation and separation techniques for extracellular vesicles. J. Chromatogr. A 2021, 1636, 461773. [Google Scholar] [CrossRef]
- De Sousa, K.P.; Rossi, I.; Abdullahi, M.; Ramirez, M.I.; Stratton, D.; Inal, J.M. Isolation and characterization of extracellular vesicles and future directions in diagnosis and therapy. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2023, 15, e1835. [Google Scholar] [CrossRef]
- Ying, S.Y.; Chang, D.C.; Lin, S.L. The MicroRNA (miRNA): Overview of the RNA Genes that Modulate Gene Function. Mol. Biotechnol. 2008, 38, 257. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Zheng, D.; Huo, M.; Li, B.; Wang, W.; Piao, H.; Wang, Y.; Zhu, Z.; Li, D.; Wang, T.; Liu, K. The Role of Exosomes and Exosomal MicroRNA in Cardiovascular Disease. Front. Cell Dev. Biol. 2021, 8, 616161. [Google Scholar] [CrossRef]
- Hu, G.; Drescher, K.M.; Chen, X.M. Exosomal miRNAs: Biological properties and therapeutic potential. Front. Genet. 2012, 3, 22036. [Google Scholar] [CrossRef]
- Vaka, R.; Parent, S.; Risha, Y.; Khan, S.; Courtman, D.; Stewart, D.J.; Davis, D.R. Extracellular vesicle microRNA and protein cargo profiling in three clinical-grade stem cell products reveals key functional pathways. Mol. Ther.Nucleic Acids 2023, 32, 80–93. [Google Scholar] [CrossRef]
- MicroRNA-193b Is a Regulator of Amyloid Precursor Protein in the Blood and Cerebrospinal Fluid Derived Exosomal microRNA-193b Is a Biomarker of Alzheimer’s Disease [Online]. Available online: https://www.spandidos-publications.com/mmr/10/5/2395 (accessed on 21 December 2023).
- Lizarraga-Valderrama, L.R.; Sheridan, G.K. Extracellular vesicles and intercellular communication in the central nervous system. FEBS Lett. 2021, 595, 1391–1410. [Google Scholar] [CrossRef] [PubMed]
- Ogaki, A.; Ikegaya, Y.; Koyama, R. Extracellular Vesicles Taken up by Astrocytes. Int. J. Mol. Sci. 2021, 22, 10553. [Google Scholar] [CrossRef] [PubMed]
- Wortzel, I.; Dror, S.; Kenific, C.M.; Lyden, D. Exosome-Mediated Metastasis: Communication from a Distance. Dev. Cell 2019, 49, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yu, D. Exosomes in cancer development, metastasis, and immunity. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Tang, Y.; Liu, Y.; Zhang, P.; Lv, L.; Zhang, X.; Jia, L.; Zhou, Y. Exosomes derived from miR-375-overexpressing human adipose mesenchymal stem cells promote bone regeneration. Cell Prolif. 2019, 52, e12669. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.; Hu, R.; Runtsch, M.C.; Kagele, D.A.; Mosbruger, T.L.; Tolmachova, T.; Seabra, M.C.; Round, J.L.; Ward, D.M.; O’Connell, R.M. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun. 2015, 6, 7321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- de los Santos, M.C.; Dragomir, M.P.; Calin, G.A. The role of exosomal long non-coding RNAs in cancer drug resistance. Cancer Drug Resist. 2019, 2, 1178. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.L.; Liu, Y.; Jiang, J.; Tang, Y.J.; Tang, Y.L.; Liang, X.H. Extracellular vesicle long non–coding RNA-mediated crosstalk in the tumor microenvironment: Tiny molecules, huge roles. Cancer Sci. 2020, 111, 2726. [Google Scholar] [CrossRef] [PubMed]
- Kenneweg, F.; Bang, C.; Xiao, K.; Boulanger, C.M.; Loyer, X.; Mazlan, S.; Schroen, B.; Hermans-Beijnsberger, S.; Foinquinos, A.; Hirt, M.N.; et al. Long Noncoding RNA-Enriched Vesicles Secreted by Hypoxic Cardiomyocytes Drive Cardiac Fibrosis. Mol. Ther. Nucleic Acids 2019, 18, 363. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Chen, J.; Yang, L.; Liu, J.; Zhang, X.; Zhang, Y.; Tu, Q.; Yin, D.; Lin, D.; Wong, P.P.; et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat. Cell Biol. 2019, 21, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Vila, M.; Yoshioka, Y.; Ochiya, T. Biological Functions Driven by mRNAs Carried by Extracellular Vesicles in Cancer. Front. Cell Dev. Biol. 2021, 9, 620498. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell. Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef] [PubMed]
- Skog, J.; Würdinger, T.; Van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry Jr, W.T.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- De La Cuesta, F.; Passalacqua, I.; Rodor, J.; Bhushan, R.; Denby, L.; Baker, A.H. Extracellular vesicle cross-talk between pulmonary artery smooth muscle cells and endothelium during excessive TGF-β signalling: Implications for PAH vascular remodelling. Cell Commun. Signal. 2019, 17, 143. [Google Scholar] [CrossRef]
- Usman, W.M.; Pham, T.C.; Kwok, Y.Y.; Vu, L.T.; Ma, V.; Peng, B.; Chan, Y.S.; Wei, L.; Chin, S.M.; Azad, A.; et al. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat. Commun. 2018, 9, 2359. [Google Scholar] [CrossRef] [PubMed]
- Quah, B.J.C.; O’Neill, H.C. The immunogenicity of dendritic cell-derived exosomes. Blood Cells Mol. Dis. 2005, 35, 94–110. [Google Scholar] [CrossRef] [PubMed]
- Mallegol, J.; Van Niel, G.; Lebreton, C.; Lepelletier, Y.; Candalh, C.; Dugave, C.; Heath, J.K.; Raposo, G.; Cerf–Bensussan, N.; Heyman, M. T84-Intestinal Epithelial Exosomes Bear MHC Class II/Peptide Complexes Potentiating Antigen Presentation by Dendritic Cells. Gastroenterology 2007, 132, 1866–1876. [Google Scholar] [CrossRef] [PubMed]
- Shenoda, B.B.; Ajit, S.K. Modulation of Immune Responses by Exosomes Derived from Antigen-Presenting Cells. Clin. Med. Insights Pathol. 2016, 9 (Suppl. S1), CPath-S39925. [Google Scholar] [CrossRef] [PubMed]
- Rimer, J.M.; Lee, J.; Holley, C.L.; Crowder, R.J.; Chen, D.L.; Hanson, P.I.; Ory, D.S.; Schaffer, J.E. Long-range function of secreted small nucleolar RNAs that direct 2′-O-methylation. J. Biol. Chem. 2018, 293, 13284–13296. [Google Scholar] [CrossRef]
- Chiou, N.T.; Kageyama, R.; Ansel, K.M. Selective Export into Extracellular Vesicles and Function of tRNA Fragments during T Cell Activation. Cell Rep. 2018, 25, 3356–3370.e4. [Google Scholar] [CrossRef] [PubMed]
- Maute, R.L.; Schneider, C.; Sumazin, P.; Holmes, A.; Califano, A.; Basso, K.; Dalla-Favera, R. TRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 1404–1409. [Google Scholar] [CrossRef]
- Zhao, C.; Sun, X.; Li, L. Biogenesis and function of extracellular mirnas. ExRNA 2019, 1, 38. [Google Scholar] [CrossRef]
- Sobolewski, C.; Dubuquoy, L.; Legrand, N. MicroRNAs, Tristetraprolin Family Members and HuR: A Complex Interplay Controlling Cancer-Related Processes. Cancers 2022, 14, 3516. [Google Scholar] [CrossRef]
- Ahmad, M.; Weiswald, L.B.; Poulain, L.; Denoyelle, C.; Meryet-Figuiere, M. Involvement of lncRNAs in cancer cells migration, invasion and metastasis: Cytoskeleton and ECM crosstalk. J. Exp. Clin. Cancer Res. 2023, 42, 173. [Google Scholar] [CrossRef]
- Pandey, K.K.; Madhry, D.; Kumar, Y.R.; Malvankar, S.; Sapra, L.; Srivastava, R.K.; Bhattacharyya, S.; Verma, B. Regulatory roles of tRNA-derived RNA fragments in human pathophysiology. Mol. Ther. Nucleic Acids 2021, 26, 161–173. [Google Scholar] [CrossRef]
- Yu, X.; Xie, Y.; Zhang, S.; Song, X.; Xiao, B.; Yan, Z. tRNA-derived fragments: Mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections. Theranostics 2021, 11, 461. [Google Scholar] [CrossRef]
- Lu, Y.; Huang, W.; Li, M.; Zheng, A. Exosome-Based Carrier for RNA Delivery: Progress and Challenges. Pharmaceutics 2023, 15, 598. [Google Scholar] [CrossRef]
- Aslan, C.; Kiaie, S.H.; Zolbanin, N.M.; Lotfinejad, P.; Ramezani, R.; Kashanchi, F.; Jafari, R. Exosomes for mRNA delivery: A novel biotherapeutic strategy with hurdles and hope. BMC Biotechnol. 2021, 21, 20. [Google Scholar] [CrossRef]
- Yang, J.; Xue, F.T.; Li, Y.Y.; Liu, W.; Zhang, S. Exosomal piRNA sequencing reveals differences between heart failure and healthy patients. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7952–7961. [Google Scholar] [CrossRef]
- Zhang, B.; Zhao, N.; Jia, L.; Che, J.; He, X.; Liu, K.; Bao, B. Identification and application of piwi-interacting RNAs from seminal plasma exosomes in Cynoglossus semilaevis. BMC Genom. 2020, 21, 302. [Google Scholar] [CrossRef]
- Rui, T.; Wang, K.; Xiang, A.; Guo, J.; Tang, N.; Jin, X.; Lin, Y.; Liu, J.; Zhang, X. Serum Exosome-Derived piRNAs Could Be Promising Biomarkers for HCC Diagnosis. Int. J. Nanomed. 2023, 18, 1989. [Google Scholar] [CrossRef]
- Allmang, C.; Kufel, J.; Chanfreau, G.; Mitchell, P.; Petfalski, E.; Tollervey, D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999, 18, 5399–5410. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Driedonks, T.A.P.; Nolte-T’Hoen, E.N.M. Circulating Y-RNAs in Extracellular Vesicles and Ribonucleoprotein Complexes; Implications for the Immune System. Front. Immunol. 2018, 9, 3164. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Maugeri, M.; Garre, E.; Nawaz, M.; Wahlgren, J.; Papadimitriou, A.; Lundqvist, C.; Lindfors, L.; Collen, A.; Sunnerhagen, P.; et al. Identification of RNA-binding proteins in exosomes capable of interacting with different types of RNA: RBP-facilitated transport of RNAs into exosomes. PLoS ONE 2018, 13, e0195969. [Google Scholar] [CrossRef]
- Fabbiano, F.; Corsi, J.; Gurrieri, E.; Trevisan, C.; Notarangelo, M.; D’Agostino, V.G. RNA packaging into extracellular vesicles: An orchestra of RNA-binding proteins? J. Extracell. Vesicles 2020, 10, e12043. [Google Scholar] [CrossRef]
- Ortiz-Quintero, B. Extracellular MicroRNAs as Intercellular Mediators and Noninvasive Biomarkers of Cancer. Cancers 2020, 12, 3455. [Google Scholar] [CrossRef] [PubMed]
- Souza-Schorey, C.; Schorey, J.S. Regulation and mechanisms of extracellular vesicle biogenesis and secretion. Essays Biochem. 2018, 62, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Tosar, J.P.; Witwer, K.; Cayota, A. Revisiting extracellular RNA release, processing, and function. Trends Biochem. Sci. 2021, 46, 438. [Google Scholar] [CrossRef] [PubMed]
- Sadik, N.; Cruz, L.; Gurtner, A.; Rodosthenous, R.S.; Dusoswa, S.A.; Ziegler, O.; Van Solinge, T.S.; Wei, Z.; Salvador-Garicano, A.M.; Gyorgy, B.; et al. Extracellular RNAs: A New Awareness of Old Perspectives. Methods Mol. Biol. 2018, 1740, 1–15. [Google Scholar] [CrossRef] [PubMed]
- He, A.T.; Liu, J.; Li, F.; Yang, B.B. Targeting circular RNAs as a therapeutic approach: Current strategies and challenges. Signal Transduct. Target. Ther. 2021, 6, 185. [Google Scholar] [CrossRef] [PubMed]
- Gatta, A.T.; Carlton, J.G. The ESCRT-machinery: Closing holes and expanding roles. Curr. Opin. Cell Biol. 2019, 59, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.; Bai, H.; Ren, L.; Zhang, L. The Role of Exosome and the ESCRT Pathway on Enveloped Virus Infection. Int. J. Mol. Sci. 2021, 22, 9060. [Google Scholar] [CrossRef]
- Juan, T.; Fürthauer, M. Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin. Cell Dev. Biol. 2018, 74, 66–77. [Google Scholar] [CrossRef]
- Sudhakaran, M.; Doseff, A.I. Role of Heterogeneous Nuclear Ribonucleoproteins in the Cancer-Immune Landscape. Int. J. Mol. Sci. 2023, 24, 5086. [Google Scholar] [CrossRef]
- Geuens, T.; Bouhy, D.; Timmerman, V. The hnRNP family: Insights into their role in health and disease. Hum. Genet. 2016, 135, 851–867. [Google Scholar] [CrossRef]
- Weaver, A.M.; Patton, J.G. Argonautes in Extracellular Vesicles: Artifact or Selected Cargo? Cancer Res. 2020, 80, 379–381. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef]
- Yao, B.; Li, S.; Chan, E.K.L. Function of GW182 and GW bodies in siRNA and miRNA pathways. Adv. Exp. Med. Biol. 2013, 768, 71–96. [Google Scholar] [CrossRef]
- Yao, B.; La, L.B.; Chen, Y.C.; Chang, L.J.; Chan, E.K.L. Defining a new role of GW182 in maintaining miRNA stability. EMBO Rep. 2012, 13, 1102–1108. [Google Scholar] [CrossRef]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Doehle, B.P.; Qin, Y.; Macara, I.G.; Cullen, B.R. Overexpression of Exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA 2005, 11, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, B.; Zhang, Z.; Raitskin, O.; Hiller, M.; Benderska, N.; Hartmann, A.M.; Bracco, L.; Elliott, D.; Ben-Ari, S.; Soreq, H.; et al. Heterogeneous Nuclear Ribonucleoprotein G Regulates Splice Site Selection by Binding to CC(A/C)-rich Regions in Pre-mRNA. J. Biol. Chem. 2009, 284, 14303–14315. [Google Scholar] [CrossRef] [PubMed]
- Shurtleff, M.J.; Yao, J.; Qin, Y.; Nottingham, R.M.; Temoche-Diaz, M.M.; Schekman, R.; Lambowitz, A.M. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl. Acad. Sci. USA 2017, 114, E8987–E8995. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, Z.; Gao, Q. Transfer of microRNA-25 by colorectal cancer cell-derived extracellular vesicles facilitates colorectal cancer development and metastasis. Mol. Ther. Nucleic Acids 2021, 23, 552–564. [Google Scholar] [CrossRef]
- Mukherjee, K.; Ghoshal, B.; Ghosh, S.; Chakrabarty, Y.; Shwetha, S.; Das, S.; Bhattacharyya, S.N. Reversible HuR-microRNA binding controls extracellular export of miR-122 and augments stress response. EMBO Rep. 2016, 17, 1184. [Google Scholar] [CrossRef]
- Han, Y.; Jones, T.W.; Dutta, S.; Zhu, Y.; Wang, X.; Narayanan, S.P.; Fagan, S.C.; Zhang, D. Overview and Update on Methods for Cargo Loading into Extracellular Vesicles. Processes 2021, 9, 356. [Google Scholar] [CrossRef] [PubMed]
- Corley, M.; Burns, M.C.; Yeo, G.W. How RNA-Binding Proteins Interact with RNA: Molecules and Mechanisms. Mol. Cell 2020, 78, 9–29. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Chen, Q.; Lin, L.; Sha, C.; Li, T.; Liu, Y.; Yin, X.; Xu, Y.; Chen, L.; Gao, W.; et al. Regulation of exosome production and cargo sorting. Int. J. Biol. Sci. 2020, 17, 163–177. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Baixauli, F.; Gutiérrez-Vázquez, C.; Sánchez-Madrid, F.; Mittelbrunn, M. Sorting it out: Regulation of exosome loading. Semin, Cancer Biol. 2014, 28, 3–13. [Google Scholar] [CrossRef]
- Wang, Q.; Yu, J.; Kadungure, T.; Beyene, J.; Zhang, H.; Lu, Q. ARMMs as a versatile platform for intracellular delivery of macromolecules. Nat. Commun. 2018, 9, 960. [Google Scholar] [CrossRef]
- György, B.; Hung, M.E.; Breakefield, X.O.; Leonard, J.N. Therapeutic applications of extracellular vesicles: Clinical promise and open questions. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 439–464. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.M.; Andreu, Z.; Bedina Zavec, A.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Yang, Y.; Hong, Y.; Cho, E.; Kim, G.B.; Kim, I.S. Extracellular vesicles as a platform for membrane-associated therapeutic protein delivery. J. Extracell. Vesicles 2018, 7, 1440131. [Google Scholar] [CrossRef]
- Janas, T.; Janas, M.M.; Sapoń, K.; Janas, T. Mechanisms of RNA loading into exosomes. FEBS Lett. 2015, 589, 1391–1398. [Google Scholar] [CrossRef]
- Komuro, H.; Aminova, S.; Lauro, K.; Harada, M. Advances of engineered extracellular vesicles-based therapeutics strategy. Sci. Technol. Adv. Mater. 2022, 23, 655. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Dominguez, M.V.; Zottel, A.; Šamec, N.; Jovčevska, I.; Dincer, C.; Kahlert, U.D.; Nickel, A.C. Current Technologies for RNA-Directed Liquid Diagnostics. Cancers 2021, 13, 5060. [Google Scholar] [CrossRef]
- Reimers, N.; Pantel, K. Liquid biopsy: Novel technologies and clinical applications. Clin. Chem. Lab. Med. 2019, 57, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Lunavat, T.R.; Nieland, L.; Vrijmoet, A.B.; Zargani-Piccardi, A.; Samaha, Y.; Breyne, K.; Breakefield, X.O. Roles of extracellular vesicles in glioblastoma: Foes, friends and informers. Front. Oncol. 2023, 13, 1291177. [Google Scholar] [CrossRef]
- Zanganeh, S.; Abbasgholinejad, E.; Doroudian, M.; Esmaelizad, N.; Farjadian, F.; Benhabbour, S.R. The Current Landscape of Glioblastoma Biomarkers in Body Fluids. Cancers 2023, 15, 3804. [Google Scholar] [CrossRef]
- Roy, B.; Lee, E.; Li, T.; Rampersaud, M. Role of miRNAs in Neurodegeneration: From Disease Cause to Tools of Biomarker Discovery and Therapeutics. Genes 2022, 13, 425. [Google Scholar] [CrossRef] [PubMed]
- Makowska, M.; Smolarz, B.; Romanowicz, H. microRNAs (miRNAs) in Glioblastoma Multiforme (GBM)—Recent Literature Review. Int. J. Mol. Sci. 2023, 24, 3521. [Google Scholar] [CrossRef] [PubMed]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef]
- Ismail, N.; Abdullah, N.; Murad, N.A.A.; Jamal, R.; Sulaiman, S.A. Long Non-Coding RNAs (lncRNAs) in Cardiovascular Disease Complication of Type 2 Diabetes. Diagnostics 2021, 11, 145. [Google Scholar] [CrossRef]
- López-Martínez, A.; Soblechero-Martín, P.; De-La-puente-ovejero, L.; Nogales-Gadea, G.; Arechavala-Gomeza, V. An Overview of Alternative Splicing Defects Implicated in Myotonic Dystrophy Type I. Genes 2020, 11, 1109. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Bajo-Santos, C.; Brokāne, A.; Zayakin, P.; Endzeliņš, E.; Soboļevska, K.; Belovs, A.; Jansons, J.; Sperga, M.; Llorente, A.; Radoviča-Spalviņa, I.; et al. Plasma and urinary extracellular vesicles as a source of RNA biomarkers for prostate cancer in liquid biopsies. Front. Mol. Biosci. 2023, 10, 980433. [Google Scholar] [CrossRef]
- Irmer, B.; Chandrabalan, S.; Maas, L.; Bleckmann, A.; Menck, K. Extracellular Vesicles in Liquid Biopsies as Biomarkers for Solid Tumors. Cancers 2023, 15, 1307. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Garrastacho, M.; Bajo-Santos, C.; Line, A.; Martens-Uzunova, E.S.; de la Fuente, J.M.; Moros, M.; Soekmadji, C.; Tasken, K.A.; Llorente, A. Extracellular vesicles as a source of prostate cancer biomarkers in liquid biopsies: A decade of research. Br. J. Cancer 2021, 126, 331–350. [Google Scholar] [CrossRef] [PubMed]
- Dang, X.T.T.; Kavishka, J.M.; Zhang, D.X.; Pirisinu, M.; Le, M.T.N. Extracellular Vesicles as an Efficient and Versatile System for Drug Delivery. Cells 2020, 9, 2191. [Google Scholar] [CrossRef]
- Ferreira, D.; Moreira, J.N.; Rodrigues, L.R. New advances in exosome-based targeted drug delivery systems. Crit. Rev. Oncol. Hematol. 2022, 172, 103628. [Google Scholar] [CrossRef]
- Song, H.; Chen, X.; Hao, Y.; Wang, J.; Xie, Q.; Wang, X. Nanoengineering facilitating the target mission: Targeted extracellular vesicles delivery systems design. J. Nanobiotechnol. 2022, 20, 431. [Google Scholar] [CrossRef]
- Pocsfalvi, G.; Turiák, L.; Ambrosone, A.; Del Gaudio, P.; Puska, G.; Fiume, I.; Silvestre, T.; Vékey, K. Protein biocargo of citrus fruit-derived vesicles reveals heterogeneous transport and extracellular vesicle populations. J. Plant Physiol. 2018, 229, 111–121. [Google Scholar] [CrossRef]
- Tang, T.T.; Wang, B.; Lv, L.L.; Liu, B.C. Extracellular vesicle-based Nanotherapeutics: Emerging frontiers in anti-inflammatory therapy. Theranostics 2020, 10, 8111. [Google Scholar] [CrossRef]
- Joshi, B.S.; Ortiz, D.; Zuhorn, I.S. Converting extracellular vesicles into nanomedicine: Loading and unloading of cargo. Mater. Today Nano 2021, 16, 100148. [Google Scholar] [CrossRef]
- Ma, Y.; Brocchini, S.; Williams, G.R. Extracellular vesicle-embedded materials. J. Control. Release 2023, 361, 280–296. [Google Scholar] [CrossRef]
- Cone, A.S.; Yuan, X.; Sun, L.; Duke, L.C.; Vreones, M.P.; Carrier, A.N.; Kenyon, S.M.; Carver, S.R.; Benthem, S.D.; Stimmell, A.C.; et al. Mesenchymal stem cell-derived extracellular vesicles ameliorate Alzheimer’s disease-like phenotypes in a preclinical mouse model. Theranostics 2021, 11, 8129–8142. [Google Scholar] [CrossRef]
- Kim, S.G.; George, N.P.; Hwang, J.S.; Park, S.; Kim, M.O.; Lee, S.H.; Lee, G. Human Bone Marrow-Derived Mesenchymal Stem Cell Applications in Neurodegenerative Disease Treatment and Integrated Omics Analysis for Successful Stem Cell Therapy. Bioengineering 2023, 10, 621. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Wang, M.; Jia, H.; Wu, P. Extracellular vesicles: Emerging anti-cancer drugs and advanced functionalization platforms for cancer therapy. Drug Deliv. 2022, 29, 2513. [Google Scholar] [CrossRef]
- Nieland, L.; Mahjoum, S.; Grandell, E.; Breyne, K.; Breakefield, X.O. Engineered EVs designed to target diseases of the CNS. J. Control. Release 2023, 356, 493–506. [Google Scholar] [CrossRef]
- Shahjin, F.; Chand, S.; Yelamanchili, S.V. Extracellular Vesicles as Drug Delivery Vehicles to the Central Nervous System. J. Neuroimmune Pharmacol. 2020, 15, 443–458. [Google Scholar] [CrossRef] [PubMed]
- Elsharkasy, O.M.; Nordin, J.Z.; Hagey, D.W.; de Jong, O.G.; Schiffelers, R.M.; Andaloussi, S.E.; Vader, P. Extracellular vesicles as drug delivery systems: Why and how? Adv. Drug Deliv. Rev. 2020, 159, 332–343. [Google Scholar] [CrossRef]
- Giacobino, C.; Canta, M.; Fornaguera, C.; Borrós, S.; Cauda, V. Extracellular Vesicles and Their Current Role in Cancer Immunotherapy. Cancers 2021, 13, 2280. [Google Scholar] [CrossRef] [PubMed]
- Najafi, S.; Majidpoor, J.; Mortezaee, K. Extracellular vesicle–based drug delivery in cancer immunotherapy. Drug Deliv. Transl. Res. 2023, 13, 2790–2806. [Google Scholar] [CrossRef] [PubMed]
- Schlake, T.; Thess, A.; Fotin-Mleczek, M.; Kallen, K.J. Developing mRNA-vaccine technologies. RNA Biol. 2012, 9, 1319. [Google Scholar] [CrossRef]
- Santos, P.; Almeida, F. Exosome-Based Vaccines: History, Current State, and Clinical Trials. Front. Immunol. 2021, 12, 711565. [Google Scholar] [CrossRef] [PubMed]
- Huda, M.N.; Nurunnabi, M. Potential Application of Exosomes in Vaccine Development and Delivery. Pharm. Res. 2022, 39, 2635. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).