Peptide Multimerization as Leads for Therapeutic Development
Abstract
:1. Introduction
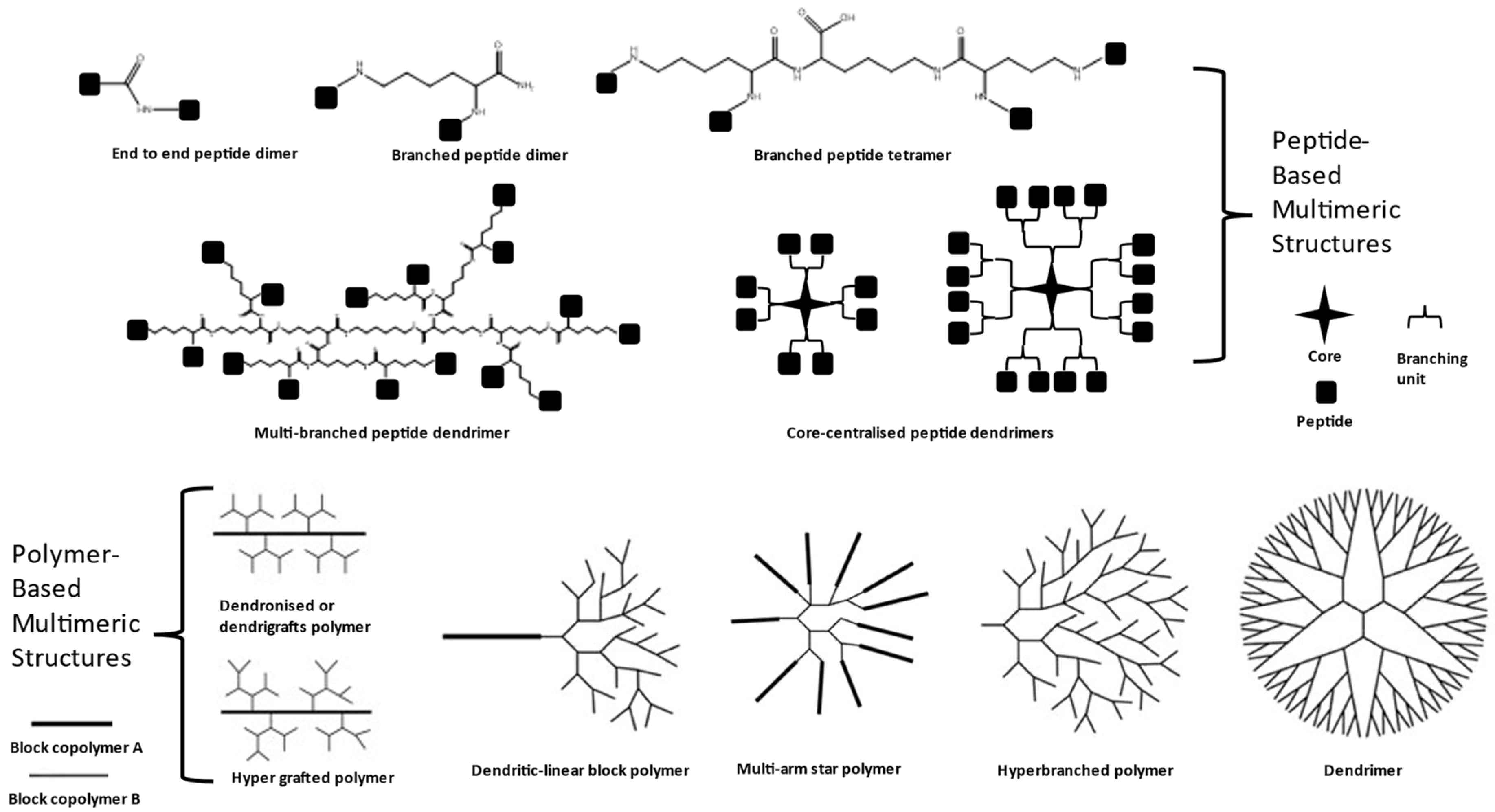
2. Overview of Multimerization and Peptide Dendrimers
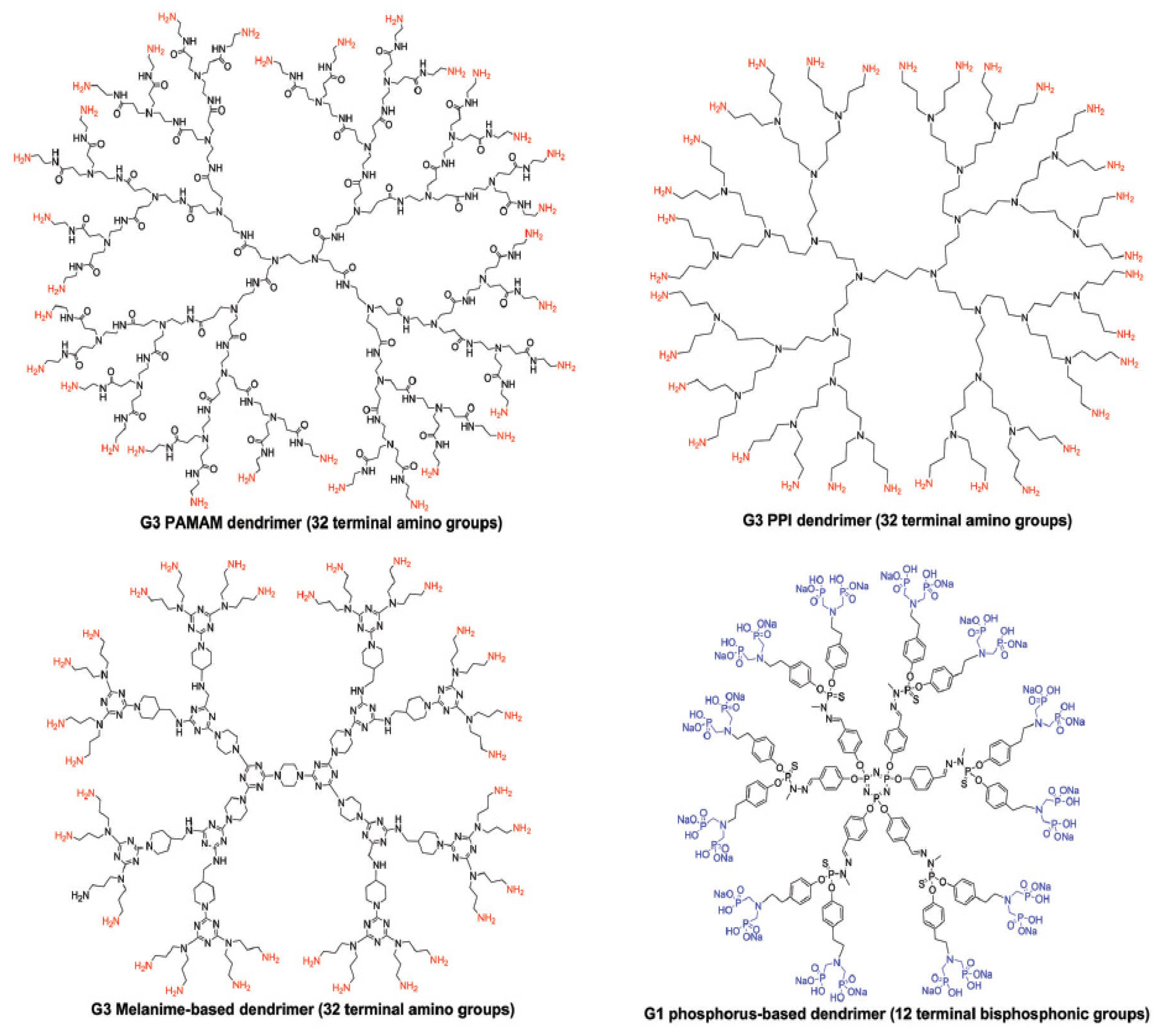
3. Overview of Dendrimers
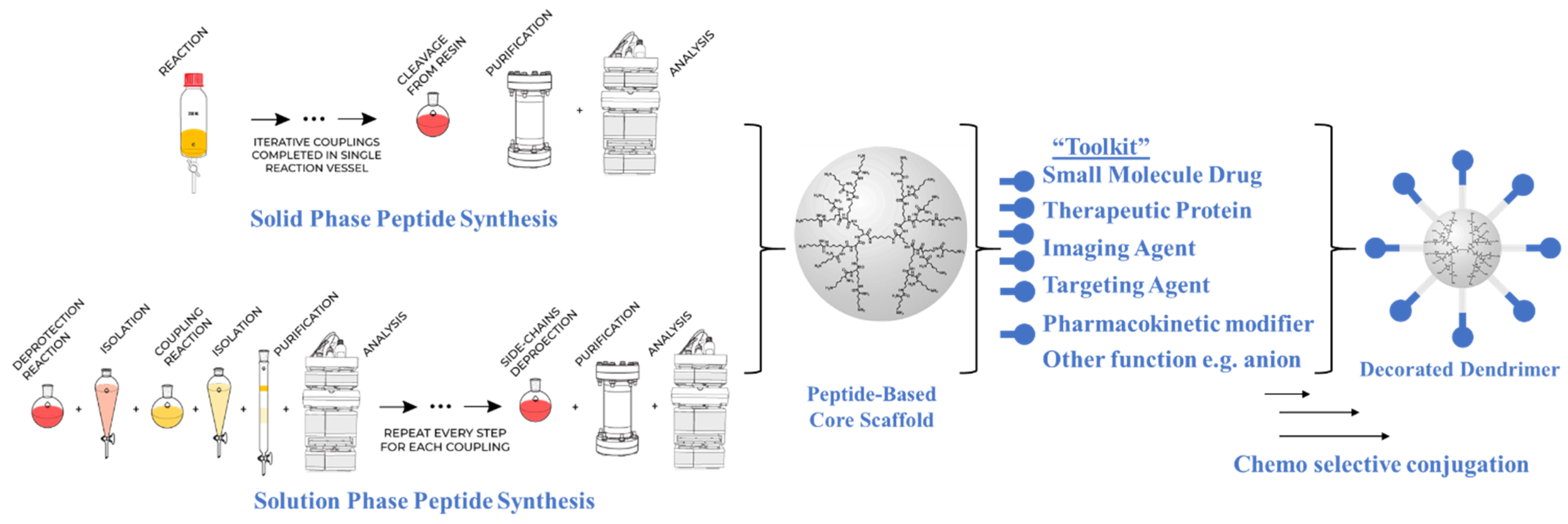
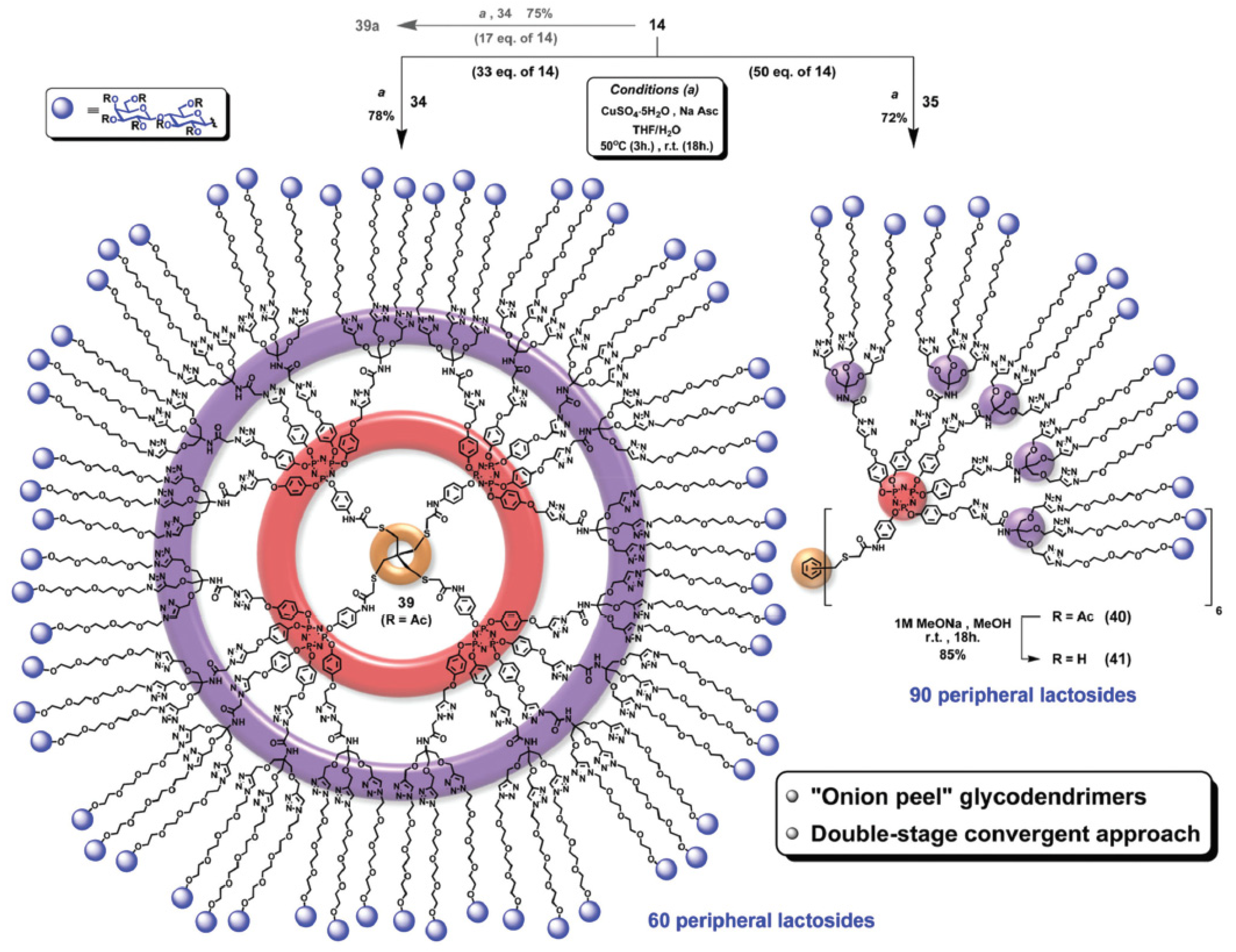
4. Innovations in Multimerization Synthetic Methodology
5. Multimeric and Dimeric Peptide Applications
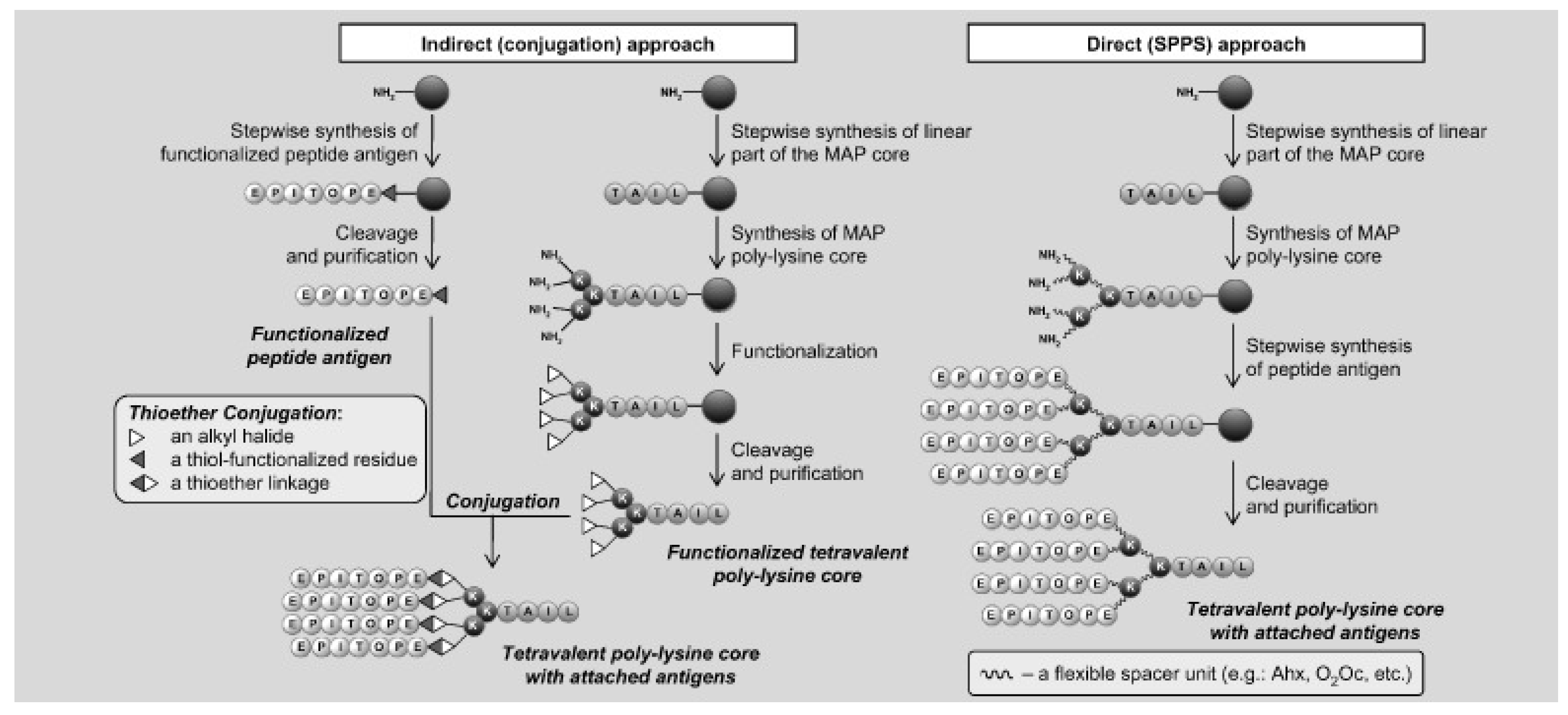
5.1. Multiple Antigenic Peptide (MAP) Systems and Their Use as Immunogens and Vaccines
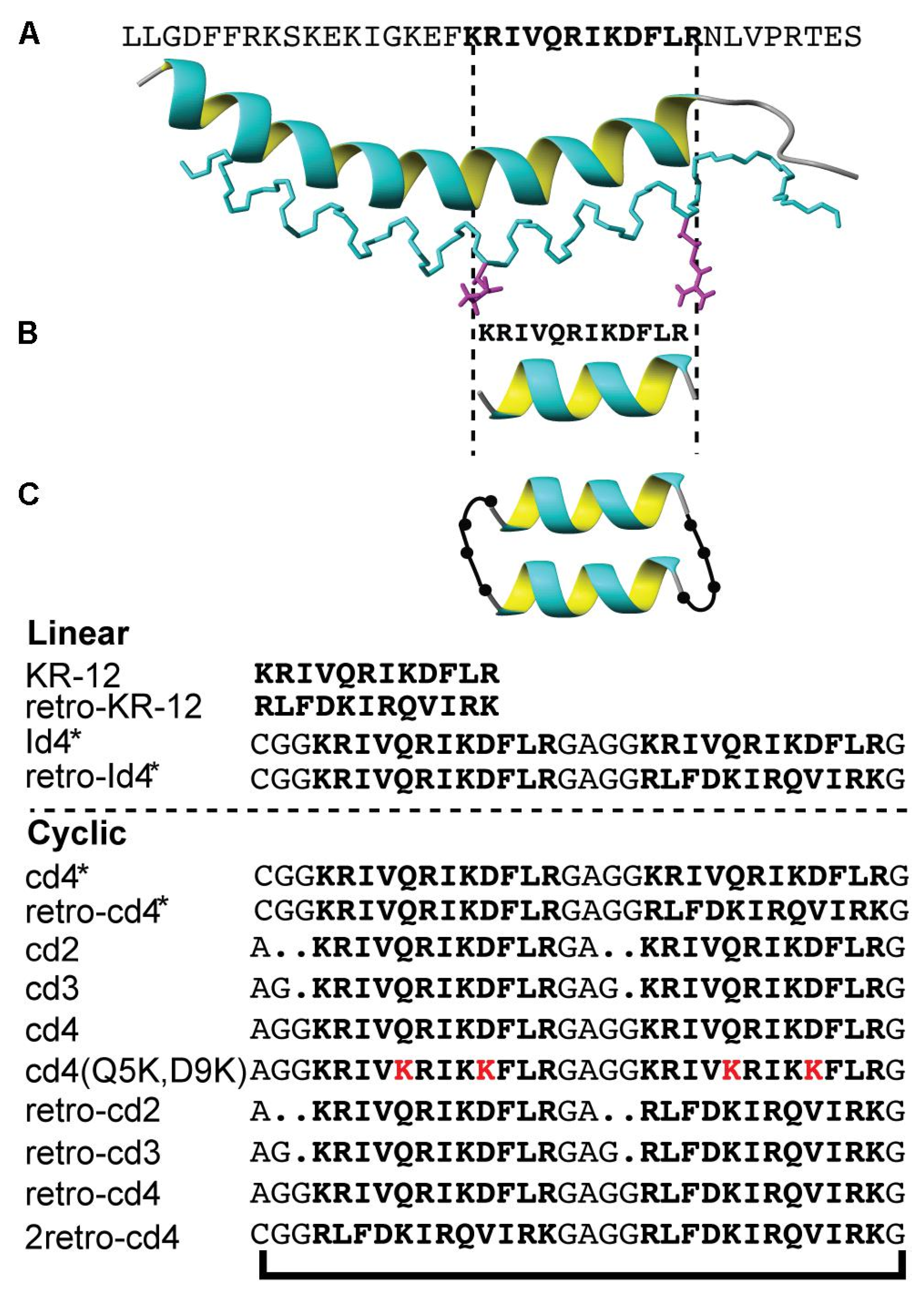
5.2. Peptide Multimers as Antimicrobial and Antiviral Agents
5.3. Multimers as Inhibitors and Mimetics
5.4. Radio-Labelled Bioactive Multimeric Peptides
5.5. Peptide-Based Polymers: Dendrimer Systems
6. Perspective of Multimeric Delivery Systems
7. Future Directions/Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
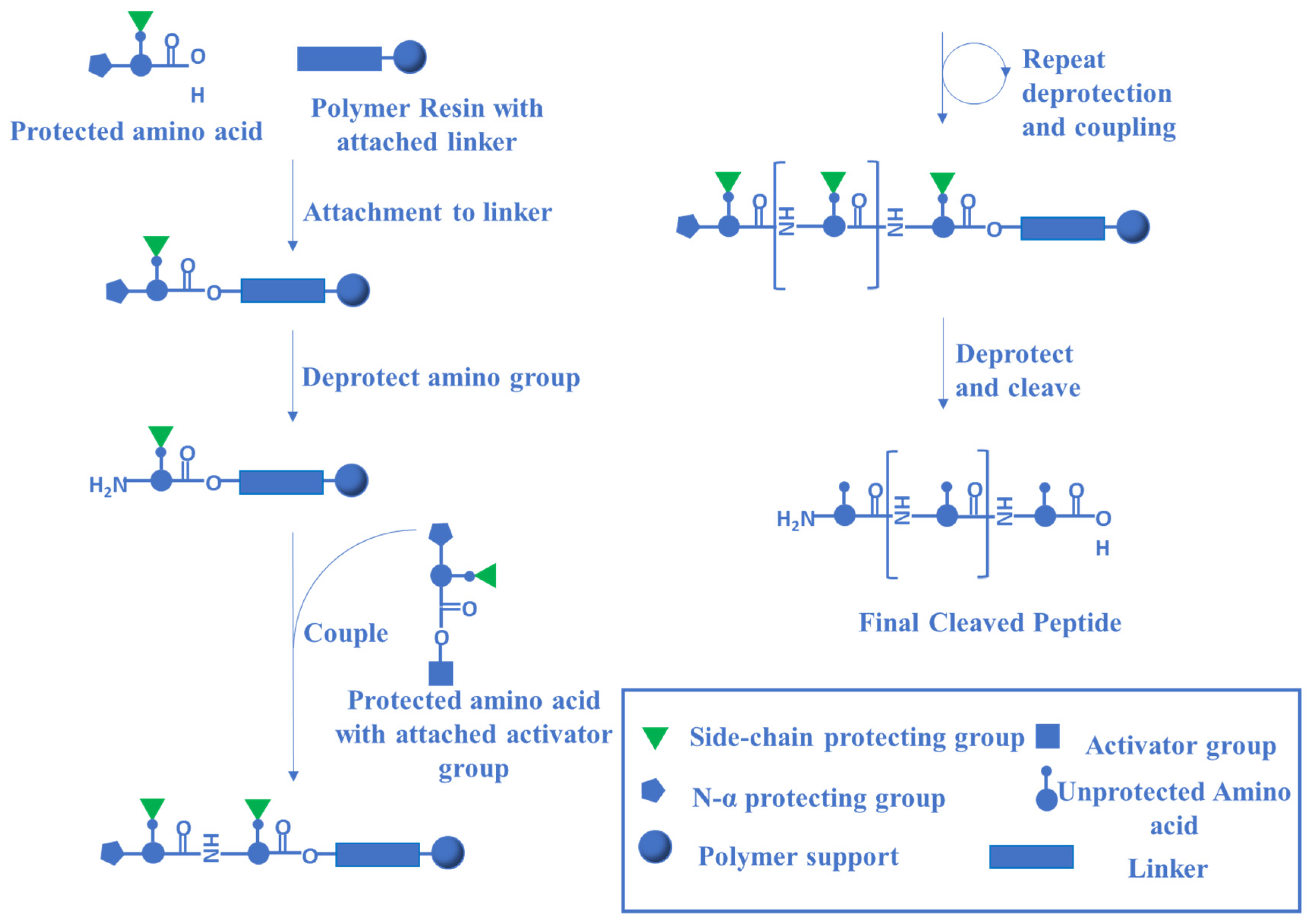
- Behrendt, R.; White, P.; Offer, J. Advances in Fmoc solid-phase peptide synthesis. J. Pept. Sci. 2016, 22, 4–27. [Google Scholar] [CrossRef] [Green Version]
- Kalia, J.; Raines, R.T. Advances in Bioconjugation. Curr. Org. Chem. 2010, 14, 138–147. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Separovic, F.; O’Brien-Simpson, N.M.; Wade, J.D. Chemically modified and conjugated antimicrobial peptides against superbugs. Chem. Soc. Rev. 2021, 50, 4932–4973. [Google Scholar] [CrossRef]
- Ornelas, C. Brief Timelapse on Dendrimer Chemistry: Advances, Limitations, and Expectations. Macromol. Chem. Phys. 2016, 217, 149–174. [Google Scholar] [CrossRef]
- Rasines Mazo, A.; Allison-Logan, S.; Karimi, F.; Chan, N.J.-A.; Qiu, W.; Duan, W.; O’Brien-Simpson, N.M.; Qiao, G.G. Ring opening polymerization of α-amino acids: Advances in synthesis, architecture and applications of polypeptides and their hybrids. Chem. Soc. Rev. 2020, 49, 4737. [Google Scholar] [CrossRef]
- Floudas, C.A.; Fung, H.K.; McAllister, S.R.; Mönnigmann, M.; Rajgaria, R. Advances in protein structure prediction and de novo protein design: A review. Chem. Eng. Sci. 2006, 61, 966–988. [Google Scholar] [CrossRef]
- Mochly-Rosen, D.; Das, K.; Grimes, K.V. Protein kinase C, an elusive therapeutic target? Nat. Rev. Drug Discov. 2012, 11, 937–957. [Google Scholar] [CrossRef] [Green Version]
- Murakami, S.; Nakashima, R.; Yamashita, E.; Matsumoto, T.; Yamaguchi, A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 2006, 443, 173–179. [Google Scholar] [CrossRef]
- Sathish Sundar, D.; Antoniraj, M.G.; Kumar, C.S.; Mohapatra, S.S.; Houreld, N.N.; Ruckmani, K. Recent Trends of Biocompatible and Biodegradable Nanoparticles in Drug Delivery: A Review. Curr. Med. Chem. 2016, 23, 3730–3751. [Google Scholar] [CrossRef]
- Sachdeva, S. Peptides as ‘drugs’: The journey so far. Int. J. Pept. Res. Ther. 2017, 23, 49–60. [Google Scholar] [CrossRef]
- Rosenfeld, L. Insulin: Discovery and controversy. Clin. Chem. 2002, 48, 2270–2288. [Google Scholar] [CrossRef] [Green Version]
- Lefèbvre, P.J. The Discovery of Glucagon and Glucagon-Related Peptides. In Unveiling Diabetes-Historical Milestones in Diabetology; Karger Publishers: Basel, Switzerland, 2020; pp. 191–201. [Google Scholar]
- Lau, J.; Bloch, P.; Schäffer, L.; Pettersson, I.; Spetzler, J.; Kofoed, J.; Madsen, K.; Knudsen, L.B.; McGuire, J.; Steensgaard, D.B. Discovery of the once-weekly glucagon-like peptide-1 (GLP-1) analogue semaglutide. J. Med. Chem. 2015, 58, 7370–7380. [Google Scholar] [CrossRef]
- Raedler, L.A. Tresiba (Insulin Degludec Injection) and Ryzodeg 70/30 (Insulin Degludec and Insulin Aspart Injection): Two New Insulin Analogs for Glycemic Control in Diabetes Mellitus. Am. Health Drug Benefits 2016, 9, 144–148. [Google Scholar] [PubMed]
- Karas, J.A.; Wade, J.D.; Hossain, M.A. The Chemical Synthesis of Insulin: An Enduring Challenge. Chem. Rev. 2021, 121, 4531–4560. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-G.; Baggio, L.; Bridon, D.; Castaigne, J.-P.; Robitaille, M.; Jetté, L.; Benquet, C.; Drucker, D. Development and characterization of a glucagon-like peptide 1-albumin conjugate: The ability to activate the glucagon-like peptide 1 receptor in vivo. Diabetes 2003, 52, 751–759. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Li, L.; Deng, K.; Xu, C.; Busse, J.; Vandvik, P.; Li, S.; Guyatt, G.; Sun, X. Incretin based treatments and mortality in patients with type 2 diabetes: Systematic review and meta-analysis. BMJ Br. Med. J. 2017, 357, j2499. [Google Scholar] [CrossRef] [Green Version]
- Cheang, J.Y.; Moyle, P.M. Glucagon-Like Peptide-1 (GLP-1)-Based Therapeutics: Current Status and Future Opportunities beyond Type 2 Diabetes. ChemMedChem 2018, 13, 662–671. [Google Scholar] [CrossRef]
- Usmani, S.S.; Bedi, G.; Samuel, J.S.; Singh, S.; Kalra, S.; Kumar, P.; Ahuja, A.A.; Sharma, M.; Gautam, A.; Raghava, G.P.S. THPdb: Database of FDA-approved peptide and protein therapeutics. PLoS ONE 2017, 12, e0181748. [Google Scholar] [CrossRef] [Green Version]
- Lau, J.L.; Dunn, M.K. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Biorg. Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef]
- Santos, G.; Ganesan, A.; Emery, F. Oral Administration of Peptide-Based Drugs: Beyond Lipinski’s Rule. ChemMedChem 2016, 11, 2245–2251. [Google Scholar] [CrossRef]
- Otvos, L.; Wade, J.D. Current challenges in peptide-based drug discovery. Front. Chem. 2014, 2, 62. [Google Scholar] [CrossRef]
- Tabatabaei Mirakabad, F.S.; Khoramgah, M.S.; Keshavarz, F.K.; Tabarzad, M.; Ranjbari, J. Peptide dendrimers as valuable biomaterials in medical sciences. Life Sci. 2019, 233, 116754. [Google Scholar] [CrossRef] [PubMed]
- Sapra, R.; Verma, R.P.; Maurya, G.P.; Dhawan, S.; Babu, J.; Haridas, V. Designer Peptide and Protein Dendrimers: A Cross-Sectional Analysis. Chem. Rev. 2019, 119, 11391–11441. [Google Scholar] [CrossRef]
- Niederhafner, P.; Šebestík, J.; Ježek, J. Peptide dendrimers. J. Pept. Sci. 2005, 11, 757–788. [Google Scholar] [CrossRef] [PubMed]
- González, T.; González Toro, D.; Thayumanavan, S. Advances in polymer and polymeric nanostructures for protein conjugation. Eur. Polym. J. 2013, 49, 2906–2918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.-L.; Kim, S.-Y.; Kim, T.; Kim, K.-H.; Park, J.-W. The role of terminal groups in dendrimer systems for the treatment of organic contaminants in aqueous environments. J. Clean. Prod. 2020, 250, 119494. [Google Scholar] [CrossRef]
- Hagner, K.; Setayeshgar, S.; Lynch, M. Stochastic protein multimerization, activity, and fitness. Phys. Rev. E 2018, 98, 062401. [Google Scholar] [CrossRef] [Green Version]
- Gunasekera, S.; Muhammad, T.; Strömstedt, A.A.; Rosengren, K.J.; Göransson, U. Backbone Cyclization and Dimerization of LL-37-Derived Peptides Enhance Antimicrobial Activity and Proteolytic Stability. Front. Microbiol. 2020, 11, 168. [Google Scholar] [CrossRef] [Green Version]
- Carlucci, G.; Ananias, H.J.; Yu, Z.; Van de Wiele, C.; Dierckx, R.A.; de Jong, I.J.; Elsinga, P.H. Multimerization improves targeting of peptide radio-pharmaceuticals. Curr. Pharm. Des. 2012, 18, 2501–2516. [Google Scholar] [CrossRef]
- Bracci, L.; Falciani, C.; Lelli, B.; Lozzi, L.; Runci, Y.; Pini, A.; De Montis, M.; Tagliamonte, A.; Neri, P. Synthetic peptides in the form of dendrimers become resistant to protease activity. J. Biol. Chem. 2003, 278, 46590–46595. [Google Scholar] [CrossRef] [Green Version]
- Sadler, K.; Tam, J.P. Peptide dendrimers: Applications and synthesis. Rev. Mol. Biotechnol. 2002, 90, 195–229. [Google Scholar] [CrossRef]
- Marsden, H.; Owsianka, A.; Graham, S.; McLean, G.; Robertson, C.; Subak-Sharpe, J. Advantages of branched peptides in serodiagnosis: Detection of HIV-specific antibodies and the use of glycine spacers to increase sensitivity. J. Immunol. Methods 1992, 147, 65–72. [Google Scholar] [CrossRef]
- Han, S.; Yoshida, T.; Uryu, T. Synthesis of a new polylysine-dendritic oligosaccharide with alkyl spacer having peptide linkage. Carbohydr. Polym. 2007, 69, 436–444. [Google Scholar] [CrossRef]
- Sheveleva, N.N.; Markelov, D.A.; Vovk, M.A.; Mikhailova, M.E.; Tarasenko, I.I.; Tolstoy, P.M.; Neelov, I.M.; Lähderanta, E. Lysine-based dendrimer with double arginine residues. RSC Adv. 2019, 9, 18018–18026. [Google Scholar] [CrossRef] [Green Version]
- Fields, G.B.; Noble, R.L. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int. J. Pept. Protein Res. 1990, 35, 161–214. [Google Scholar] [CrossRef] [PubMed]
- Mahindra, A.; Sharma, K.K.; Jain, R. Rapid microwave-assisted solution-phase peptide synthesis. Tetrahedron Lett. 2012, 53, 6931–6935. [Google Scholar] [CrossRef]
- Kowalczyk, W.; Monsó, M.; de la Torre, B.G.; Andreu, D. Synthesis of multiple antigenic peptides (MAPs)—Strategies and limitations. J. Pept. Sci. 2011, 17, 247–251. [Google Scholar] [CrossRef]
- Coin, I.; Beyermann, M.; Bienert, M. Solid-phase peptide synthesis: From standard procedures to the synthesis of difficult sequences. Nat. Protoc. 2007, 2, 3247–3256. [Google Scholar] [CrossRef]
- Okarvi, S.M. Solution-phase vs solid-phase peptide synthesis methods to prepare peptide radiopharmaceuticals. Nucl. Med. Commun. 2000, 21, 592. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhao, L.; Li, Y.; Xu, T. Design of biocompatible dendrimers for cancer diagnosis and therapy: Current status and future perspectives. Chem. Soc. Rev. 2011, 40, 2673–2703. [Google Scholar] [CrossRef]
- Giuliani, A.; Rinaldi, A. Beyond natural antimicrobial peptides: Multimeric peptides and other peptidomimetic approaches. Cell. Mol. Life Sci. 2011, 68, 2255–2266. [Google Scholar] [CrossRef]
- Borbély, A.; Thoreau, F.; Figueras, E.; Kadri, M.; Coll, J.-L.; Boturyn, D.; Sewald, N. Synthesis and Biological Characterization of Monomeric and Tetrameric RGD-Cryptophycin Conjugates. Chem. Eur. J. 2020, 26, 2602–2605. [Google Scholar] [CrossRef]
- Ekladious, I.; Colson, Y.L.; Grinstaff, M.W. Polymer–drug conjugate therapeutics: Advances, insights and prospects. Nat. Rev. Drug Discov. 2019, 18, 273–294. [Google Scholar] [CrossRef]
- Hussein, W.M.; Liu, T.-Y.; Jia, Z.; McMillan, N.A.J.; Monteiro, M.J.; Toth, I.; Skwarczynski, M. Multiantigenic peptide–polymer conjugates as therapeutic vaccines against cervical cancer. Biorg. Med. Chem. 2016, 24, 4372–4380. [Google Scholar] [CrossRef] [Green Version]
- Skwarczynski, M.; Toth, I. Peptide-based synthetic vaccines. Chem. Sci. 2016, 7, 842–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filipe, L.C.S.; Machuqueiro, M.; Baptista, A.M. Unfolding the Conformational Behavior of Peptide Dendrimers: Insights from Molecular Dynamics Simulations. J. Am. Chem. Soc. 2011, 133, 5042–5052. [Google Scholar] [CrossRef]
- Javor, S.; Natalello, A.; Doglia, S.M.; Reymond, J.-L. α-Helix Stabilization within a Peptide Dendrimer. J. Am. Chem. Soc. 2008, 130, 17248–17249. [Google Scholar] [CrossRef] [PubMed]
- Bastings, M.M.C.; Helms, B.A.; van Baal, I.; Hackeng, T.M.; Merkx, M.; Meijer, E.W. From Phage Display to Dendrimer Display: Insights into Multivalent Binding. J. Am. Chem. Soc. 2011, 133, 6636–6641. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, S.; Gupta, L.; Rani, S.; Dave, K.; Gupta, U. Impact of Dendrimers on Solubility of Hydrophobic Drug Molecules. Front. Pharmacol. 2017, 8, 261. [Google Scholar] [CrossRef] [Green Version]
- Bansal, R.; Dhawan, S.; Chattopadhyay, S.; Maurya, G.P.; Haridas, V.; Rathore, A.S. Peptide Dendrons as Thermal-Stability Amplifiers for Immunoglobulin G1 Monoclonal Antibody Biotherapeutics. Bioconjug. Chem. 2017, 28, 2549–2559. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Jain, K.; Mehra, N.K.; Jain, N.K. Dendrimers in anticancer drug delivery: Mechanism of interaction of drug and dendrimers. Artif. Cells Nanomed. Biotechnol. 2016, 44, 1626–1634. [Google Scholar] [CrossRef]
- Sebestik, J.; Niederhafner, P.; Jezek, J. Peptide and glycopeptide dendrimers and analogous dendrimeric structures and their biomedical applications. Amino Acids 2010, 40, 301–370. [Google Scholar] [CrossRef] [PubMed]
- Medina, S.H.; El-Sayed, M.E.H. Dendrimers as Carriers for Delivery of Chemotherapeutic Agents. Chem. Rev. 2009, 109, 3141–3157. [Google Scholar] [CrossRef]
- Blencowe, A.; Tan, J.F.; Goh, T.K.; Qiao, G.G. Core cross-linked star polymers via controlled radical polymerisation. Polymer 2009, 50, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Ishizu, K.; Taiichi, F.; Ochi, K. Architecture of star and hyperbranched polymers. In Focus on Polymeric Materials Research; Nova Science Publishers: Hauppauge, NY, USA, 2006; pp. 129–156. [Google Scholar]
- Olofsson, K.; Andrén, O.C.J.; Malkoch, M. Recent advances on crosslinked dendritic networks. J. Appl. Polym. Sci. 2014, 131, 39876. [Google Scholar] [CrossRef]
- Tomalia, D.A.; Huang, B.; Swanson, D.R.; Brothers, H.M.; Klimash, J.W. Structure control within poly(amidoamine) dendrimers: Size, shape and regio-chemical mimicry of globular proteins. Tetrahedron 2003, 59, 3799–3813. [Google Scholar] [CrossRef]
- Galliot, C.; Larré, C.; Caminade, A.-M.; Majoral, J.-P. Regioselective Stepwise Growth of Dendrimer Units in the Internal Voids of a Main Dendrimer. Science 1997, 277, 1981–1984. [Google Scholar] [CrossRef]
- Ciolkowski, M.; Petersen, J.F.; Ficker, M.; Janaszewska, A.; Christensen, J.B.; Klajnert, B.; Bryszewska, M. Surface modification of PAMAM dendrimer improves its biocompatibility. Nanomed. Nanotechnol. Biol. Med. 2012, 8, 815–817. [Google Scholar] [CrossRef] [PubMed]
- Hecht, S.; Fréchet, J.M.J. An Alternative Synthetic Approach toward Dendritic Macromolecules: Novel Benzene-Core Dendrimers via Alkyne Cyclotrimerization. J. Am. Chem. Soc. 1999, 121, 4084. [Google Scholar] [CrossRef]
- Smith, R.J.; Gorman, C.; Menegatti, S. Synthesis, structure, and function of internally functionalized dendrimers. J. Polym. Sci. 2021, 59, 10–28. [Google Scholar] [CrossRef]
- Kharwade, R.; More, S.; Warokar, A.; Agrawal, P.; Mahajan, N. Starburst pamam dendrimers: Synthetic approaches, surface modifications, and biomedical applications. Arab. J. Chem. 2020, 13, 6009–6039. [Google Scholar] [CrossRef]
- Goyal, P.; Yoon, K.; Weck, M. Multifunctionalization of Dendrimers through Orthogonal Transformations. Chem. Eur. J. 2007, 13, 8801–8810. [Google Scholar] [CrossRef] [PubMed]
- Vogtle, F.; Buhleier, E.; Wehner, W. Cascade and nonskid-chain-like syntheses of molecular cavity topologies. Synthesis 1978, 2, 155–158. [Google Scholar]
- Tomalia, D.A.; Baker, H.; Dewald, J.; Hall, M.; Kallos, G.; Martin, S.; Roeck, J.; Ryder, J.; Smith, P. A New Class of Polymers: Starburst-Dendritic Macromolecules. Polym. J. 1985, 17, 117–132. [Google Scholar] [CrossRef] [Green Version]
- Tomalia, D.A.; Baker, H.; Dewald, J.; Hall, M.; Kallos, G.; Martin, S.; Roeck, J.; Ryder, J.; Smith, P. Dendritic macromolecules: Synthesis of starburst dendrimers. Macromolecules 1986, 19, 2466–2468. [Google Scholar] [CrossRef]
- Tomalia, D.A.; Hall, M.; Hedstrand, D.M. Starburst dendrimers. III. The importance of branch junction symmetry in the development of topological shell molecules. J. Am. Chem. Soc. 1987, 109, 1601–1603. [Google Scholar] [CrossRef]
- de Brabander-van den Berg, E.M.M.; Meijer, E.W. Poly(propylene imine) Dendrimers: Large-Scale Synthesis by Hetereogeneously Catalyzed Hydrogenations. Angew. Chem. Int. Ed. 1993, 32, 1308–1311. [Google Scholar] [CrossRef] [Green Version]
- Newkome, G.R.; Yao, Z.; Baker, G.R.; Gupta, V.K.; Russo, P.S.; Saunders, M.J. Chemistry of micelles series. Part 2. Cascade molecules. Synthesis and characterization of a benzene[9]3-arborol. J. Am. Chem. Soc. 1986, 108, 849–850. [Google Scholar] [CrossRef]
- Mekelburger, H.-B.; Vögtle, F.; Jaworek, W. Dendrimers, Arborols, and Cascade Molecules: Breakthrough into Generations of New Materials. Angew. Chem. Int. Ed. Engl. 1992, 31, 1571–1576. [Google Scholar] [CrossRef]
- Denkewalter, R.G.; Kolc, J.; Lukasavage, W.J. Macromolecular Highly Branched Homogeneous Compound Based on Lysine Units. U.S. Patent 4289872A; Multiple antigen peptide system, 15 September 1981. [Google Scholar]
- Chai, S.K.; Clavijo, P.; Tam, J.P.; Zavala, F. Immunogenic properties of multiple antigen peptide systems containing defined T and B epitopes. J. Immunol. 1992, 149, 2385–2390. [Google Scholar]
- Tam, J.P. Multiple Antigen Peptide System. U.S. Patent 5229490A, 20 July 1993. [Google Scholar]
- Esfand, R.; Tomalia, D.A. Poly(amidoamine) (PAMAM) dendrimers: From biomimicry to drug delivery and biomedical applications. Drug Discov. Today 2001, 6, 427–436. [Google Scholar] [CrossRef]
- Nigam, S.; Chandra, S.; Bahadur, D. Dendrimers based electrochemical biosensors. J. Biomed. Res. 2015, 2, 21. [Google Scholar]
- Skwarczynski, M.; Zaman, M.; Urbani, C.N.; Lin, I.-C.; Jia, Z.; Batzloff, M.R.; Good, M.F.; Monteiro, M.J.; Toth, I. Polyacrylate Dendrimer Nanoparticles: A Self-Adjuvanting Vaccine Delivery System. Angew. Chem. Int. Ed. 2010, 49, 5742–5745. [Google Scholar] [CrossRef] [PubMed]
- Zechel, S.; Hager, M.D.; Schubert, S.; Manners, I.; Schubert, U.S. From Dendrimers to Macrocycles: 80 Years George R. Newkome—Milestones of a Gentleman Scientist. Macromol. Chem. Phys. 2018, 219, 1800269. [Google Scholar] [CrossRef]
- Ahlborg, N. Synthesis of a diepitope multiple antigen peptide containing sequences from two malaria antigens using Fmoc chemistry. J. Immunol. Methods 1995, 179, 269–275. [Google Scholar] [CrossRef]
- Caminade, A.-M.; Majoral, J.-P. Which Dendrimer to Attain the Desired Properties? Focus on Phosphorhydrazone Dendrimers. Molecules 2018, 23, 622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gajbhiye, V.; Palanirajan, V.K.; Tekade, R.K.; Jain, N.K. Dendrimers as therapeutic agents: A systematic review. J. Pharm. Pharmacol. 2009, 61, 989–1003. [Google Scholar] [CrossRef] [PubMed]
- Madaan, K.; Kumar, S.; Poonia, N.; Lather, V.; Pandita, D. Dendrimers in drug delivery and targeting: Drug-dendrimer interactions and toxicity issues. J. Pharm. Bioallied Sci. 2014, 6, 139–150. [Google Scholar]
- Starpharma. VivaGel®. 2020. Available online: https://www.starpharma.com/vivagel (accessed on 23 December 2021).
- Bernstein, D.I.; Stanberry, L.R.; Sacks, S.; Ayisi, N.K.; Gong, Y.H.; Ireland, J.; Mumper, R.J.; Holan, G.; Matthews, B.; McCarthy, T.; et al. Evaluations of Unformulated and Formulated Dendrimer-Based Microbicide Candidates in Mouse and Guinea Pig Models of Genital Herpes. Antimicrob. Agents Chemother. 2003, 47, 3784–3788. [Google Scholar] [CrossRef] [Green Version]
- Starpharma. Okamoto Licenses VivaGel® Antiviral Condom in Asian Countries. 2020. Available online: https://starpharma.com/news/story/okamoto-licenses-vivagel-antiviral-condom-in-asian-countries#_ftn1 (accessed on 23 December 2021).
- Starpharma. VivaGel® for Bacterial Vaginosis. 2020. Available online: https://starpharma.com/vivagel_bv#article1799 (accessed on 23 December 2021).
- Merrifield, R.B. Solid phase peptide synthesis. I. The synthesis of a tetrapeptide. J. Am. Chem. Soc. 1963, 85, 2149–2154. [Google Scholar] [CrossRef]
- Merrifield, R.B. Automated peptide synthesis. In Hypotensive Peptides; Springer: New York, NY, USA, 1966; pp. 1–13. [Google Scholar]
- Merrifield, R.B. Solid-phase peptide synthesis. III. An improved synthesis of bradykinin. Biochemistry 1964, 3, 1385–1390. [Google Scholar] [CrossRef]
- Merrifield, R.B. Solid phase peptide synthesis. II. The synthesis of bradykinin. J. Am. Chem. Soc. 1964, 86, 304–305. [Google Scholar] [CrossRef]
- Stewart, J.M. [3] Cleavage methods following Boc-based solid-phase peptide synthesis. Methods Enzymol. 1997, 289, 29–44. [Google Scholar] [PubMed]
- Li, W.; O’Brien-Simpson, N.M.; Hossain, M.A.; Wade, J.D. The 9-Fluorenylmethoxycarbonyl (Fmoc) Group in Chemical Peptide Synthesis–Its Past, Present, and Future. Aust. J. Chem. 2020, 73, 271–276. [Google Scholar] [CrossRef] [Green Version]
- Collins, J.M.; Porter, K.A.; Singh, S.K.; Vanier, G.S. High-Efficiency Solid Phase Peptide Synthesis (HE-SPPS). Org. Lett. 2014, 16, 940–943. [Google Scholar] [CrossRef]
- Mäde, V.; Els-Heindl, S.; Beck-Sickinger, A.G. Automated solid-phase peptide synthesis to obtain therapeutic peptides. Beilstein J. Org. Chem. 2014, 10, 1197–1212. [Google Scholar] [CrossRef] [Green Version]
- El-Faham, A.; Albericio, F. Peptide Coupling Reagents, More than a Letter Soup. Chem. Rev. 2011, 111, 6557–6602. [Google Scholar] [CrossRef]
- Amblard, M.; Fehrentz, J.-A.; Martinez, J.; Subra, G. Methods and protocols of modern solid phase peptide synthesis. Mol. Biotechnol. 2006, 33, 239–254. [Google Scholar] [CrossRef]
- Itoh, H.; Inoue, M. Full solid-phase total synthesis of macrocyclic natural peptides using four-dimensionally orthogonal protective groups. Org. Biomol. Chem. 2019, 17, 6519–6527. [Google Scholar] [CrossRef]
- Xu, W.-L.; Cui, A.L.; Hu, X.-X.; You, X.-F.; Li, Z.-R.; Zheng, J.-S. A new strategy for total solid-phase synthesis of polymyxins. Tetrahedron Lett. 2015, 56, 4796–4799. [Google Scholar] [CrossRef]
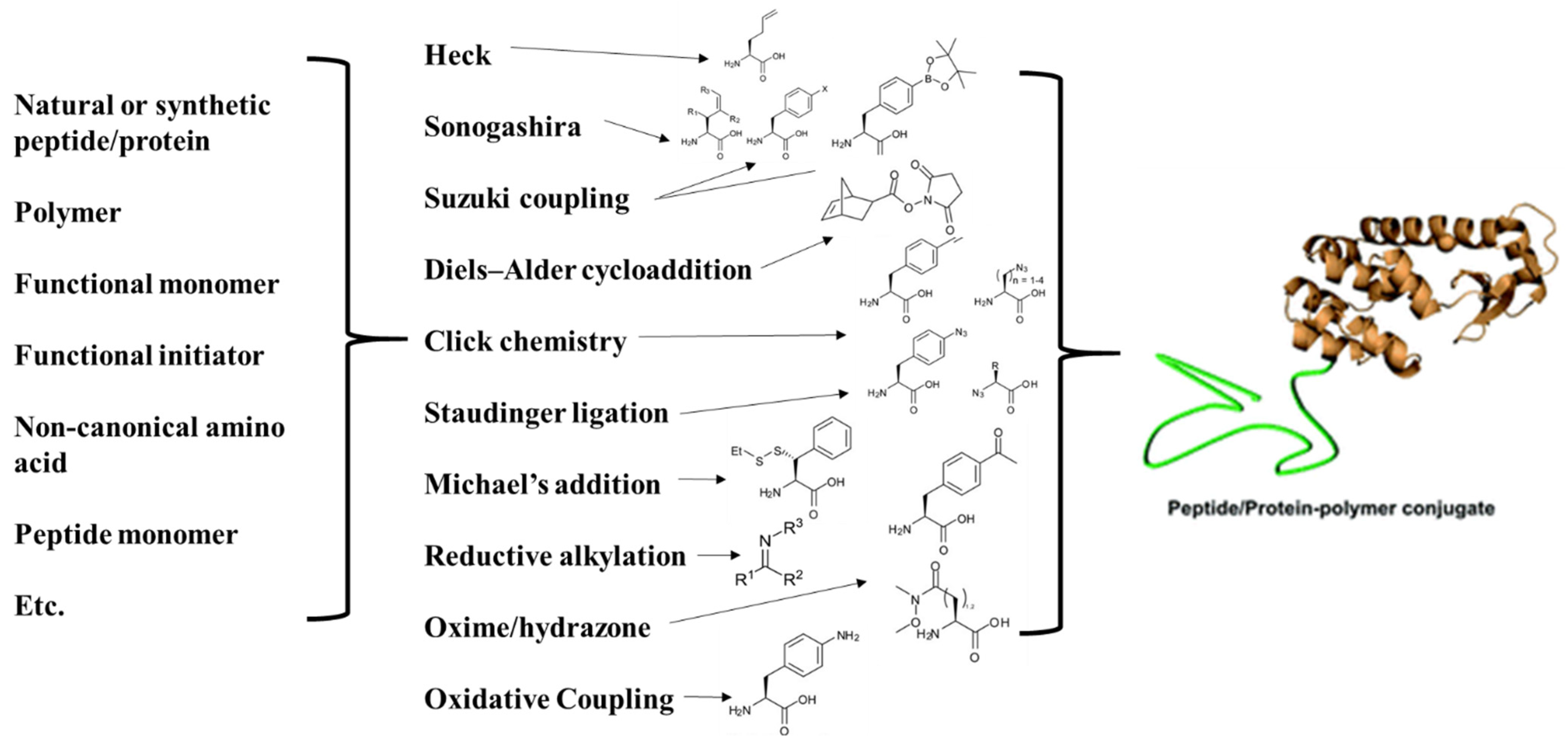
- Gauthier, M.A.; Klok, H.-A. Peptide/protein–polymer conjugates: Synthetic strategies and design concepts. Chem. Commun. 2008, 23, 2591–2611. [Google Scholar] [CrossRef] [PubMed]
- Dawson, P.E.; Kent, S.B.H. Synthesis of Native Proteins by Chemical Ligation. Annu. Rev. Biochem. 2000, 69, 923. [Google Scholar] [CrossRef] [Green Version]
- Dawson, P.E.; Muir, T.W.; Clark Lewis, I.; Kent, S.B. Synthesis of proteins by native chemical ligation. Science 1994, 266, 776–779. [Google Scholar] [CrossRef] [PubMed]
- Conibear, A.C.; Watson, E.; Payne, R.; Becker, C.F.W. Native chemical ligation in protein synthesis and semi-synthesis. Chem. Soc. Rev. 2018, 47, 9046–9068. [Google Scholar] [CrossRef]
- Muir, T.W.; Sondhi, D.; Cole, P.A. Expressed protein ligation: A general method for protein engineering. Proc. Natl. Acad. Sci. USA 1998, 95, 6705–6710. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, S.S.; Watson, E.E.; Premdjee, B.; Conde-Frieboes, K.W.; Payne, R.J. Diselenide–selenoester ligation for chemical protein synthesis. Nat. Protoc. 2019, 14, 2229–2257. [Google Scholar] [CrossRef]
- Majumdar, S.; Siahaan, T.J. Peptide-mediated targeted drug delivery. Med. Res. Rev. 2012, 32, 637–658. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; Wang, M.; Qi, W.; Su, R.; He, Z. Solid-Phase Enzymatic Peptide Synthesis to Produce an Antioxidant Dipeptide. Trans. Tianjin Univ. 2019, 25, 276–282. [Google Scholar] [CrossRef]
- Nuijens, T.; Toplak, A.; Schmidt, M.; Ricci, A.; Cabri, W. Natural Occurring and Engineered Enzymes for Peptide Ligation and Cyclization. Front. Chem. 2019, 7, 829. [Google Scholar] [CrossRef]
- Cao, Y.; Nguyen, G.K.T.; Chuah, S.; Tam, J.P.; Liu, C.-F. Butelase-Mediated Ligation as an Efficient Bioconjugation Method for the Synthesis of Peptide Dendrimers. Bioconjugate Chem. 2016, 27, 2592–2596. [Google Scholar] [CrossRef]
- Kolb, H.; Finn, M.G.; Sharpless, K.B. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chem. Int. Ed. 2001, 40, 2004–2021. [Google Scholar] [CrossRef]
- Hein, C.D.; Liu, X.-M.; Wang, D. Click Chemistry, A Powerful Tool for Pharmaceutical Sciences. Pharm. Res. 2008, 25, 2216–2230. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.; Becker, M.L. “Click” reactions: A versatile toolbox for the synthesis of peptide-conjugates. Chem. Soc. Rev. 2014, 43, 7013–7039. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, F.; Ahmad Fuaad, A.; Azmi, F.; Skwarczynski, M.; Toth, I. Peptide Conjugation via CuAAC ‘Click’ Chemistry. Mol. Online 2013, 18, 13148–13174. [Google Scholar] [CrossRef] [Green Version]
- Rostovtsev, V.V.; Green, L.G.; Fokin, V.V.; Sharpless, K.B. A stepwise huisgen cycloaddition process: Copper (I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. 2002, 114, 2708–2711. [Google Scholar] [CrossRef]
- Tornøe, C.W.; Christensen, C.; Meldal, M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper (I)-catalyzed 1, 3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 2002, 67, 3057–3064. [Google Scholar] [CrossRef] [PubMed]
- Agard, N.J.; Prescher, J.A.; Bertozzi, C.R. A strain-promoted [3 + 2] azide−alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 2004, 126, 15046–15047. [Google Scholar] [CrossRef]
- Codelli, J.A.; Baskin, J.M.; Agard, N.J.; Bertozzi, C.R. Second-generation difluorinated cyclooctynes for copper-free click chemistry. J. Am. Chem. Soc. 2008, 130, 11486–11493. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Hao, X.; Jing, L.; Wu, G.; Kang, D.; Liu, X.; Zhan, P. Recent applications of click chemistry in drug discovery. Expert Opin. Drug Discov. 2019, 14, 779–789. [Google Scholar] [CrossRef] [PubMed]
- Posnett, D.N.; Tam, J.P. [46] Multiple antigenic peptide method for producing antipeptide site-specific antibodies, Antibodies, Antigens, and Molecular Mimicry. Methods Enzymol. 1989, 178, 739–746. [Google Scholar]
- Tam, J.P. Recent advances in multiple antigen peptides. J. Immunol. Methods 1996, 196, 17–32. [Google Scholar] [CrossRef]
- Tam, J.P. Synthetic peptide vaccine design: Synthesis and properties of a high-density multiple antigenic peptide system. Proc. Natl. Acad. Sci. USA 1988, 85, 5409–5413. [Google Scholar] [CrossRef] [Green Version]
- Azmi, F.; Fuaad, A.A.H.A.; Skwarczynski, M.; Toth, I. Recent progress in adjuvant discovery for peptide-based subunit vaccines. Hum. Vaccin. Immunother. 2013, 10, 778–796. [Google Scholar] [CrossRef] [Green Version]
- Ciesielski, M.J.; Latif Kazim, A.; Barth, R.; Fenstermaker, R.A. Cellular antitumor immune response to a branched lysine multiple antigenic peptide containing epitopes of a common tumor-specific antigen in a rat glioma model. Cancer Immunol. Immunother. 2005, 54, 107–119. [Google Scholar] [CrossRef]
- Tam, J.P.; Clavijo, P.; Lu, Y.A.; Nussenzweig, V.; Nussenzweig, R.; Zavala, F. Incorporation of T and B epitopes of the circumsporozoite protein in a chemically defined synthetic vaccine against malaria. J. Exp. Med. 1990, 171, 299–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.-Z.; Tang, X.-D.; Lü, M.-H.; Gao, J.-H.; Liang, G.-P.; Li, N.; Li, C.-Z.; Wu, Y.-Y.; Chen, L.; Cao, Y.-L.; et al. Multiple antigenic peptides of human heparanase elicit a much more potent immune response against tumors. Cancer Prev. Res. 2011, 4, 1285–1295. [Google Scholar] [CrossRef] [Green Version]
- Neefjes, J.; Jongsma, M.L.M.; Paul, P.; Bakke, O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 2011, 11, 823–836. [Google Scholar] [CrossRef]
- Girard, J.-P.; Moussion, C.; Förster, R. HEVs, lymphatics and homeostatic immune cell trafficking in lymph nodes. Nat. Rev. Immunol. 2012, 12, 762–773. [Google Scholar] [CrossRef] [PubMed]
- Bray, B.L. Large-scale manufacture of peptide therapeutics by chemical synthesis. Nat. Rev. Drug Discov. 2003, 2, 587–593. [Google Scholar] [CrossRef]
- Nordström, T.; Pandey, M.; Calcutt, A.; Powell, J.; Phillips, Z.N.; Yeung, G.; Giddam, A.K.; Shi, Y.; Haselhorst, T.; von Itzstein, M.; et al. Enhancing Vaccine Efficacy by Engineering a Complex Synthetic Peptide to Become a Super Immunogen. J. Immunol. 2017, 199, 2794–2802. [Google Scholar] [CrossRef]
- Grau Argente, B. Understanding Membrane Protein Folding and Interfaciality. Ph.D. Thesis, Universitat de València, Valencia, Spain, 2021. [Google Scholar]
- Sundberg, E.J.; Mariuzza, R.A. Molecular recognition in antibody-antigen complexes. Adv. Protein Chem. 2002, 61, 119–160. [Google Scholar]
- Cooper, J.A.; Hayman, W.; Reed, C.; Kagawa, H.; Good, M.F.; Saul, A. Mapping of conformational B cell epitopes within alpha-helical coiled coil proteins. Mol. Immunol. 1997, 34, 433–440. [Google Scholar] [CrossRef]
- Bergmann, C.C.; Yao, Q.; Ho, C.K.; Buckwold, S.L. Flanking residues alter antigenicity and immunogenicity of multi-unit CTL epitopes. J. Immunol. 1996, 157, 3242. [Google Scholar]
- Xing, L.; Fan, Y.-T.; Zhou, T.-J.; Gong, J.-H.; Cui, L.-H.; Cho, K.-H.; Choi, Y.-J.; Jiang, H.-L.; Cho, C.-S. Chemical modification of chitosan for efficient vaccine delivery. Molecules 2018, 23, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bird, G.H.; Irimia, A.; Ofek, G.; Kwong, P.D.; Wilson, I.A.; Walensky, L.D. Stapled HIV-1 peptides recapitulate antigenic structures and engage broadly neutralizing antibodies. Nat. Struct. Mol. Biol. 2014, 21, 1058–1067. [Google Scholar] [CrossRef]
- Li, W.; Joshi, M.D.; Singhania, S.; Ramsey, K.H.; Murthy, A.K. Peptide Vaccine: Progress and Challenges. Vaccines 2014, 2, 515–536. [Google Scholar] [CrossRef] [Green Version]
- TopuzoĞullari, M.; Acar, T.; Arayici, P.P.; Uçar, B.; Uğurel, E.; Abamor, E.Ş.; Arasoğlu, T.; BALIK, D.; Derman, S. An insight into the epitope-based peptide vaccine design strategy and studies against COVID-19. Turk. J. Biol. 2020, 44, 215–227. [Google Scholar] [CrossRef]
- Ayala, A.J.; Dimitrov, K.M.; Becker, C.R.; Goraichuk, I.V.; Arns, C.W.; Bolotin, V.I.; Ferreira, H.L.; Gerilovych, A.P.; Goujgoulova, G.V.; Martini, M.C.; et al. Presence of Vaccine-Derived Newcastle Disease Viruses in Wild Birds. PLoS ONE 2016, 11, e0162484. [Google Scholar] [CrossRef] [Green Version]
- Wallach, J.C.; Ferrero, M.C.; Victoria Delpino, M.; Fossati, C.A.; Baldi, P.C. Occupational infection due to Brucella abortus S19 among workers involved in vaccine production in Argentina. Clin. Microbiol. Infect. 2008, 14, 805–807. [Google Scholar] [CrossRef] [Green Version]
- Lin, Q.; Zhao, Q.; Lev, B. Cold chain transportation decision in the vaccine supply chain. Eur. J. Oper. Res. 2020, 283, 182–195. [Google Scholar] [CrossRef]
- Costerton, J.W.; Irvin, R.T.; Cheng, K.J.; Sutherland, I.W. The Role of Bacterial Surface Structures in Pathogenesis. CRC Crit. Rev. Microbiol. 1981, 8, 303–338. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; He, Y.; Zhou, Y.; Liu, S.; Zheng, B.-J.; Jiang, S. The spike protein of SARS-CoV—A target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009, 7, 226–236. [Google Scholar] [CrossRef]
- Butz, S.; Rawer, S.; Rapp, W.; Birsner, U. Immunization and affinity purification of antibodies using resin-immobilized lysine-branched synthetic peptides. Pept. Res. 1994, 7, 20–23. [Google Scholar]
- Nwe, K.; Brechbiel, M.W. Growing Applications of “Click Chemistry” for Bioconjugation in Contemporary Biomedical Research. Cancer Biother. Radiopharm. 2009, 24, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Otvos, L.; Otvos, L.; Wade, J.; Lin, F.; Condie, B.; Hanrieder, J.; Hoffmann, R. Designer Antibacterial Peptides Kill Fluoroquinolone-Resistant Clinical Isolates. J. Med. Chem. 2005, 48, 5349–5359. [Google Scholar] [CrossRef]
- Li, W.; Tailhades, J.; O’Brien-Simpson, N.M.; Separovic, F.; Otvos, L.; Hossain, M.A.; Wade, J.D. Proline-rich antimicrobial peptides: Potential therapeutics against antibiotic-resistant bacteria. Amino Acids 2015, 46, 2287–2294. [Google Scholar] [CrossRef] [PubMed]
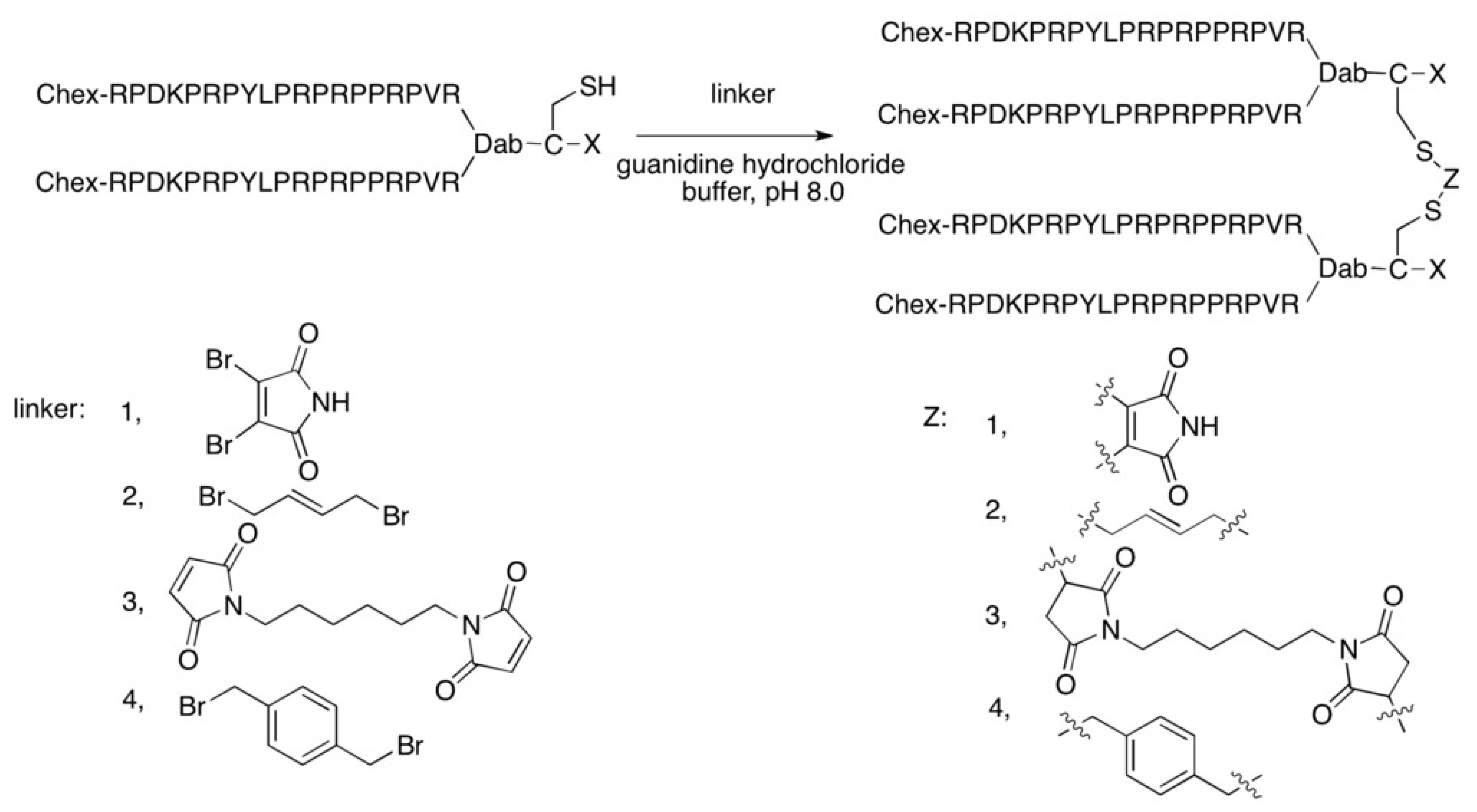
- Li, W.; O’Brien-Simpson, N.M.; Yao, S.; Tailhades, J.; Reynolds, E.C.; Dawson, R.M.; Otvos, L.J.; Hossain, M.A.; Separovic, F.; Wade, J.D. C-Terminal Modification and Multimerization Increase the Efficacy of a Proline-Rich Antimicrobial Peptide. Chem. Eur. J. 2017, 23, 390–396. [Google Scholar] [CrossRef]
- Arrevus. Aceragen Announces Acquisition of Arrevus. 2021. Available online: https://www.pharmtech.com/view/aceragen-acquires-arrevus-expands-late-stage-rare-disease-pipeline (accessed on 23 December 2021).
- Li, W.; O’Brien-Simpson, N.; Tailhades, J.; Pantarat, N.; Dawson, R.; Otvos, L.; Reynolds, E.; Separovic, F.; Hossain, M.; Wade, J. Multimerization of a Proline-Rich Antimicrobial Peptide, Chex-Arg20, Alters Its Mechanism of Interaction with the Escherichia coli Membrane. Chem. Biol. 2015, 22, 1250–1258. [Google Scholar] [CrossRef]
- Sani, M.-A.; Separovic, F. How Membrane-Active Peptides Get into Lipid Membranes. Acc. Chem. Res. 2016, 49, 1130–1138. [Google Scholar] [CrossRef]
- Lee, T.-H.; Hall, K.N.; Aguilar, M.-I. Antimicrobial Peptide Structure and Mechanism of Action: A Focus on the Role of Membrane Structure. Curr. Top. Med. Chem. 2016, 16, 25–39. [Google Scholar] [CrossRef]
- Arnusch, C.; Pieters, R.; Breukink, E.; Williams, S. Enhanced Membrane Pore Formation through High-Affinity Targeted Antimicrobial Peptides. PLoS ONE 2012, 7, e39768. [Google Scholar] [CrossRef]
- Bai, Y.; Liu, S.; Li, J.; Lakshminarayanan, R.; Sarawathi, P.; Tang, C.; Ho, D.; Verma, C.; Beuerman, R.W.; Pervushin, K. Progressive structuring of a branched antimicrobial peptide on the path to the inner membrane target. J. Biol. Chem. 2012, 287, 26606–26617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakshminarayanan, R.; Liu, S.; Li, J.; Nandhakumar, M.; Aung, T.T.; Goh, E.; Chang, J.Y.T.; Saraswathi, P.; Tang, C.; Safie, S.R.B.; et al. Synthetic Multivalent Antifungal Peptides Effective against Fungi. PLoS ONE 2014, 9, e87730. [Google Scholar]
- Xiao, J.; Tolbert, T.J. Modular assembly of dimeric HIV fusion inhibitor peptides with enhanced antiviral potency. Bioorg. Med. Chem. Lett. 2013, 23, 6046–6051. [Google Scholar] [CrossRef] [Green Version]
- Budge, P.J.; Lebowitz, J.; Graham, B.S. Antiviral Activity of RhoA-Derived Peptides against Respiratory Syncytial Virus Is Dependent on Formation of Peptide Dimers. Antimicrob. Agents Chemother. 2003, 47, 3470–3477. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, S.M.; Talukder, S.A.; Khan, A.M.; Afrin, N.; Ali, M.A.; Islam, R.; Parves, R.; Al Mamun, A.; Sufian, M.A.; Hossain, M.N.; et al. Antiviral Peptides as Promising Therapeutics against SARS-CoV-2. J. Phys. Chem. B 2020, 124, 9785–9792. [Google Scholar] [CrossRef]
- Cossarizza, A.; De Biasi, S.; Guaraldi, G.; Girardis, M.; Mussini, C.; Group, M.C.W. SARS-CoV-2, the virus that causes COVID-19: Cytometry and the new challenge for global health. Cytometry 2020, 97, 340. [Google Scholar] [CrossRef]
- Starpharma. SPL7013 COVID-19 Nasal Spray Virucidal against SARS-CoV-2. 2020. Available online: https://starpharma.com/news/story/spl7013-covid-19-nasal-spray-virucidal-against-sars-cov-2 (accessed on 23 December 2021).
- Starpharma. VIRALEZE SPL7013 Virucidal (>99.99%) against Delta Variant (ASX Announcement). 2021. Available online: https://starpharma.com/news/609 (accessed on 23 December 2021).
- Paull, J.R.A.; Heery, G.P.; Bobardt, M.D.; Castellarnau, A.; Luscombe, C.A.; Fairley, J.K.; Gallay, P.A. Virucidal and antiviral activity of astodrimer sodium against SARS-CoV-2 in vitro. Antivir. Res. 2021, 191, 105089. [Google Scholar] [CrossRef]
- Starpharma. VIRALEZE™ Antiviral Nasal Spray. 2021. Available online: https://starpharma.com/viraleze/spl7013 (accessed on 23 December 2021).
- Burki, T.K. Challenges in the rollout of COVID-19 vaccines worldwide. Lancet Respir. Med. 2021, 9, e42–e43. [Google Scholar] [CrossRef]
- Mammen, M.; Choi, S.-K.; Whitesides, G.M. Polyvalent Interactions in Biological Systems: Implications for Design and Use of Multivalent Ligands and Inhibitors. Angew. Chem. Int. Ed. 1998, 37, 2754–2794. [Google Scholar] [CrossRef]
- Yahi, N.; Sabatier, J.-M.; Baghdiguian, S.; Gonzalez-Scarano, F.; Fantini, J. Synthetic multimeric peptides derived from the principal neutralization domain (V3 loop) of human immunodeficiency virus type 1 (HIV-1) gp120 bind to galactosylceramide and block HIV-1 infection in a human CD4-negative mucosal epithelial cell line. J. Virol. 1995, 69, 320–325. [Google Scholar] [CrossRef] [Green Version]
- Luganini, A.; Giuliani, A.; Pirri, G.; Pizzuto, L.; Landolfo, S.; Gribaudo, G. Peptide-derivatized dendrimers inhibit human cytomegalovirus infection by blocking virus binding to cell surface heparan sulfate. Antivir. Res. 2010, 85, 532–540. [Google Scholar] [CrossRef]
- Wan, J.; Mobli, M.; Brust, A.; Muttenthaler, M.; Andersson, Å.; Ragnarsson, L.; Castro, J.; Vetter, I.; Huang, J.X.; Nilsson, M.; et al. Synthesis of Multivalent [Lys8]-Oxytocin Dendrimers that Inhibit Visceral Nociceptive Responses. Aust. J. Chem. 2017, 70, 162–171. [Google Scholar] [CrossRef]
- Nomizu, M.; Yamamura, K.; Kleinman, H.K.; Yamada, Y. Multimeric Forms of Tyr-Ile-Gly-Ser-Arg (YIGSR) Peptide Enhance the Inhibition of Tumor Growth and Metastasis. Cancer Res. 1993, 53, 3459. [Google Scholar]
- Fassina, G.; Verdoliva, A.; Odierna, M.R.; Ruvo, M.; Cassini, G. Protein a mimetic peptide ligand for affinity purification of antibodies. J. Mol. Recognit. 1996, 9, 564–569. [Google Scholar] [CrossRef]
- Quinlan, B.D.; Joshi, V.R.; Gardner, M.R.; Ebrahimi, K.H.; Farzan, M. A double-mimetic peptide efficiently neutralizes HIV-1 by bridging the CD4- and coreceptor-binding sites of gp120. J. Virol. 2014, 88, 3353–3358. [Google Scholar] [CrossRef] [Green Version]
- Putterman, C.; Diamond, B. Immunization with a Peptide Surrogate for Double-stranded DNA (dsDNA) Induces Autoantibody Production and Renal Immunoglobulin Deposition. J. Exp. Med. 1998, 188, 29–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tam, J.P.; Spetzler, J.C. Synthesis and Application of Peptide Dendrimers as Protein Mimetics. Curr. Protoc. Immunol. 1999, 34, 9.6.1–9.6.36. [Google Scholar]
- Rezazadeh, F.; Sadeghzadeh, N. Tumor targeting with 99mTc radiolabeled peptides: Clinical application and recent development. Chem. Biol. Drug Des. 2019, 93, 205–221. [Google Scholar] [CrossRef] [PubMed]
- Khoshbakht, S.; Beiki, D.; Geramifar, P.; Kobarfard, F.; Sabzevari, O.; Amini, M.; Bolourchian, N.; Shamshirian, D.; Shahhosseini, S. Design, synthesis, radiolabeling, and biologic evaluation of three 18F-FDG-radiolabeled targeting peptides for the imaging of apoptosis. Cancer Biother. Radiopharm. 2019, 34, 271–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aweda, T.A.; Muftuler, Z.F.B.; Massicano, A.V.F.; Gadhia, D.; McCarthy, K.A.; Queern, S.; Bandyopadhyay, A.; Gao, J.; Lapi, S.E. Radiolabeled Cationic Peptides for Targeted Imaging of Infection. Contrast Media Mol. Imaging 2019, 2019, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haubner, R.; Wester, H.J.; Burkhart, F.; Senekowitsch-Schmidtke, R.; Weber, W.; Goodman, S.L.; Kessler, H.; Schwaiger, M. Glycosylated RGD-containing peptides: Tracer for tumor targeting and angiogenesis imaging with improved biokinetics. J. Nucl. Med. 2001, 42, 326. [Google Scholar] [PubMed]
- Dijkraaf, I.; Wester, H.J. Peptides, multimers and polymers. Handb. Exp. Pharmacol. 2008, 185, 61–92. [Google Scholar]
- Mittra, E.; Goris, M.; Iagaru, A.; Kardan, A.; Burton, L.; Berganos, R.; Chang, E.; Liu, S.; Shen, B.; Chin, F.; et al. Pilot pharmacokinetic and dosimetric studies of (18)F-FPPRGD2: A PET radiopharmaceutical agent for imaging α(v)β(3) integrin levels. Radiology 2011, 260, 182–191. [Google Scholar] [CrossRef] [Green Version]
- Liolios, C.; Buchmuller, B.; Bauder Wüst, U.; Schäfer, M.; Leotta, K.; Haberkorn, U.; Eder, M.; Kopka, K. Monomeric and Dimeric 68Ga-Labeled Bombesin Analogues for Positron Emission Tomography (PET) Imaging of Tumors Expressing Gastrin-Releasing Peptide Receptors (GRPrs). J. Med. Chem. 2018, 61, 2062–2074. [Google Scholar] [CrossRef]
- Fournier, P.; Dumulon-Perreault, V.; Ait-Mohand, S.; Langlois, R.; Bénard, F.; Lecomte, R.; Guérin, B. Comparative study of 64Cu/NOTA-[D-Tyr6,βAla11,Thi13,Nle14]BBN(6-14) monomer and dimers for prostate cancer PET imaging. EJNMMI Res. 2012, 2, 8. [Google Scholar] [CrossRef] [Green Version]
- Fani, M.; Maecke, H.R. Radiopharmaceutical development of radiolabelled peptides. Eur. J. Nucl. Med. Mol. Imag. 2012, 39, 11–30. [Google Scholar] [CrossRef]
- Wang, J.; Kim, Y.-S.; Liu, S. 99mTc-Labeling of HYNIC-Conjugated Cyclic RGDfK Dimer and Tetramer Using EDDA as Coligand. Bioconjugate Chem. 2008, 19, 634–642. [Google Scholar] [CrossRef]
- Yan, Y.; Chen, X. Peptide heterodimers for molecular imaging. Amino Acids 2011, 41, 1081–1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, H.-J.; Huynh, T.T.; Rogers, B.E.; Mirica, L.M. Design of a multivalent bifunctional chelator for diagnostic 64Cu PET imaging in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2020, 117, 30928–30933. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.; O’Brien Simpson, N.; Pantarat, N.; Sulistio, A.; Wong, E.H.H.; Chen, Y.-Y.; Lenzo, J.; Holden, J.; Blencowe, A.; Reynolds, E.; et al. Combating multidrug-resistant Gram-negative bacteria with structurally nanoengineered antimicrobial peptide polymers. Nat. Microbiol. 2016, 1, 16162. [Google Scholar] [CrossRef]
- Borase, T.; Heise, A. Hybrid Nanomaterials by Surface Grafting of Synthetic Polypeptides Using N-Carboxyanhydride (NCA) Polymerization. Adv. Mater. 2016, 28, 5725–5731. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.J.; Wong, E.H.H.; O’Brien-Simpson, N.M.; Pantarat, N.; Blencowe, A.; Reynolds, E.C.; Qiao, G.G. Bionano Interaction Study on Antimicrobial Star-Shaped Peptide Polymer Nanoparticles. ACS Appl. Mater. Interfaces 2016, 8, 33446–33456. [Google Scholar] [CrossRef]
- Hutnick, M.A.; Pokorski, J.K. Polymeric Interventions for Microbial Infections: A Review. Mol. Pharm. 2018, 15, 2910–2921. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.J.; Wong, E.H.H.; Boyer, C.; Qiao, G.G. Antimicrobial polymeric nanoparticles. Prog. Polym. Sci. 2018, 76, 40–64. [Google Scholar] [CrossRef]
- Sulistio, A.; Widjaya, A.; Blencowe, A.; Zhang, X.; Qiao, G. Star polymers composed entirely of amino acid building blocks: A route towards stereospecific, biodegradable and hierarchically functionalized stars. Chem. Commun. 2011, 47, 1151–1153. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Wang, W.; Li, J. Star polymers: Advances in biomedical applications. Prog. Polym. Sci. 2015, 46, 55–85. [Google Scholar] [CrossRef]
- Mignani, S.; Rodrigues, J.; Tomas, H.; Zablocka, M.; Shi, X.; Caminade, A.-M.; Majoral, J.-P. Dendrimers in combination with natural products and analogues as anti-cancer agents. Chem. Soc. Rev. 2018, 47, 514–532. [Google Scholar] [CrossRef]
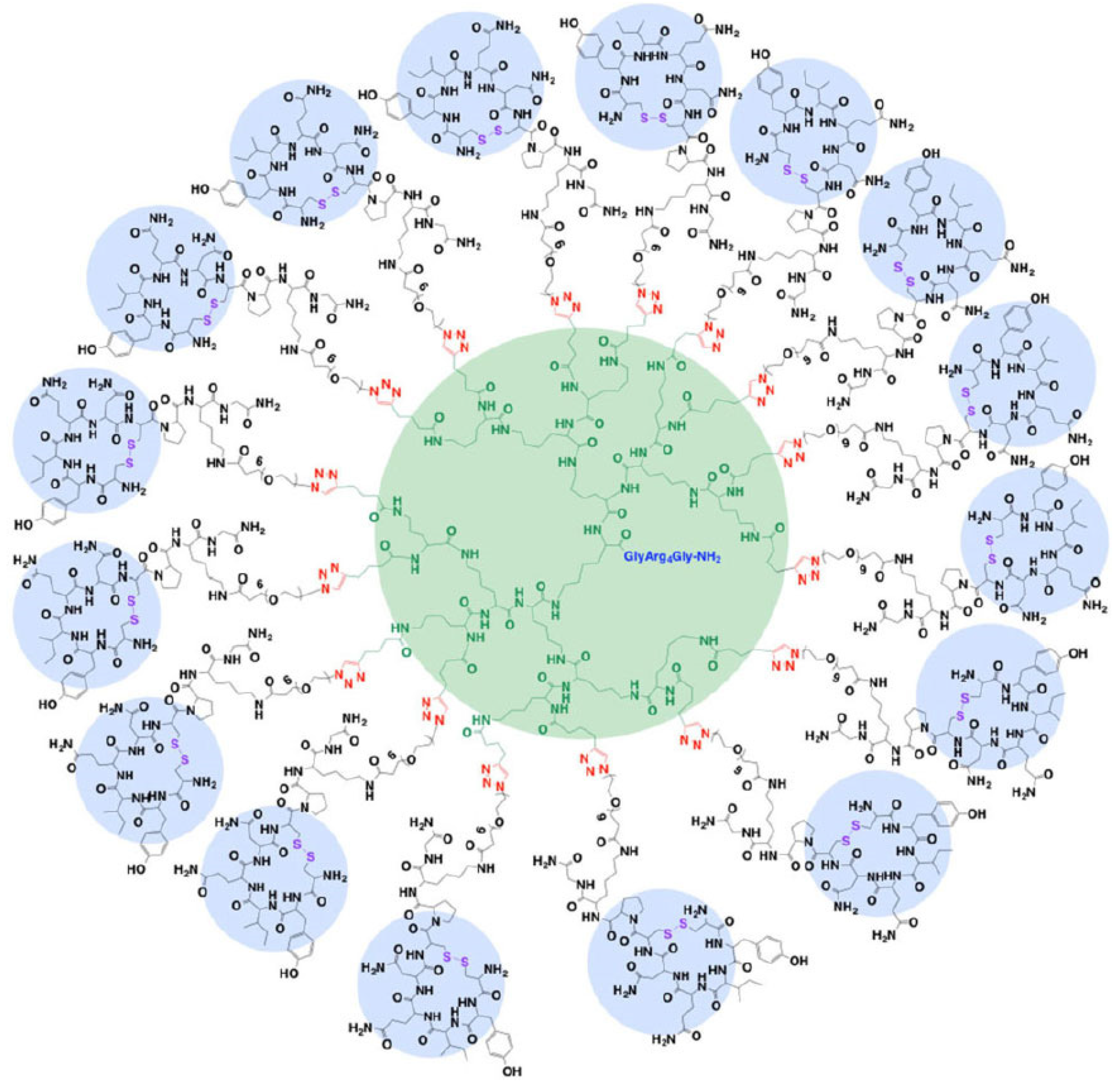
- Kokil, G.R.; Veedu, R.N.; Le, B.T.; Ramm, G.A.; Parekh, H.S. Self-assembling asymmetric peptide-dendrimer micelles—A platform for effective and versatile in vitro nucleic acid delivery. Sci. Rep. 2018, 8, 4832. [Google Scholar] [CrossRef] [PubMed]
- Arseneault, M.; Wafer, C.; Morin, J.-F. Recent Advances in Click Chemistry Applied to Dendrimer Synthesis. Molecules 2015, 20, 9263–9294. [Google Scholar] [CrossRef] [Green Version]
- Klajnert, B.; Bryszewska, M.B. Dendrimers: Properties and applications. Acta Biochim. Pol. 2001, 48, 199–208. [Google Scholar] [CrossRef] [Green Version]
- Wijagkanalan, W.; Kawakami, S.; Hashida, M. Designing Dendrimers for Drug Delivery and Imaging: Pharmacokinetic Considerations. Pharm. Res. 2011, 28, 1500–1519. [Google Scholar] [CrossRef]
- Munder, A.; Moskovitz, Y.; Meir, A.; Kahremany, S.; Levy, L.; Kolitz-Domb, M.; Cohen, G.; Shtriker, E.; Viskind, O.; Lellouche, J.-P.; et al. Neuroligin-2-derived peptide-covered polyamidoamine-based (PAMAM) dendrimers enhance pancreatic β-cells’ proliferation and functions. MedChemComm 2019, 10, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, A.; Mihov, G.; Vandermeulen, G.W.M.; Klok, H.-A.; Müllen, K. Peptide-functionalized polyphenylene dendrimers. Tetrahedron 2003, 59, 3925–3935. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Hossain, M.D.; Jia, Z.; Monteiro, M.J. One-Pot Orthogonal Copper-Catalyzed Synthesis and Self-Assembly of l-Lysine-Decorated Polymeric Dendrimers. Macromolecules 2015, 48, 1688–1702. [Google Scholar] [CrossRef]
- Sun, H.; Dong, Y.; Feijen, J.; Zhong, Z. Peptide-decorated polymeric nanomedicines for precision cancer therapy. J. Control. Release 2018, 290, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Alewood, P.F. Peptide-Decorated Dendrimers and Their Bioapplications. Angew. Chem. Int. Ed. 2016, 55, 5124–5134. [Google Scholar] [CrossRef]
- Liu, J.; Gray, W.D.; Davis, M.E.; Luo, Y. Peptide- and saccharide-conjugated dendrimers for targeted drug delivery: A concise review. Interface Focus 2012, 2, 307–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Liu, C.; Chen, D.; Madrid, K.; Peng, S.; Dong, X.; Zhang, M.; Gu, Y. Bacteria-Targeting Conjugates Based on Antimicrobial Peptide for Bacteria Diagnosis and Therapy. Mol. Pharm. 2015, 12, 2505–2516. [Google Scholar] [CrossRef]
- Ogura, A.; Tahara, T.; Nozaki, S.; Morimoto, K.; Kizuka, Y.; Kitazume, S.; Hara, M.; Kojima, S.; Onoe, H.; Kurbangalieva, A.; et al. Visualizing Trimming Dependence of Biodistribution and Kinetics with Homo- and Heterogeneous N-Glycoclusters on Fluorescent Albumin. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Abbassi, L.; Chabre, Y.; Kottari, N.; Arnold, A.; André, S.; Josserand, J.; Gabius, H.-J.; Roy, R. Multifaceted glycodendrimers with programmable bioactivity through convergent, divergent, and accelerated approaches using polyfunctional cyclotriphosphazenes. Polym. Chem. 2015, 6, 7666–7683. [Google Scholar] [CrossRef]
- Ochiai, H.; Yoshida, K.; Shibutani, H.; Kanatani, A.; Nishiuchi, Y. Spontaneously Cleavable Glycosylated Linker Capable of Extended Release of Its Conjugated Peptide. Chem. Pharm. Bull. 2019, 67, 236. [Google Scholar] [CrossRef]
- GlyTechInc. Hello Glycan. 2020. Available online: https://www.glytech-inc.com/glycan/ (accessed on 23 December 2021).
- Hagimori, M.; Fuchigami, Y.; Kawakami, S. Peptide-Based Cancer-Targeted DDS and Molecular Imaging. Chem. Pharm. Bull. 2017, 65, 618–624. [Google Scholar] [CrossRef] [Green Version]
- Sarangthem, V.; Kim, Y.; Singh, T.D.; Seo, B.-Y.; Cheon, S.-H.; Lee, Y.-J.; Lee, B.-H.; Park, R.-W. Multivalent Targeting Based Delivery of Therapeutic Peptide using AP1-ELP Carrier for Effective Cancer Therapy. Theranostics 2016, 6, 2235–2249. [Google Scholar] [CrossRef]
- Wang, Y.; Cheetham, A.G.; Angacian, G.; Su, H.; Xie, L.; Cui, H. Peptide–drug conjugates as effective prodrug strategies for targeted delivery. Adv. Drug Del. Rev. 2017, 110–111, 112–126. [Google Scholar] [CrossRef] [Green Version]
- Liang, K.; Richardson, J.J.; Ejima, H.; Such, G.K.; Cui, J.; Caruso, F. Peptide-Tunable Drug Cytotoxicity via One-Step Assembled Polymer Nanoparticles. Adv. Mater. 2014, 26, 2398–2402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christensen, S.B.; Skytte, D.M.; Denmeade, S.R.; Dionne, C.; Møller, J.V.; Nissen, P.; Isaacs, J.T. A Trojan horse in drug development: Targeting of thapsigargins towards prostate cancer cells. Anticancer Agents Med. Chem. 2009, 9, 276–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halford, B. A new generation of antibody-drug conjugates for cancer patients. Chem. Eng. News 2020, 14, 16. [Google Scholar]
- de la Torre, B.G.; Albericio, F. Peptide Therapeutics 2.0. Molecules 2020, 25, 2293. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Kaneko, R.; Kamio, T.; Kamiyama, E.; Muto, R.; Sugihara, M. Pharmacological and clinical study results of trastuzumab deruxtecan (T-DXd, ENHERTU®). Folia Pharmacol. Jpn. 2021, 156, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Schwaederle, M.; Arguello, D.; Millis, S.Z.; Gatalica, Z.; Kurzrock, R. HER2 expression status in diverse cancers: Review of results from 37,992 patients. Cancer Metastasis Rev. 2015, 34, 157–164. [Google Scholar] [CrossRef] [Green Version]
- Kamath, A.V.; Iyer, S. Challenges and advances in the assessment of the disposition of antibody-drug conjugates. Biopharm. Drug Disposition 2016, 37, 66–74. [Google Scholar] [CrossRef] [Green Version]
- Starpharma. DEP® HER2-Lutetium Outperforms in Human Breast Cancer Model. 2021. Available online: https://starpharma.com/news/story/dep-her2-lutetium-outperforms-in-human-breast-cancer-model#_ftn2 (accessed on 23 December 2021).
- Starpharma. Starpharma Signs DEP® ADC Research Agreement with MSD. 2021. Available online: https://starpharma.com/news/story/starpharma-signs-dep-adc-research-agreement-with-msd (accessed on 23 December 2021).
- Rao, M.S.; Gupta, R.; Liguori, M.J.; Hu, M.; Huang, X.; Mantena, S.R.; Mittelstadt, S.W.; Blomme, E.A.G.; Van Vleet, T.R. Novel Computational Approach to Predict Off-Target Interactions for Small Molecules. Front. Big Data 2019, 2, 25. [Google Scholar] [CrossRef] [Green Version]
- Gilad, Y.; Firer, M.; Gellerman, G. Recent Innovations in Peptide Based Targeted Drug Delivery to Cancer Cells. Biomedicines 2016, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Shi, X. Dendrimer-based nanodevices for targeted drug delivery applications. J. Mater. Chem. B Mater. Biol. Med. 2013, 1, 4199–4211. [Google Scholar] [CrossRef] [PubMed]
- Woller, E.K. The lectin-binding properties of six generations of mannose-functionalized dendrimers. Org. Lett. 2002, 4, 7–10. [Google Scholar] [CrossRef]
- Gajbhiye, V.; Vijayaraj Kumar, P.; Kumar Tekade, R.; Jain, N.V. Pharmaceutical and Biomedical Potential of PEGylated Dendrimers. Curr. Pharm. Des. 2007, 13, 415–429. [Google Scholar] [CrossRef]
- Agrawal, P.; Gupta, U.; Jain, N.K. Glycoconjugated peptide dendrimers-based nanoparticulate system for the delivery of chloroquine phosphate. Biomaterials 2007, 28, 3349–3359. [Google Scholar] [CrossRef]
- Florence, A.T.; Sakthivel, T.; Toth, I. Oral uptake and translocation of a polylysine dendrimer with a lipid surface. J. Controlled Release 2000, 65, 253–259. [Google Scholar] [CrossRef]
- Bernkop-Schnurch, A.; Clausen, A.E. Biomembrane Permeability of Peptides: Strategies to Improve Their Mucosal Uptake. Mini Rev. Med. Chem. 2002, 2, 295–305. [Google Scholar] [CrossRef]
- Siriwardena, T.N.; Stach, M.; He, R.; Gan, B.-H.; Javor, S.; Heitz, M.; Ma, L.; Cai, X.; Chen, P.; Wei, D.; et al. Lipidated Peptide Dendrimers Killing Multidrug-Resistant Bacteria. J. Am. Chem. Soc. 2018, 140, 423–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novartis. Novartis Annual Report 2019. 2019. Available online: https://www.novartis.com/sites/novartis_com/files/novartis-annual-report-2019.pdf (accessed on 23 December 2021).
- Takeda Pharmaceutical Company Limited, Annual report 2019. 2019. Available online: https://www.takeda.com/4afd73/siteassets/system/investors/report/sec-filings/20-f_2020-06-24.pdf (accessed on 23 December 2021).
- CSL. Momenta and CSL Announce Collaboration. 2019. Available online: https://www.csl.com/news/2017/20170105-momenta-collaboration (accessed on 23 December 2021).
- Starpharma. Dendrimer Drug Delivery (DEP®). 2019. Available online: https://starpharma.com/drug_delivery (accessed on 23 December 2021).
- Starpharma. Partnered-DEP® Products—AZD0466. 2019. Available online: https://starpharma.com/drug_delivery/dep-azd0466 (accessed on 23 December 2021).
- Starpharma. DEP® Docetaxel Summary and Commercial Opportunity. 2019. Available online: https://starpharma.com/drug_delivery/dep_docetaxel (accessed on 23 December 2021).
- Starpharma. DEP® Cabazitaxel. 2019. Available online: https://starpharma.com/drug_delivery/dep_cabazitaxel (accessed on 23 December 2021).



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheard, D.E.; Li, W.; O’Brien-Simpson, N.M.; Separovic, F.; Wade, J.D. Peptide Multimerization as Leads for Therapeutic Development. Biologics 2022, 2, 15-44. https://doi.org/10.3390/biologics2010002
Sheard DE, Li W, O’Brien-Simpson NM, Separovic F, Wade JD. Peptide Multimerization as Leads for Therapeutic Development. Biologics. 2022; 2(1):15-44. https://doi.org/10.3390/biologics2010002
Chicago/Turabian StyleSheard, Dean E., Wenyi Li, Neil M. O’Brien-Simpson, Frances Separovic, and John D. Wade. 2022. "Peptide Multimerization as Leads for Therapeutic Development" Biologics 2, no. 1: 15-44. https://doi.org/10.3390/biologics2010002
APA StyleSheard, D. E., Li, W., O’Brien-Simpson, N. M., Separovic, F., & Wade, J. D. (2022). Peptide Multimerization as Leads for Therapeutic Development. Biologics, 2(1), 15-44. https://doi.org/10.3390/biologics2010002







