Phenotypic Characterization and Prevalence of Carbapenemase-Producing Pseudomonas aeruginosa Isolates in Six Health Facilities in Cameroon
Abstract
1. Introduction
2. Materials and Methods
2.1. Type, Site, and Duration of Study
2.2. Sampling Method and Selection Criteria
2.3. Re-Identification and Samples Processing
2.4. Data Evaluation and Analysis
2.5. Ethical Considerations
3. Results
3.1. Description of the Source Characteristics of the Isolates Collected
3.2. Susceptibility of Pseudomonas aeruginosa Isolates to Beta-Lactams
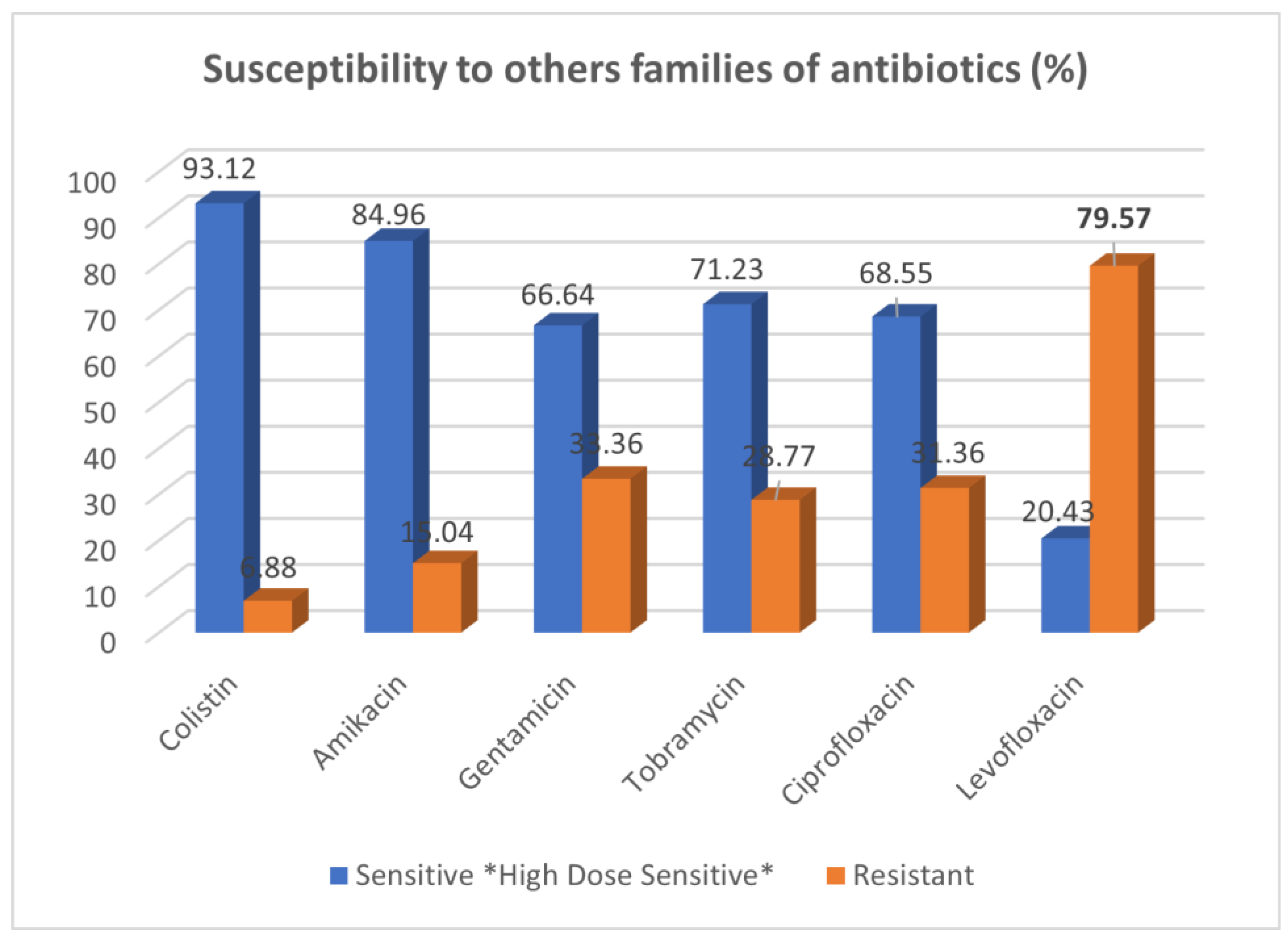
3.3. Resistance Profile of Pseudomonas aeruginosa to Other Families of Antibiotics
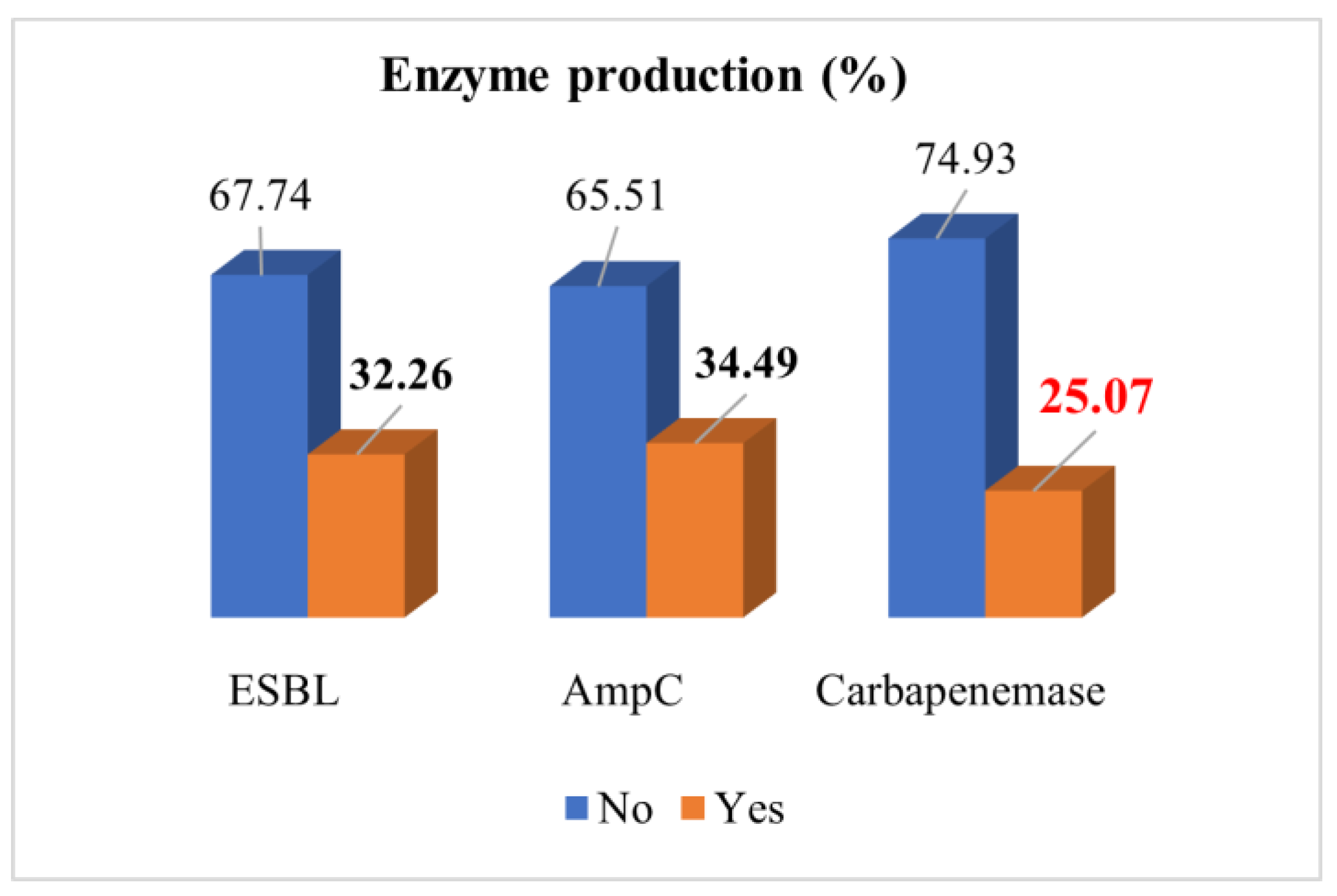
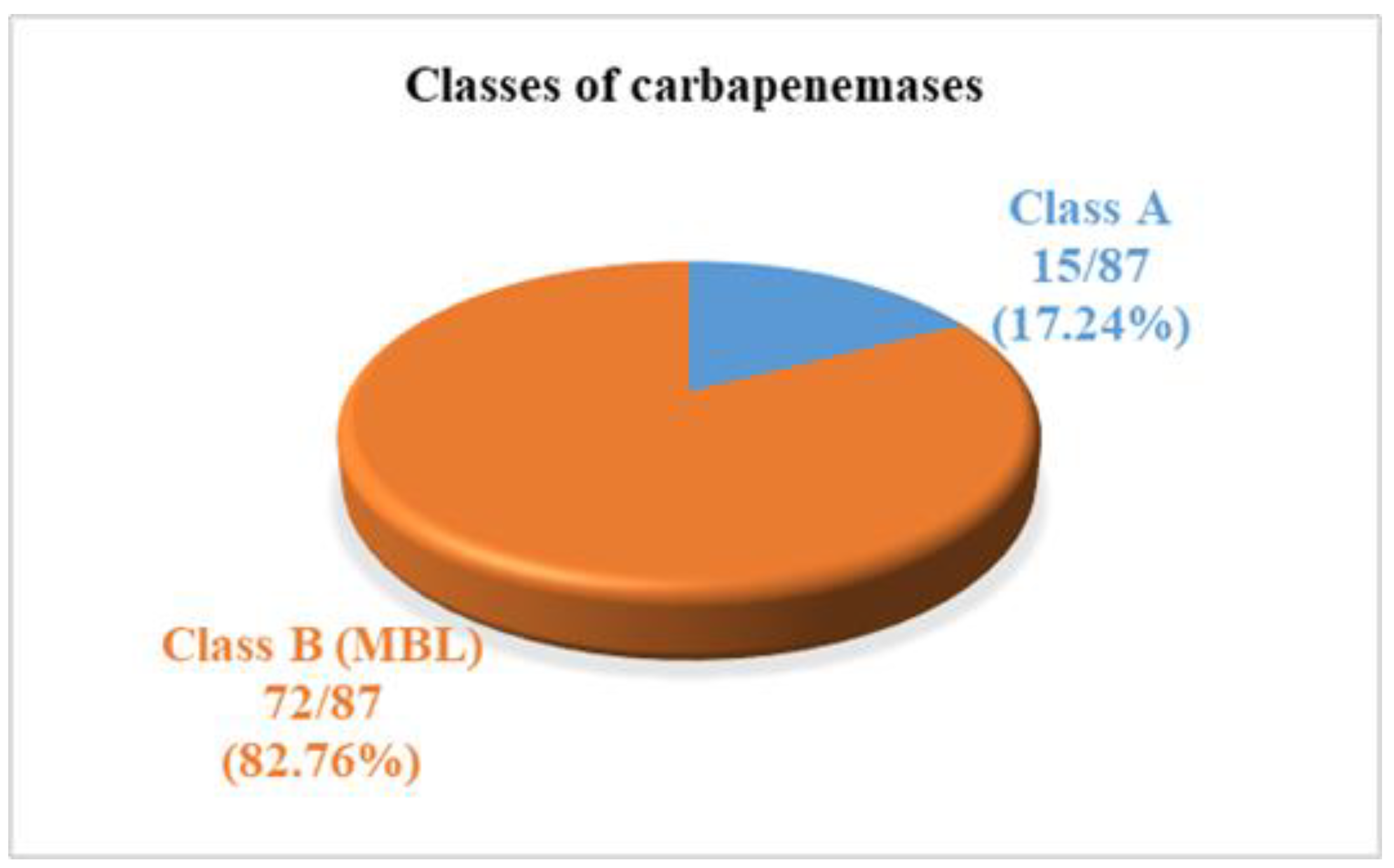
3.4. Distribution of Pseudomonas aeruginosa Isolates According to Enzyme Production and Carbapenemase Classification
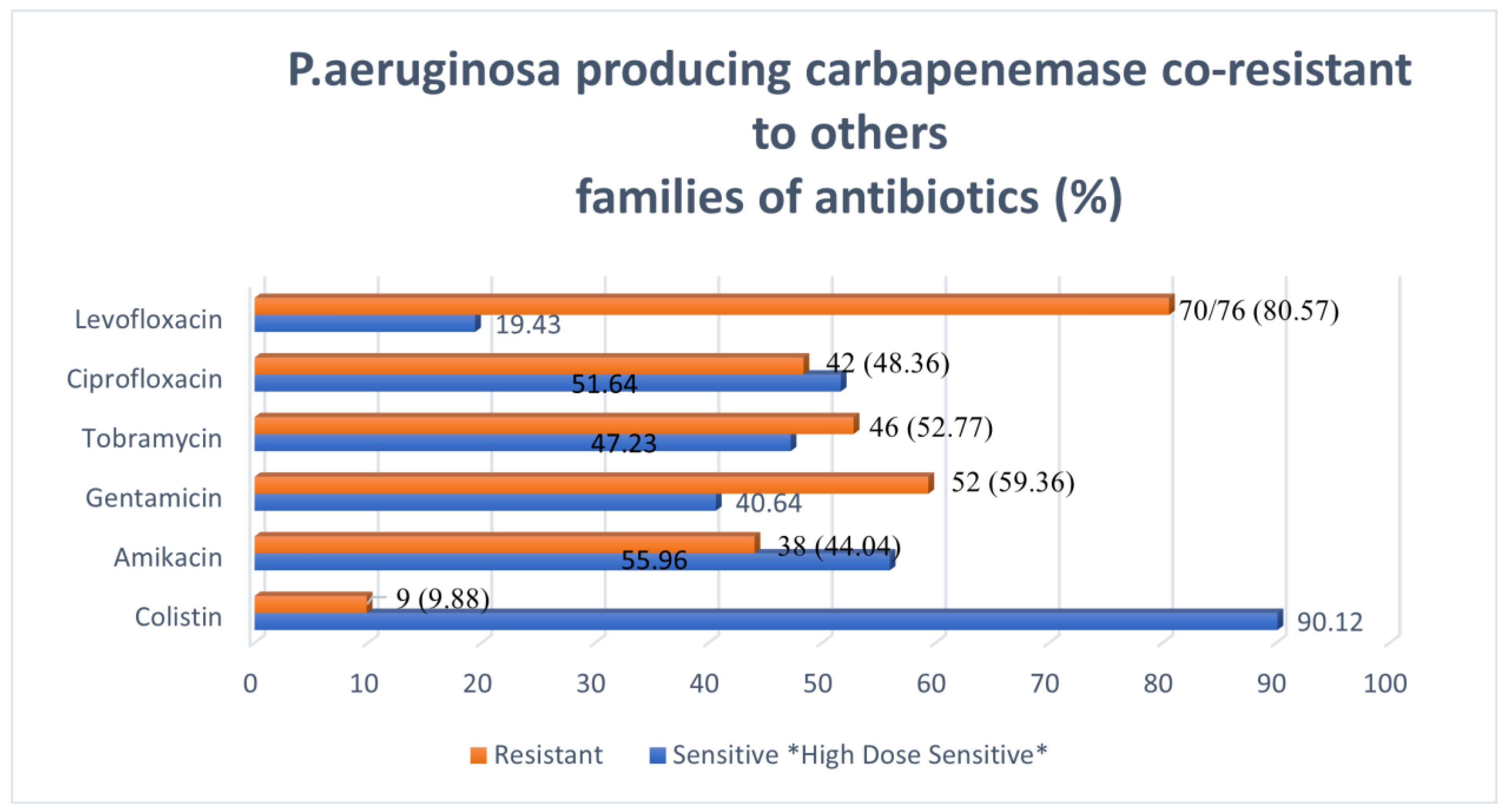
3.5. Co-Resistance Profile of Pseudomonas aeruginosa Producing Carbapenemase to Various Families of Antibiotics
3.6. Association between Risk Factors and Carbapenemase-Producing Pseudomonas aeruginosa
4. Discussion
5. Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Consent for Publication
References
- Callejas-Díaz, A.; Fernández-Pérez, C.; Ramos-Martínez, A.; Múñez-Rubio, E.; Sánchez-Romero, I.; Núñez, J.A.V. Impact of Pseudomonas aeruginosa bacteraemia in a tertiary hospital: Mortality and prognostic factors. Med. Clín. 2019, 152, 83–89. [Google Scholar] [CrossRef]
- Bodro, M.; Sabé, N.; Tubau, F.; Lladó, L.; Baliellas, C.; González-Costello, J.; Cruzado, J.M.; Carratalà, J. Extensively Drug-Resistant Pseudomonas aeruginosa Bacteremia in Solid Organ Transplant Recipients. Transplantation 2015, 99, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Kara Ali, R.; Surme, S.; Balkan, I.I.; Salihoglu, A.; Sahin Ozdemir, M.; Ozdemir, Y.; Mete, B.; Can, G.; Ar, M.C.; Tabak, F.; et al. An eleven-year cohort of bloodstream infections in 552 febrile neutropenic patients: Resistance profiles of Gram-negative bacteria as a predictor of mortality. Ann. Hematol. 2020, 99, 1925–1932. [Google Scholar] [CrossRef]
- Righi, E.; Peri, A.M.; Harris, P.N.A.; Wailan, A.; Liborio, M.; Lane, S.W.; Paterson, D.L. Global prevalence of carbapenem resistance in neutropenic patients and association with mortality and carbapenem use: Systematic review and meta-analysis. J. Antimicrob. Chemother. 2017, 72, 668–677. [Google Scholar] [CrossRef] [PubMed]
- Koujane, L. Epidemiology and Resistance of Multiresistant Bacteria at the Mohammed V Military Instruction Hospital in Rabat. Doctoral Thesis, Mohammed V University Faculty of Medicine and Pharmacy, Rabat, Morocco, 2011. [Google Scholar]
- Brunet, P.; Tsimaratos, M.; Guys, J.M.; Chevallier, E. Urinary Tract Infections in Children and Adults; Leucocyturia, Faculty of Medicine of Marseill, France: Marseill, France, 2006. [Google Scholar]
- Wang, M.-G.; Liu, Z.-Y.; Liao, X.-P.; Sun, R.-Y.; Li, R.-B.; Liu, Y.; Fang, L.-X.; Sun, J.; Liu, Y.-H.; Zhang, R.-M. Retrospective Data Insight into the Global Distribution of Carbapenemase-Producing Pseudomonas aeruginosa. Antibiotics 2021, 10, 548. [Google Scholar] [CrossRef]
- Zowawi, H.M.; Balkhy, H.H.; Walsh, T.R.; Paterson, D.L. β-Lactamase production in key gram-negative pathogenisolates from the Arabian peninsula. Clin. Microbiol. Rev. 2013, 26, 361–380. [Google Scholar] [CrossRef]
- Escandón-Vargas, K.; Reyes, S.; Gutiérrez, S.; Villegas, M.V. The epidemiology of carbapenemases in Latin America and the Caribbean. Expert Rev. Anti-Infect. Ther. 2017, 15, 277–297. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Nordmann, P.; Lagrutta, E.; Cleary, T.; Munoz-Price, L.S. Emergence of KPC-producing Pseudomonas aeruginosa in the United States. Antimicrob. Agents Chemother. 2010, 54, 3072. [Google Scholar] [CrossRef]
- Walters, M.S.; Grass, J.E.; Bulens, S.N.; Hancock, E.B.; Phipps, E.C.; Muleta, D.; Mounsey, J.; Kainer, M.A.; Concannon, C.; Dumyati, G.; et al. Carbapenem-resistant Pseudomonas aeruginosa at US Emerging infections program sites, 2015. Emerg. Infect. Dis. 2019, 25, 1281–1288. [Google Scholar] [CrossRef]
- Simner, P.J.; Opene, B.N.A.; Chambers, K.K.; Naumann, M.E.; Carroll, K.C.; Tamma, P.D. Carbapenemase detection among carbapenem-resist-ant glucose-nonfermenting gram-negative bacilli. J. Clin. Microbiol. 2017, 55, 2858–2864. [Google Scholar] [CrossRef]
- Mizan, K.; Lemma, D.; Baye, G.; Feleke, M. Carbapenemase-Producing Non-Glucose-Fermenting Gram Negative Bacilli in Africa, Pseudomonas aeruginosa and Acinetobacter baumanii: A systematic Review and Meta-analysis. Int. J. Microbiol. 2020, 2020, 9461901. [Google Scholar]
- World Health Organization (WHO). Antimicrobial Resistance. Global Report on Surveillance. Geneva, Switzerland, 2014. Available online: https://reliefweb.int/report/world/antimicrobial-resistance-global-report-surveillance-2014?gclid=EAIaIQobChMI1MP7hoeN_AIV1oXVCh0NGAWKEAAYASAAEgIpdfD_BwE (accessed on 18 November 2022).
- Weiner, L.M.; Webb, A.K.; Limbago, B.; Dudeck, M.A.; Patel, J.; Kallen, A.J.; Edwards, J.R.; Sievert, D.M. Antimicrobial-Resistant Pathogens Associated with Healthcare-Associated Infections: Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect. Control Hosp. Epidemiol. 2016, 37, 1288–1301. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.-L.; Sakr, Y.; Singer, M.; Martin-Loeches, I.; Machado, F.R.; Marshall, J.C.; Finfer, S.; Pelosi, P.; Brazzi, L.; Aditianingsih, D.; et al. Prevalence and Outcomes of Infection among Patients in Intensive Care Units in 2017. JAMA 2020, 323, 1478–1487. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, V.D.; Al-Abdely, H.M.; El-Kholy, A.A.; AlKhawaja, S.A.A.; Leblebicioglu, H.; Mehta, Y.; Rai, V.; Hung, N.V.; Kanj, S.S.; Salama, M.F.; et al. International Nosocomial Infection Control Consortium report, data summary of 50 countries for 2010–2015: Device-associated module. Am. J. Infect. Control. 2016, 44, 1495–1504. [Google Scholar] [CrossRef] [PubMed]
- Mittal, R.; Aggarwal, S.; Sharma, S.; Chhibber, S.; Harjai, K. Urinary tract infections caused by Pseudomonas aeruginosa: A minireview. J. Infect. Public Health 2009, 2, 101–111. [Google Scholar] [CrossRef]
- Karlowsky, J.A.; Bouchillon, S.K.; El Mahdy Kotb, R.; Mohamed, N.; Stone, G.G.; Sahm, D.F. Carbapenem-resistant Enterobacterales and Pseudomonas aeruginosa causing infection in Africa and the Middle East: A surveillance study from the ATLAS programme (2018–20). JAC Antimicrob. Resist. 2022, 4, dlac060. [Google Scholar] [CrossRef]
- Massri, A. Épidémiologie de la résistance aux carbapénèmes de Pseudomonas Aeruginosa dans 3 services de réanimation français: études des mécanismes de résistance et évaluation des tests phénotypiques de dépistage des carbapénémases. Médecine humaine et pathologie. 2016. Available online: https://dumas.ccsd.cnrs.fr/dumas-01493599 (accessed on 4 November 2022).
- Lukuke, H.M.; Kasamba, E.; Mahuridi, A.; Nlandu, R.N.; Narufumi, S.; Mukengeshayi, A.N.; Malou, V.; Makoutode, M.; Kaj, F.M. L’incidence des infections nosocomiales urinaires et des sites opératoires dans la maternité de l’Hôpital Général de Référence de Katuba à Lubumbashi en République Démocratique du Congo [Nosocomial urinary tract and surgical site infection rates in the Maternity Ward at the General Referral Hospital in Katuba, Lubumbashi, Democratic Republic of the Congo]. Pan Afr. Med. J. 2017, 28, 57, French. [Google Scholar] [CrossRef]
- CLSI Publishes M100—Performance Standards for Antimicrobial Susceptibility Testing. 31st ed. The Clinical and Laboratory Standards Institute (CLSI): Malvern, PA, USA, 2021. Available online: https://clsi.org/about/press-releases/clsi-publishes-m100-performance-standards-for-antimicrobial-susceptibility-testing-31st-edition/ (accessed on 10 October 2021).
- Madaha, E.L.; Mienie, C.; Gonsu, K.H.; Bughe, R.N.; Fonkoua, M.C. Whole-genome sequence of multi-drug resistant Pseudomonas aeruginosa strains UYPSABAL and UYPSABAL2 isolated from human broncho-alveolar lavage. PLoS ONE 2020, 15, e0238390. [Google Scholar] [CrossRef]
- Zahraa, A.F.; Saad, S.H. The frequency and sensitivity pattern of Pseudomonas aeruginosa among Ottitis Media patients in Nassirayah City. Univ. Thi-Quar J. 2019, 3, 25. [Google Scholar]
- Gonsu, K.H.; Toukam, M.; Sando, Z.; Ngamba, N.J.M.; Mbakop, D.C.; Adiogo, D. Phenotypic characterization of Pseudomonas aeruginosa isolates isolated in the city of Yaoundé (Cameroon). Afr. J. Pathol. Microbiol. 2015, 5, 1–4. [Google Scholar]
- Ankur, K.; Suprakash, D.; Nahid, A.; Viskash, O.; Sushmita, D. Antimicrobial susceptibility pattern of extended spectrum beta-lactamase (ESBL) and non ESBL producing Pseudomonas aeruginosa, isolated from pus samples from a tertiary care hospital in Bihar. Int. J. Curr. Microbiol. Appl. Sci. 2020, 9, 3646–3655. [Google Scholar]
- López-Causapé, C.; Cabot, G.; Del Barrio-Tofiño, E.; Oliver, A. The Versatile Mutational Resistome of Pseudomonas aeruginosa. Front. Microbiol. 2018, 9, 685. [Google Scholar] [CrossRef] [PubMed]
- Hernando-Amado, S.; Sanz-Garcia, F.; Blanco, P.; Martinez, J.L. Fitness costs associated with the acquisition of antibiotic resistance. Essays Biochem. 2017, 61, 37–48. [Google Scholar]
- Zheng, P.; Renee, R.; Glick, B.R.; Lin, T.-J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef]
- Breidenstein, E.B.; de la Fuente-Nunez, C.; Hancock, R.E. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef]
- Drenkard, E. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. 2003, 5, 1213–1219. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.R.; Burns, J.L.; Lory, S.; Lewis, K. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 2010, 192, 6191–6199. [Google Scholar] [CrossRef]
- Moctar, M.; Moffo, F.; Kihla, J. Antimicrobial resistance from a one health perspective in Cameroon: A systematic review and meta-analysid. BMC Public Health 2019, 19, 1135. [Google Scholar]
- Deplano, A.; Denis, O.; Poirel, L.; Hocquet, D.; Nonhoff, C.; Byl, B.; Nordmann, P.; Vincent, J.L.; Struelens, M.J. Molecular characterization of an epidemic clone of panantibiotic-resistant Pseudomonas aeruginosa. J. Clin. Microbiol. 2005, 43, 1198–1204. [Google Scholar] [CrossRef]
- Suárez, C.; Peña, C.; Arch, O.; Domínguez, M.A.; Tubau, F.; Juan, C.; Gavaldá, L.; Sora, M.; Oliver, A.; Pujol, M.; et al. A large sustained endemic outbreak of multiresistant Pseudomonas aeruginosa: A new epidemiological scenario for nosocomial acquisition. BMC Infect. Dis. 2011, 11, 272. [Google Scholar] [CrossRef]
- Berrazeg, M.; Jeannot, K.; Ntsogo Enguéné, V.Y.; Broutin, I.; Loeffert, S.; Fournier, D.; Plésiat, P. Mutations in β-Lactamase AmpC Increase Resistance of Pseudomonas aeruginosa Isolates to Antipseudomonal Cephalosporins. Antimicrob. Agents Chemother. 2015, 59, 6248–6255. [Google Scholar] [CrossRef]
- Castanhiera, M.; Deshpande, L.M.; Costello, A.; Davies, T.A.; Jones, R.N. Epidemiology and carbapenem resistance mechanisms of carbapenem-non-susceptible Pseudomonas aeruginosa collected during 2009–2011 in 14 European and Mediterranean countries. J. Antimicrob. Chemother. 2014, 69, 1804–1814. [Google Scholar] [CrossRef] [PubMed]
- Al-Khudhairy, M.K.; Al-Shammari, M.M.M. Prevalence of metallo-β-lactamase producing Pseudomonas aeruginosa isolated from diabetic foot infections in Iraq. New Microbes New Infect. 2020, 35, 100661. [Google Scholar] [CrossRef] [PubMed]
- Stije, J.L.; Leth, F.V.; Tarekegn, H.; Constance, S. Antimicrobial drug resistance among clinically relevant bacterial isolates in sub-Saharan African. J. Antimicrob. Chemother. 2014, 69, 2337–2353. [Google Scholar]
- Magoue, L.C.; Merlin, P.; Gangoue, P.J.; Okomo, A.M.C.; Boreux, R.; De Mol, P. Prevalence and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae in Ngaoundéré, Cameroon. Clin. Microbiol. Infect. 2013, 19, 416–420. [Google Scholar] [CrossRef]

| Characteristics | Effective (n) | Percentage (%) |
|---|---|---|
| Collection Sites | ||
| Center region | 89 | 25.64 |
| Littoral region | 102 | 29.40 |
| West region | 156 | 44.96 |
| Age (in years) | ||
| Mean ± Standard Deviation | 34.38 ± 20.67 | |
| Minimum; Maximum | 2 days; 91 years | |
| Sex | ||
| Male | 187 | 53.89 |
| Female | 160 | 46.11 |
| Sex ratio | 1.2 | |
| Healthcare unit | ||
| Surgery | 143 | 41.21 |
| Intensive care | 78 | 22.48 |
| Medicine | 56 | 16.14 |
| Pediatrics | 36 | 10.37 |
| Outpatient | 19 | 5.48 |
| Maternity | 15 | 4.32 |
| Hospitalization | ||
| Yes | 243 | 70.03 |
| No | 104 | 29.97 |
| Samples type | ||
| Pus | 142 | 40.92 |
| Wounds | 113 | 32.56 |
| Probe tip | 45 | 12.96 |
| Urine | 26 | 7.49 |
| Blood | 16 | 4.61 |
| Effusion fluid | 4 | 1.15 |
| Endocervical swab | 1 | 0.31 |
| Treatment | ||
| Yes | 219 | 59.65 |
| No | 128 | 36.88 |
| Variables | Total (n) | Carbapenemase Productionn (%) | OR | 95% CI | p-Value | |
|---|---|---|---|---|---|---|
| Sampling sites | ||||||
| Rural (West region) | 156 | 16 (10.44%) | Ref | |||
| Urban (Center and Littoral region) | 191 | 54 (28.48%) | 1.31 | 0.66–2.59 | 0.0467 | |
| Gender | ||||||
| Female | 160 | 30 (18.84%) | Ref | |||
| Male | 187 | 41 (21.74%) | 1.20 | 0.68–2.11 | 0.5355 | |
| Hospitalization | ||||||
| No | 104 | 15 (14.44%) | Ref | |||
| Yes | 243 | 56 (22.96%) | 0.94 | 0.51–1.73 | 0.1114 | |
| Treatment | ||||||
| No | 128 | 27 (20.91%) | Ref | |||
| Yes | 219 | 46 (20.11%) | 0.95 | 0.53–1.70 | 0.2480 | |
| Healthcare unit | ||||||
| External | 19 | 1 (33.56%) | Ref | |||
| Maternity | 15 | 4 (28.57%) | 1.84 | 0.89–4.45 | 0.5785 | |
| Medicine | 56 | 11 (18.75%) | 0.75 | 0.32–1.73 | 0.4960 | |
| Pediatrics | 36 | 3 (9.68%) | 0.35 | 0.10–1.23 | 0.1003 | |
| Surgery | 143 | 48 (6.25%) | 1.88 | 0.99–2.41 | 0.0432 | |
| Intensive care | 78 | 17 (22.39%) | 1.4 | 0.04–1.90 | 0.0457 | |
| Sample type | ||||||
| probe tip | 45 | 12 (25.64%) | Ref | |||
| Blood | 16 | - | 7.24 × 10−7 | - | 0.9789 | |
| ascites fluid | 4 | 1 (25%) | 0.97 | 0.19–2.57 | 0.9935 | |
| PCV | 1 | - | 7.24 × 10−7 | - | 0.9944 | |
| Wounds | 113 | 20 (17.53%) | 0.62 | 0.25–1.50 | 0.2859 | |
| Pus | 142 | 34 (23.77%) | 0.90 | 0.39–2.08 | 0.8124 | |
| Urine | 26 | 5 (18.18%) | 0.64 | 0.18–2.37 | 0.5078 | |
| Variables | Adjusted OR | 95% CI | p-Value | |
|---|---|---|---|---|
| Sampling sites | ||||
| Rural (West) | Ref | |||
| Urban (Center and Littoral) | 1.39 | 0.52–3.68 | 0.5116 | |
| Hospitalization | ||||
| No | Ref | |||
| Yes | 1.45 | 0.03–1.73 | 0.0456 | |
| Treatment | ||||
| No | Ref | |||
| Yes | 0.86 | 0.47–1.60 | 0.6435 | |
| Ward | ||||
| External | Ref | |||
| Medicine | 1.30 | 0.38–4.45 | 0.6795 | |
| Maternity | 1.75 | 0.02–1.73 | 0.0365 | |
| Pediatrics | 0.35 | 0.10–1.23 | 0.1003 | |
| Surgery | 1.41 | 0.32–1.71 | 0.0434 | |
| Intensive care | 1.29 | 0.05–1.45 | 0.0484 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Djuikoue, C.I.; Djouela Djoulako, P.D.; Same Njanjo, H.V.; Kiyang, C.P.; Djantou Biankeu, F.L.; Guegang, C.; Tchouotou, A.S.D.; Wouambo, R.K.; Thumamo Pokam, B.D.; Apalata, T.; et al. Phenotypic Characterization and Prevalence of Carbapenemase-Producing Pseudomonas aeruginosa Isolates in Six Health Facilities in Cameroon. BioMed 2023, 3, 77-88. https://doi.org/10.3390/biomed3010006
Djuikoue CI, Djouela Djoulako PD, Same Njanjo HV, Kiyang CP, Djantou Biankeu FL, Guegang C, Tchouotou ASD, Wouambo RK, Thumamo Pokam BD, Apalata T, et al. Phenotypic Characterization and Prevalence of Carbapenemase-Producing Pseudomonas aeruginosa Isolates in Six Health Facilities in Cameroon. BioMed. 2023; 3(1):77-88. https://doi.org/10.3390/biomed3010006
Chicago/Turabian StyleDjuikoue, Cecile Ingrid, Paule Dana Djouela Djoulako, Hélène Valérie Same Njanjo, Christiane Possi Kiyang, Feline Leina Djantou Biankeu, Celianthe Guegang, Andrea Sheisley Didi Tchouotou, Rodrigue Kamga Wouambo, Benjamin D. Thumamo Pokam, Teke Apalata, and et al. 2023. "Phenotypic Characterization and Prevalence of Carbapenemase-Producing Pseudomonas aeruginosa Isolates in Six Health Facilities in Cameroon" BioMed 3, no. 1: 77-88. https://doi.org/10.3390/biomed3010006
APA StyleDjuikoue, C. I., Djouela Djoulako, P. D., Same Njanjo, H. V., Kiyang, C. P., Djantou Biankeu, F. L., Guegang, C., Tchouotou, A. S. D., Wouambo, R. K., Thumamo Pokam, B. D., Apalata, T., & Jeannot, K. (2023). Phenotypic Characterization and Prevalence of Carbapenemase-Producing Pseudomonas aeruginosa Isolates in Six Health Facilities in Cameroon. BioMed, 3(1), 77-88. https://doi.org/10.3390/biomed3010006







