1. Introduction
Inborn errors of metabolism (IEMs) are a diverse group of genetic disorders resulting from defects in biochemical pathways critical for the synthesis, breakdown, or transport of essential metabolites or nutrients. Organic acidurias, also known as organic acidemias, represent a subset of IEMs characterized by the abnormal accumulation of organic acids (OAs) in urine and/or blood [
1,
2,
3]. These disorders can lead to a broad spectrum of symptoms, including metabolic acidosis, seizure, poor feeding, vomiting, developmental delay, irreversible neurological injury, and, in some cases, death [
4]. Although symptoms mostly present in the first few weeks of life, late-onset or milder forms may emerge during adolescence or adulthood.
Individually, organic acidurias are rare, with incidences ranging from 1 in 10,000 to over 1 in 1,000,000 live births. Collectively, however, their incidence is estimated to be 1 in 3000 live births [
5]. To date, more than 65 distinct organic acidurias have been described [
5], and the Society for the Study of Inborn Errors of Metabolism (SSIEM) classifies them into 19 major groups.
Chemically, OAs are non-amine-containing, low-molecular-weight molecules [
1] containing one or more carboxylic acid groups [
4]. They may be linear or branched with a varying number of carbon chain lengths and may carry a variety of functional groups such as keto, hydroxy, and phenyl groups.
Table 1 shows the basic nomenclature of common monocarboxylic and dicarboxylic acids with varying numbers of carbon atoms, and
Figure 1 illustrates a subset of common OAs in blood and urine
Physiologically, OAs are intermediates or end products of a large number of metabolic pathways, including those involved in the metabolism of amino acids, carbohydrates, vitamins, neurotransmitters, lipids, sterols, and nucleic acids [
1,
4]. Consequently, a detailed evaluation of the OA profile is necessary to identify the abnormal accumulation of one or more compounds due to enzymatic or transporter defects, supporting the diagnosis of not only organic acidurias but also a wide range of IEMs.
Importantly, while many organic acidurias are associated with irreversible damage and can be fatal if untreated, they can be effectively managed with relatively simple and cost-effective interventions such as dietary restrictions or supplements. As such, early diagnosis and prompt treatments are critical, and several organic acidurias have been incorporated into mandated newborn screening programs.
Urine is the preferred specimen for OA analysis because most OAs are hydrophilic and are rapidly excreted into urine. Urine organic acids (UOA) analysis evaluates hundreds of substances for patterns indicative of disrupted metabolic pathways [
1]. Paired with other clinical tests such as the plasma acylcarnitine test, plasma amino acid test, and molecular sequencing, UOA analysis plays a critical role not only in the differential diagnosis of positive newborn screening results but also in identifying organic acidurias and IEMs not captured by the current newborn screening programs.
Gas Chromatography-Mass Spectrometry (GC-MS) has served as the gold standard method for UOA analysis for several decades and continues to be widely used in most clinical laboratories. However, the inherent limitations, such as labor-intensive sample preparation and limited availability of instruments, have motivated the development of novel Liquid Chromatography-Mass Spectrometry (LC-MS) methods. These emerging approaches offer simplified workflows and reduced sample volume requirements while providing comparable analytical performance.
In this review, we provide a historical perspective on the emergence of UOA analysis for the diagnosis of IEMs, with a focus on urine organic acidurias. We review the principles and procedures of classical GC-MS methods and recent advancements in LC-MS methods, as well as targeted and untargeted applications for expanded diagnostic capabilities [
6].
2. Historical Milestones in Urine Organic Acid Testing
Before UOA testing became available, the diagnosis of IEMs relied primarily on clinical observation. In neonates and infants, symptoms such as persistent vomiting, metabolic acidosis, lethargy, unusual odors, and developmental delays often serve as the earliest clues to an underlying metabolic disorder. Alkaptonuria, considered the first described organic aciduria, was documented as early as 1858 when Bödeker isolated a compound he named “alkapton.” [
7]. This substance was later identified as homogentisic acid in 1891 and is known for causing urine to darken upon standing [
8]. In 1902, alkaptonuria was formally recognized as an IEM [
9]. At the time, diagnosis relied on qualitative chemical tests—such as reactions with ferric chloride and silver nitrate—to detect specific chemical properties of homogentisic acid [
9]. These classical approaches were labor-intensive, lacked specificity, and were susceptible to interference from other urinary compounds.
Following early chemical tests, more refined chromatographic and bioanalytical techniques emerged. For example, paper chromatography separates non-volatile OAs in urine based on their differential solubility in acidic and basic solvent systems [
10]. In the first dimension, the sample is run in an acidic solvent system, followed by a second perpendicular run in a basic solvent, allowing improved resolution of overlapping compounds [
10]. After chromatographic separation, the dried paper is visualized under UV light to detect fluorescent or light-absorbing spots and then treated with various spray reagents (such as ninhydrin, silver nitrate, and potassium ferrocyanide) to develop color reactions specific to different organic acids [
10]. The position of each spot is measured as an Rf value (the ratio of distance traveled by the compound to that of the solvent front) in both dimensions [
10]. These two Rf values, combined with characteristic color reactions, allow for the tentative identification of specific organic acids present in the urine sample [
10]. The major limitations of this method are its inability to support quantitative analysis, its restricted identification of only a limited range of OAs, and the complexity of the spray reagents required for detection.
A quantitative colorimetric method was first introduced in 1968, in which methylmalonic acid reacts with diazotized
p-nitroaniline to produce a green-colored compound measurable at 620 nm [
11]. The main advantage of this colorimetric approach is its speed. To enhance specificity, later adaptations involved measuring absorbance spectra across multiple wavelengths [
12]. However, interference from background absorbance due to other urinary components remained a challenge. To address this, researchers incorporated solvent extraction and ion-exchange chromatography [
13]. By 1979, a significant improvement was achieved through the extraction of the colored product into amyl alcohol, which greatly minimized nonspecific absorbance [
13]. In 1978, Auray-Blais and colleagues developed a micro–thin-layer chromatography (TLC) method for methylmalonic acid that overcame the interference problems often encountered with colorimetric assays [
14].
Certain OAs in urine can be detected using enzymatic assays. For instance, propionic acid can be measured through a reaction involving acetyl coenzyme A [
15]. In the same study, methylmalonic acid was indirectly quantified by first converting it to propionic acid, which was then measured using the same enzymatic approach [
15].
The development of GC-MS in the mid-20th century revolutionized the diagnosis of organic acidurias. The first application of GC-MS for UOA analysis was reported by Tanaka et al. in 1966, when they detected elevated isovaleric acid in a patient with isovaleryl-CoA dehydrogenase deficiency [
16]. Isovaleric acid was identified and confirmed by matching the retention time and mass spectra of the reference standard on two different GC columns [
16].
Recognizing the ability of mass spectrometry to detect a wide range of compounds, researchers aimed at addressing the need for a comprehensive screening method capable of analyzing all relevant urinary OAs in a single run. Initially, GC-MS screening of infant urine primarily focused on detecting hyperaminoacidurias caused by inborn errors of amino acid metabolism or defects in renal amino acid transport [
17]. Methylmalonic aciduria was the only organic aciduria routinely screened, and other organic acidurias were not included in these early screening programs. In 1980, Tanaka et al. developed a GC-MS method that enabled the analysis of multiple OAs in a single run and established retention indices for 155 metabolically important compounds, laying the foundation for efficient metabolic screening [
18]. More targeted methods began to be implemented in clinical settings in the late 1980s, specifically designed to routinely screen for a broader range of organic acidurias using automated, computerized GC-MS systems [
19,
20]. Reference levels, normalized to urinary creatinine concentration, were established for these analytes.
3. Urine Organic Acid Testing by GC-MS
Today, UOA analysis is a routine procedure in biochemical genetics laboratories, with GC-MS recognized as the gold standard. Among the 65 U.S. laboratories participating in the CAP Biochemical Genetics proficiency testing program, 64 reported using GC-MS for this purpose [
21]. To enable consistent comparison of results, the volume of urine tested is normalized to a fixed equivalent of creatinine to correct for the hydration status of the individual [
1]. Internal standards are added before liquid-liquid extraction (LLE) to compensate for sample variability and losses during sample preparation, while enriching target organic acids and removing interfering biological materials. Prior to LLE, samples are acidified to neutralize OAs, improving their partitioning into the organic solvents [
1]. To prepare compounds for GC-MS analysis, trimethylsilyl (TMS) derivatization converts reactive groups such as hydroxyl and carboxyl groups into more volatile TMS esters. After derivatization, samples are injected into the GC-MS system, where compounds are first separated in the gas chromatograph based on volatility and polarity. The mass spectrometer then ionizes the separated compounds, detecting the intact molecules and characteristic fragments, which generate unique patterns enabling accurate compound identification. Depending on the specific method, analytical run times for GC-MS methods can vary widely, ranging from as short as 15 min to as long as 60 min per injection (
Table 2).
3.1. Sample Handling and Pre-Analytical Considerations
Random urine samples are typically used where no additional patient preparation is required [
4,
23,
24]. The best specimens for the diagnosis of IEMs are those collected during acute illness or metabolic decompensation, where the UOA profile is most altered and therefore can be more easily detected [
1,
4,
24]. Although the required urine volume varies based on its creatinine concentration and sensitivity of the testing method, 2–10 mL is generally required by most reference laboratories [
4,
24]. Dilute urine specimens with lower creatinine concentrations can still be tested; however, analytical sensitivity will be compromised if extraction is not performed using larger urine volumes normalized to the regular amount of creatinine used for other samples [
1,
4].
Urine is collected in sterile, preservative-free containers. While short-term refrigeration may be acceptable for local transportation, urine specimens are best stored and transported frozen to avoid the loss of unstable volatile compounds. For long-term storage, temperatures of –70 to –80 °C are preferred, as storage at –20 °C may result in degradation of oxoacids [
25]. Additionally, freeze-thaw cycles should be minimized.
3.2. Internal Standard
Historically, GC-MS methods were often constrained by the limited availability and high cost of structurally appropriate isotope-labeled internal standards. In earlier workflows, internal standards were sometimes selected more for their availability than for their chemical similarity to target analytes. However, isotope-labeled internal standards have become more widely used due to improvements in commercial availability and lower costs of isotope production. Standards labeled with 13C and/or 15N are especially popular. They behave almost exactly like the native compounds during extraction and chromatography, which makes them appealing for accurate analysis.
Besides supporting accurate quantitative analysis, internal standards can also be used for other applications. Many laboratories add another internal standard just before derivatization—such as tetracosane—to track potential variations in derivatization and instrument responses. Additionally, responses from internal standards can be used to track the imprecision of the test over a long period of time. What’s more, responses from internal standards across samples from the same batch will detect unexpected losses, matrix effects, and instrument drift that might otherwise go unnoticed. For example, given the inconsistent extraction efficiency for orotic acid, isotope-labeled orotic acid has been used as a quality indicator to identify issues during sample preparation and instrument behavior [
4].
3.3. Extraction
Modern UOA extraction usually uses LLE with water-immiscible organic solvents. Before extraction, urine is acidified to convert organic acids into their protonated forms, which aids their partition into the organic layer. Salt, like sodium chloride, is often added to improve extraction efficiency by reducing the solubility of polar compounds in water—a process called “salting out”.
LLE separates compounds based on their differential partition in water versus organic solvents. Typically, 3 to 5 mL of ethyl acetate or diethyl ether is added per 1 mg/mL of creatinine in the urine [
1,
6,
24]. After mixing and centrifugation, the top organic layer is removed, dried, and prepared for derivatization. The residue can be reconstituted in a smaller volume to improve detection. Modified LLE steps, like repeating the extraction or using different solvents, may help recover more analytes.
LLE has been widely used for sample preparation, but it is time-consuming and requires large volumes of organic solvents, with challenges like emulsion formation and wet extracts. Earlier methods using organic solvents or anion-exchange columns gave only rough estimates, which often missed mild or partial disorders, especially in fluids other than urine. Today’s GC-MS methods use more refined extraction and isotope-labeled internal standards to improve accuracy [
4,
24]. Even so, interpretation remains complex due to the number of compounds detected, which is why other tests—like acylcarnitine or acylglycine analysis by tandem mass spectrometry—are often used complementarily.
Solid-phase extraction (SPE) has emerged as an alternative. It uses less solvent, avoids emulsions, and gives cleaner extracts. However, electing suitable eluents remains a challenge. Recent advances focus on miniaturizing extraction protocols, reducing sample and solvent volumes, and automating the process with robotic systems [
26].
3.4. Derivatization
In UOA analysis, derivatization is a key step that prepares polar, less volatile compounds for GC analysis. The derivatization process replaces labile hydrogen atoms—typically found in hydroxyl, carboxyl, or amine functional groups—with TMS groups, thereby increasing molecular weight and volatility. This conversion enables non-volatile organic acids to be vaporized and separated on the GC column. In a typical TMS derivatization procedure, the sample undergoes a heating step to promote the reaction. This usually involves incubation at temperatures ranging from 65 °C to 90 °C for a duration of 10 to 30 min [
1]. The heating facilitates the formation of stable TMS derivatives, which are more volatile and thermally stable.
The volatility and thermal stability of derivatized compounds depend significantly on the functional groups involved: O-trimethylsilyl derivatives are generally stable and amenable to GC-MS analysis, while N-trimethylsilyl derivatives (e.g., from amines and amino acids) are more prone to instability, potentially leading to inconsistent quantitation across samples [
4,
24,
27].
The derivatization reaction is commonly carried out using N,O-bis (trimethylsilyl) trifluoroacetamide (BSTFA), often in the presence of a solvent such as pyridine. These conditions are chosen to strike a balance between reaction completeness and the avoidance of over-derivatization, which can lead to the formation of unstable or interfering byproducts [
4].
Reaction temperature and duration must be constant to ensure reproducible derivatization patterns when comparing retention times and spectral fragments across runs. TMS derivatives also yield distinctive fragmentation patterns upon electron impact (EI) ionization in the mass spectrometer, which is critical for accurate compound identification. To ensure effective derivatization, the organic solvent extracts must be thoroughly dried prior to reagent addition, as incomplete drying could hinder the silylation reaction, reducing derivatization completeness and contributing to inconsistent UOA profiles.
3.5. Gas Chromatography–Mass Spectrometry Analysis
GC has undergone major technological advancements since the 1950s, transitioning from low-resolution packed columns to high-efficiency fused-silica capillary columns introduced in the late 1970s, which increased theoretical plate numbers from approximately 1500 to as high as 50,000—greatly enhancing resolution [
28]. Developments from the 1980s through the 2000s, including chemically selective stationary phases, electronically controlled pneumatics, and ultra-high-resolution columns, reduced co-elution, and enabled sharper peak separation as well as faster run times without compromising analytical performance [
29]. The increased resolving power of capillary columns also addressed the issue of unresolved components in urine and facilitated the structural elucidation of OAs via their unique retention times and mass spectra.
Derivatized compounds from the urine specimen are separated based on their volatility and interactions with the stationary phase of the GC column. These factors—such as boiling point, polarity, and molecular structure—collectively determine the retention time of each analyte. A standard GC system uses an inert carrier gas, commonly helium, to transport the sample from the heated injection port through the analytical column. The injector is usually held at a high constant temperature to promote complete vaporization of the derivatized analytes, while the column temperature is controlled through a programmed gradient to facilitate sequential elution of compounds. Gradual increases in column temperature help to separate analytes that may otherwise co-elute, especially those with similar physical properties.
The chromatographic column itself is generally non-polar, often composed of methyl- or phenyl-substituted polysiloxanes. Common column configurations include 5% phenyl methylpolysiloxane stationary phases, such as HP-5ms or Ultra-2, which offer a balance between resolution and thermal stability. Column length, internal diameter, and film thickness are selected based on instrument capabilities and analytical priorities. Sample injections are commonly performed in split mode to manage the amount of material entering the column. Split ratios ranging from 1:15 to 1:70 are frequently used to avoid overloading, which can compromise peak shape and reproducibility. Injection volumes are typically in the range of 1–3 µL. Following separation in the GC column, TMS-derivatized compounds enter the mass spectrometer, where they are ionized with EI ionization mode and generate predictable fragments. As discussed earlier, these patterns are highly specific to molecular structure and provide key data for metabolite identification. Detection is often performed using single quadrupole or ion trap mass spectrometers operating in full scan mode, covering a mass-to-charge (m/z) range of approximately 50–550.
The total runtime of a GC-MS method can influence both throughput and analytical performance. Longer runs typically allow for improved resolution and more reliable metabolite identification, while abbreviated programs provide shorter turnaround times but may risk peak overlapping and difficulties in result interpretation.
3.6. Interpretation and Reporting
The interpretation of UOA profiles is a complex and nuanced task that demands significant analytical expertise, biochemical knowledge, and clinical correlation. Providing a clinically meaningful interpretation is one of the most intellectually challenging activities in the biochemical genetics laboratory. Several hundred compounds are excreted in the urine at various concentrations, and the elevation of diagnostic biomarkers could be only marginal [
3]. Consequently, specialized and focused training and hands-on experiences are critical. A technical standard for laboratory analysis of OA has been provided by organizations such as the American College of Medical Genetics and Genomics (ACMG) [
1].
3.6.1. Analytical Expertise
A thorough understanding of the analytical performance of the testing method is essential for the interpretation of UOA profiles. As GC-MS methods have been widely used for the past few decades, a substantial body of knowledge has been established regarding best practices, capabilities, and limitations [
1].
Accurate identification of chromatographic peaks depends on matching mass spectral fragments of TMS-derivatized OAs and their retention times to reputable libraries (e.g., NIST or Wiley) or self-developed databases [
1,
22]. A customized report is often generated to list all identified substances exceeding a pre-defined intensity threshold [
3]. While commercial software can suggest the most likely identity of a chromatographic peak and provide a confidence score, this automated process may be erroneous due to ions from co-eluted substances. Therefore, peak identification is best performed by a properly trained individual under the supervision of a qualified laboratory director. When necessary, the presence of diagnostic biomarkers should be confirmed by visual inspection of the spectra.
Interpretation of UOA profiles starts with reviewing the total ion chromatogram (TIC) and assessing results from quality indicators. Successful sample preparation and data acquisition can be confirmed by reviewing the intensities from internal standards. Recognition of UOA profiles from healthy individuals is the first step before abnormal patterns can be identified. In urine from healthy individuals, a group of major peaks representing the most abundant substances (e.g., urea, citric acid, hippuric acid, succinic acid) can typically be easily identified [
23]. Many of the compounds have multiple TMS derivatives and appear as multiple chromatographic peaks.
Quantitation can be accomplished by building calibration curves for each compound of interest. However, setting up calibration curves for all diagnostic biomarkers is not practical, and some of the diagnostic biomarkers are not even commercially available. Additionally, absolute quantitation offers limited added value in most scenarios. For these reasons, qualitative methods or semi-quantitative methods remain the standard practice in most laboratories.
Significant interlaboratory variability exists in UOA testing due to differences in sample preparation, extraction, use of oximation, and instrumentation. Consequently, each laboratory should independently establish reference intervals and understand the analytical limitations of its testing method. For example, the separation of the six isomers of hydroxybutyric acid is inherently challenging due to their similar physicochemical properties. Additionally, measurement of orotic acid could be hindered by its low extraction efficiency and co-eluting cis-aconitic acid at much higher intensities [
1,
3,
30]. If needed, orotic acid should be measured by its dedicated clinical test. A list of possibly overlapping peaks has been published [
3]. Lastly, D- and L-2-hydroxyglutaric acid cannot be separated or distinguished from each other.
While some urine organic acidurias and IEMs present with significantly elevated single diagnostic biomarkers, others may display only subtle elevations (e.g., mevalonate kinase deficiency and glutaric acidemia type 1) [
30]. In such cases, the analytical sensitivity of the method must be adequately validated to be sufficient before the diagnosis of those disorders can be supported.
3.6.2. Biochemical Knowledge
A thorough understanding of metabolic pathways is indispensable for interpreting complex UOA profiles. In IEMs, abnormally elevated primary metabolites may be further metabolized through alternative pathways, leading to the accumulation of secondary metabolites that are also diagnostically informative. Recognizing these patterns is essential to identifying disrupted metabolic pathways and pinpointing enzyme defects.
For example, in glutaric acidemia type 1, defects in glutaryl-CoA dehydrogenase lead to significantly accumulated glutaryl-CoA, which subsequently gives rise to elevated glutaric acid and glutarylcarnitine [
31]. Similarly, defects in propionyl-CoA carboxylase lead to significantly accumulated propionyl-CoA, which subsequently gives rise to elevated propionylcarnitine and propionylglycine. Additionally, excess propionyl-CoA can also react with oxaloacetate to produce 2-methylcitric acid, a hallmark biomarker for both propionic acidemia and methylmalonic aciduria [
31]. Elevated propionyl-CoA may also inhibit the glycine cleavage pathway and urea cycle, leading to elevated glycine and ammonia [
32,
33]. These examples underscore the complexity in interpreting UOA profiles, which reflect the crosstalk among interconnected metabolic pathways.
Interpretation of UOA profiles should always be integrated with results from other diagnostic tests, including plasma amino acids, plasma acylcarnitines, ammonia, lactate, and pyruvate, and molecular sequencing. Since many OAs are intermediates of amino acid metabolism, defects in these pathways typically result in the concurrent elevation of both corresponding amino acids and OAs. Additionally, OA coenzyme A esters may be conjugated with carnitine to regenerate free coenzyme A. Many OAs are also conjugated with glycine to form acylglycine for rapid excretion. Therefore, patterns of acylglycines from UOA analysis and acylcarnitines from plasma acylcarnitine analysis are also valuable tools for the interpretation of OAU profiles [
5].
Diagnostic biomarkers for organic acidurias, as classified by the Study of Inborn Errors of Metabolism (SSIEM), are shown in
Table 3 (Data from [
34]).
3.6.3. Clinical Correlation
Many OAs are elevated for reasons unrelated to IEMs, including dietary intake, gut microbial metabolism, medications, and environmental exposures. These confounding factors underscore the importance of clinical correlation to avoid misinterpretation, incorrect diagnosis, and unnecessary workup.
For example, adipic acid, suberic acid, and sebacic acid are elevated in medium-chain acyl-CoA dehydrogenase (MCAD) deficiency, but their elevations are also observed in individuals receiving medium-chain triglyceride (MCT) oil supplementation. In such cases, the relative concentrations of these dicarboxylic acids typically follow a characteristic pattern, with adipic acid being the lowest and sebacic acid being the highest. Similarly, methylmalonic acid is a hallmark of methylmalonic aciduria, but is also elevated in vitamin B
12 deficiency or because of bacterial metabolism in the intestine. Other metabolites may appear in the urine due to the metabolism of medications. Pyroglutamic acid (5-oxoproline), for instance, is a known biomarker of glutathione synthetase deficiency, but may also be elevated in patients treated with acetaminophen and vigabatrin [
1]. Valproic acid treatment inhibits 3-methylcrotonyl-CoA carboxylase and leads to the elevation of 3-hydroxyisovalerate, a compound that also accumulates in isovaleric acidemia. Glycerol, being a biomarker for glycerol kinase deficiency, may also appear in urine specimens from infants. Elevated medium- and long-chain monocarboxylic fatty acids may come from skin products [
1]. A comprehensive list of nutritional, iatrogenic, and artifactual sources of urinary OAs has been summarized [
6].
3.6.4. Reporting
As recommended in the most recent technical standard by ACMG, the report of UOA analysis should be clinically meaningful and should be clear to a nonspecialist. When no clinically significant abnormalities are identified, the report may include qualitative terms only. However, if the UOA profile is abnormal, the report should cover the potential clinical significance, correlation with clinical information, differential diagnosis, recommendations for additional studies, and contact information of the reporting laboratories to enable direct communications in the future [
1].
4. Emerging LC-MS Technologies for Urine Organic Acid Testing
Although GC-MS methods have been widely used for UOA analysis for the past few decades, they require labor-intensive, multi-step sample preparation procedures (i.e., LLE, TMS derivatization). Additionally, GC-MS instrumentation is not widely available in clinical laboratories due to limited clinical applications. In contrast, LC-MS instrumentation has become increasingly accessible in medical centers and reference laboratories, enabling the development of novel methods for UOA analysis.
UOA analysis using LC is challenging due to poor retention on conventional reverse-phase columns. In recent years, with improvements in both column chemistry and analytical sensitivity of modern triple quadrupole and Quadrupole Time-of-Flight (QTOF) mass spectrometers, direct LC-MS analysis of UOAs without derivatization has become feasible. This new development has led to simplified sample preparation and shorter turnaround times [
35]. Additionally, a broader metabolite coverage can be accomplished to support the diagnosis of additional IEMs, as compounds with poor recovery from LLE as well as those incompatible with TMS derivatization and GC-MS may now be reliably detected by LC-MS. Moreover, LC-MS methods support the analysis of significantly smaller urine volumes. While conventional GC-MS methods typically require approximately 2 mL of urine, LC-MS methods only require a few tens of μL, making them more suitable for neonates and infants [
36]. The key analytical features, advantages, and limitations of GC-MS, LC-MS/MS, and LC-QTOF methodologies are summarized in
Table 4.
The interpretation of UOA profiles generated by LC-MS platforms generally follows the same principles as those used for GC-MS platforms; however, platform-specific differences in analytical performance characteristics must be taken into account.
4.1. Targeted LC-MS/MS Methods
Targeted LC-MS/MS approaches for UOA analysis have been demonstrated to offer satisfactory analytical performance and clinical utility. Barbora Piskláková et al. from the University Hospital Olomouc developed an LC-MS/MS method that measures 99 OAs, 15 acylglycines, as well as 32 acylcarnitines in 26 min. Each biomarker is identified by its retention time and characteristic multiple reaction monitoring (MRM) transitions [
36]. Data acquisition was performed in both positive and negative electrospray ionization (ESI) modes using polarity switching. Chromatographic separation of many isomers was accomplished with an Acquity HSS T3 column. Sample preparation involves mixing 100 μL of creatinine-normalized urine with 10 μL of an internal standard solution with 6 isotope-labeled compounds. Despite some matrix effects, limits of quantification (LOQs) less than 0.5 mmol/mol creatinine were achieved for most analytes. This method was clinically and analytically validated with a total of 34 IEMs using leftover patient samples and external proficiency testing materials, and subsequently replaced their conventional GC-MS method.
In terms of chromatographic performance, the Acquity HSS T3 column comes with its inherent strengths and limitations. For example, it can separate 3-hydroxyglutaric acid from 2-hydroxyglutaric acid, which can be challenging using GC-MS methods [
36,
37]. However, as with GC-MS methods, L- and D-2-hydroxyglutaric remain chromatographically indistinguishable. Additionally, peak shapes for succinylacetone and oxoacids are not ideal using this column [
36].
4.2. High-Resolution LC-QTOF Methods
Similar to GC-MS methods, UOA profiling using high-resolution LC-QTOF instruments supports both targeted and untargeted analyses. Each biomarker is identified by its retention time and high-resolution mass-to-charge ratio. Researchers from Maastricht University Medical Center and Radboud University Medical Center pioneered UPLC-QTOF methods for the diagnosis of organic aciduria and other IEMs. A 35 min method developed by Irene M. L. W. Körver-Keularts et al. quantifies 71 diagnostic biomarkers in negative ESI mode, covering disorders classically identified by GC-MS methods [
38]. A ‘dilute-and-shoot’ strategy was used for sample preparation, involving diluting 25 μL of creatinine-normalized urine with 350 μL of mobile phase and 25 μL of an internal standard solution with 19 isotope-labeled compounds. Although the urine specimen was diluted more than 15 times, LOQs < 3 μM were still achieved for most biomarkers, and clinical validation demonstrated successful identification of 32 IEMs. While matrix effects were found for 4-hydroxybutyric acid and pyroglutaric acid, diagnostic accuracy was not compromised.
Building on this work, the method was further expanded to a more comprehensive platform known as “targeted urine metabolomics (TUM)”. Using both positive and negative ESI modes, the expanded method covers 258 diagnostic biomarkers, including additional classes of diagnostic biomarkers such as acylcarnitines. Clinical validation included 289 samples across 78 IEMs, with all tested organic acidurias successfully identified.
Unfortunately, high-resolution LC-QTOF instruments are less commonly found in clinical laboratories. However, compared with GC-MS instruments, high-resolution LC-QTOF instruments can support a larger variety of clinical applications. Moving forward, LC-QTOF instruments may become exceedingly appealing for chemistry and biochemical genetics laboratories [
39].
4.3. Emerging Data Analysis Tools for LC-MS Platforms
Mass spectrometry has proven to be a sufficiently stable analytical instrument. Targeted metabolite profiling of a comprehensive panel could easily quantify more than 50 biomarkers, while untargeted metabolite screening could identify as many as 10,000 features (i.e., signals with a specific mass-to-charge ratio, intensity, and retention time) per sample [
39]. This complexity increases the risk of interpretation mistakes, transcription errors, and user fatigue. To address these challenges, automated and streamlined data analysis tools have been developed to facilitate quick and easier diagnosis of organic acidurias and other IEMs.
4.3.1. Alternatives to Traditional Reference Intervals
Conventional interpretation of UOA profiles relies on age-grouped reference intervals. However, such an approach does not capture the extent of metabolic abnormality or the variation within the healthy population. To address this, statistical approaches utilizing z-score (based on population means and standard deviations) and robust standardized (RS) values (based on medians and interquartile ranges) have been adopted.
4.3.2. XCMS for Untargeted Metabolite Screening
XCMS is an open-source software package in R, widely used to process and analyze complex untargeted LC-MS metabolomics data [
40]. It can identify features with significantly altered responses in test versus control datasets by (1) identifying all features in each dataset based on pre-defined parameters; (2) matching and grouping features across datasets and correcting for minor shifts in retention times; and (3) performing statistical analysis. This approach eliminates the need for calibration curves. XCMS has been used to analyze LC-QTOF data from plasma, from which more than 10,000 features were identified [
39,
40]. With this approach, abnormally accumulated biomarkers were identified in all organic acidurias tested, as well as 42 out of 46 IEMs.
However, analysis using XCMS has a few limitations. First of all, XCMS can easily identify hundreds of features that are significantly altered, but most of those features may just represent differences between the test and control groups and carry no diagnostic value. To overcome this, it is best to introduce variations to both test and control groups. For example, multiple specimens can be collected from the same patient at different times of the day. However, this is generally not feasible in urgent clinical settings. Secondly, poorly shaped peaks could also be identified as distinct features. Lastly, minor elevation of diagnostic markers may not be identified and reported. Among the four IEMs that were missed, diagnostic markers for three (guanidinoacetate methyltransferase (GAMT) deficiency, argininosuccinic aciduria, dimethylglycine dehydrogenase deficiency) were significantly altered but filtered out by XCMS [
39].
4.3.3. Bioinformatics Pipelines and Visualization Tools
Rather than stacking z-scores and RS values from more than 100 diagnostic biomarkers, various strategies have been implemented to guide the interpreter to possible organic acidurias and IEMs.
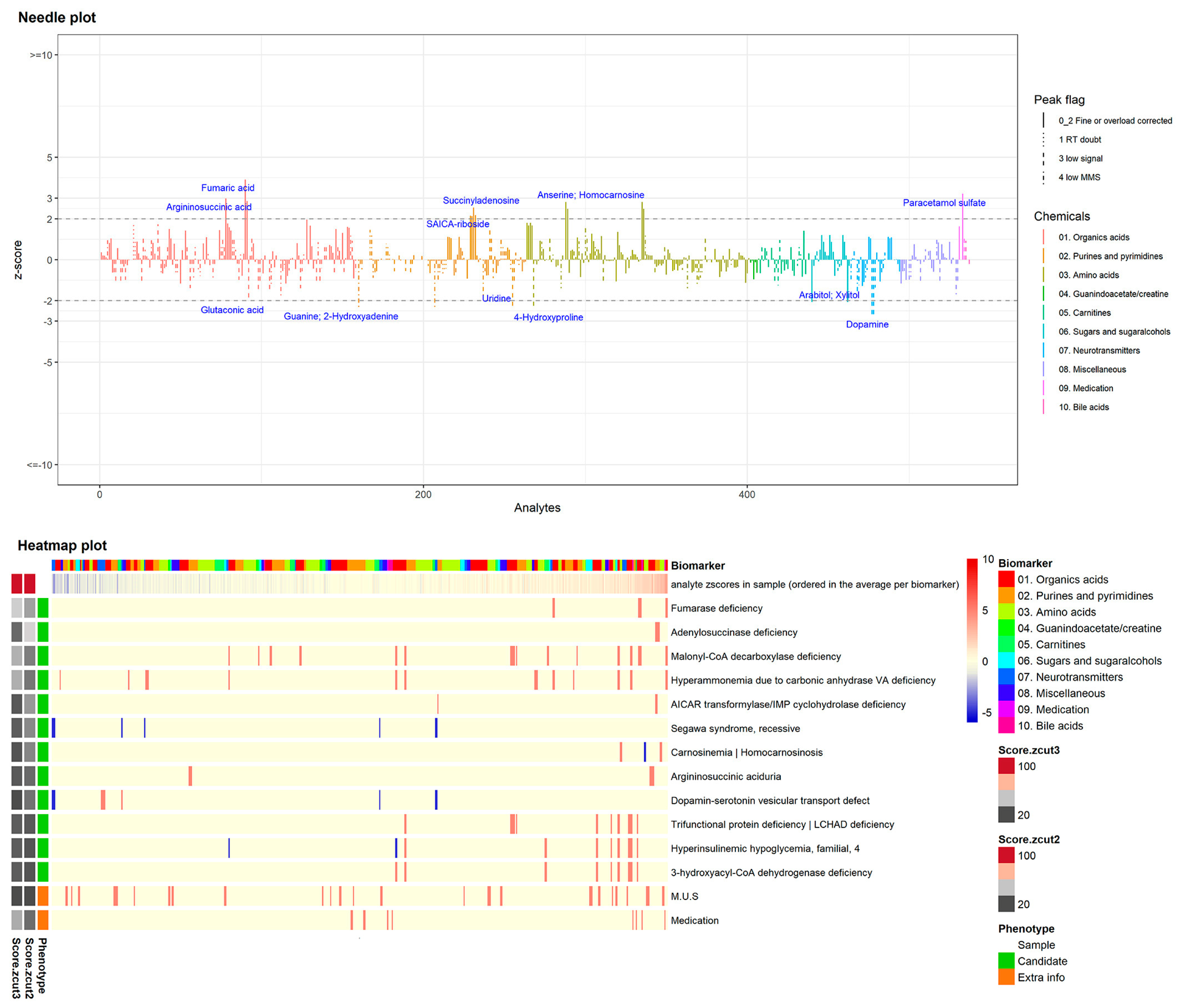
Laura K.M. Steinbusch et al. implemented a novel and automated workflow combining z-score calculation, peak quality assessment, and graphical visualization via a needle plot to highlight the significantly altered biomarkers. Additionally, the z-score profile is automatically matched to the top 10 most likely IEMs based on information from IEMbase (
Figure 2) [
41].
Similarly, Barbora Piskláková et al. created a metabolic network for over 80 IEMs in the Cytoscape software [
36]. RS values from patient specimens are mapped onto the network, providing a graphical representation of the disrupted metabolic pathway.
5. Summary and Future Perspectives
UOA testing has undergone significant evolution over the past century, from chemical tests to two-dimensional paper chromatography, then to modern MS-based methods that simultaneously measure hundreds of compounds. The introduction of GC-MS in the mid-20th century revolutionized the diagnosis of organic acidurias by enabling sensitive and multiplex analysis for a broad range of OAs and other biomarkers. Despite the labor-intensive sample preparation procedure, GC-MS remains the gold standard.
Recent advances in LC-MS platforms, including both LC-MS/MS and high-resolution LC-QTOF, have enabled simplified, derivatization-free workflows with reduced sample volume requirements and broader metabolite coverage. These methods now rival GC-MS in analytical performance, while offering expanded diagnostic capabilities across a broader array of IEMs. To better manage the increasing complexity of data from these platforms, new bioinformatics tools and automated workflows have been developed to visualize disrupted metabolic networks.
Looking ahead, the implementation of LC-MS platforms and automated and novel bioinformatics tools may reshape the landscape of IEM testing.