Superinfections in Hospitalized COVID-19 Patients (Super COVID-19): Data from the Multicentric Retrospective CH-SUR Cohort Study in Switzerland
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Setting, Design and Population
2.2. Outcome Measures
2.3. Data Collection
2.4. Data Analysis
2.5. Definitions
2.6. Statistical Analysis
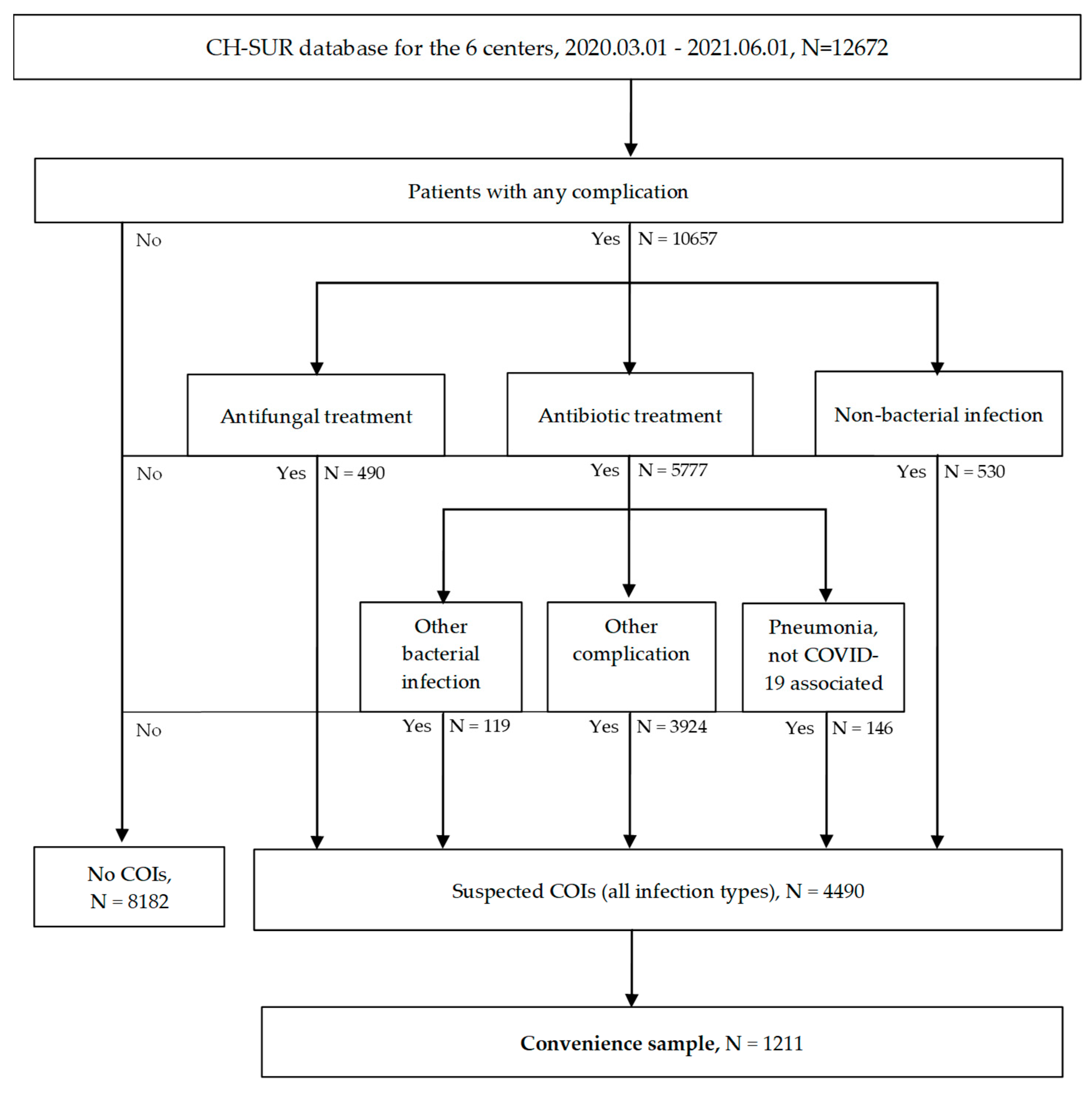
3. Results
3.1. Comparison Between Suspected COI and No COI Groups at CH-SUR Level
3.2. Survival Analysis from CH-SUR
3.3. Convenience Sample Data: Type of Infection and Microbial Spectrum
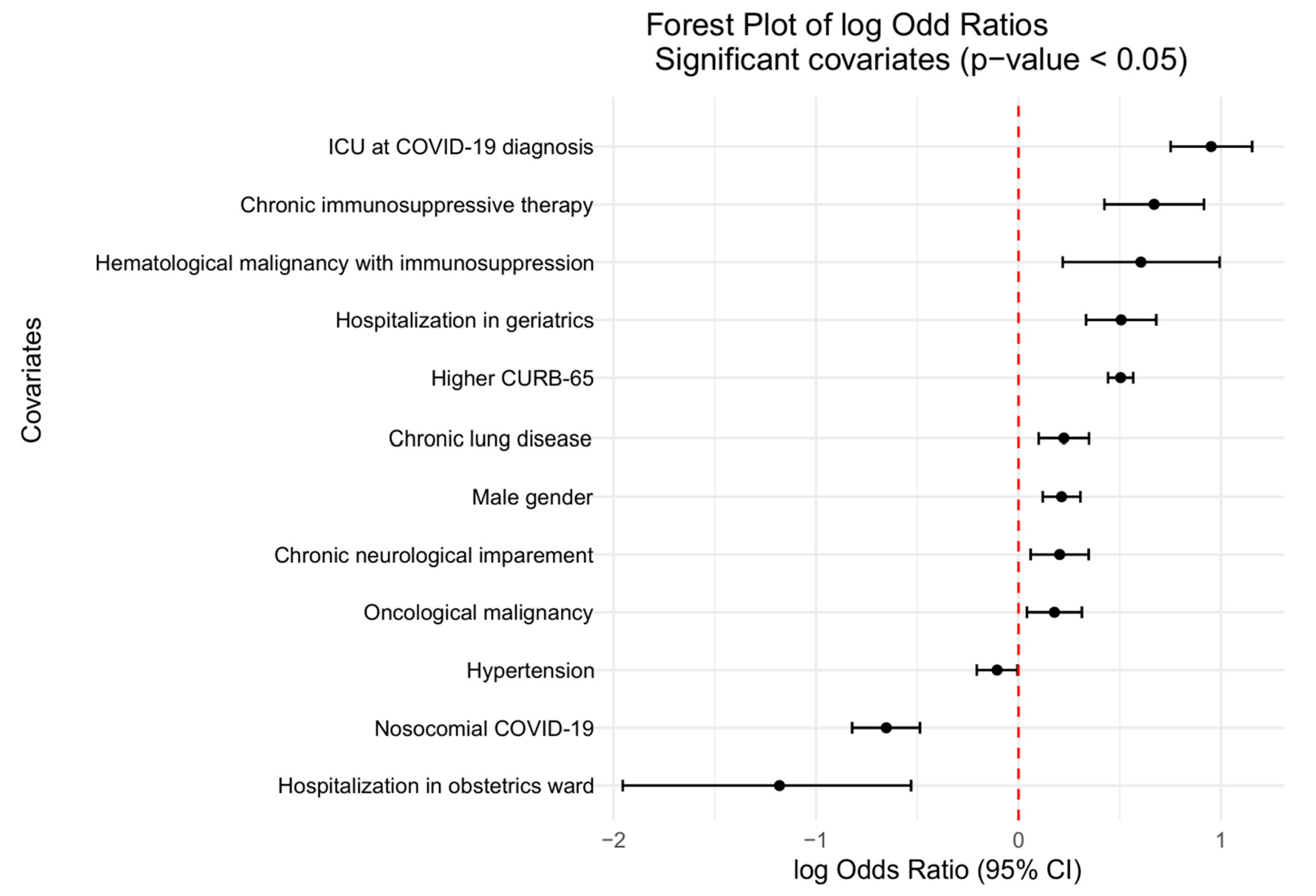
3.4. Risk Factors for COIs
3.5. Antibiotic Use
3.6. Immunomodulatory Treatment for COVID-19
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| COVID-19 | Coronavirus disease 2019 |
| COI | Coinfection |
| CH-SUR | COVID-19 Hospital-Based Surveillance |
| BSI | Bloodstream infection |
| UTI | Urinary tract infection |
| ICU | Intensive care unit |
| IFI | Invasive fungal disease |
| CAPA | COVID-19-associated aspergillosis |
| PCR | Polymerase chain reaction |
| ICU-LOS | Intensive care unit length of stay |
| LOS | Length of stay |
| BMI | Body mass index |
| HIV | Human immunodeficiency virus |
| ARDS | Acute respiratory distress syndrome |
| LTCF | Long-term care facility |
| ESBL | Extended-spectrum beta lactamase |
| ANOVA | Analysis of variance |
| GLM | Generalized linear model |
| ANRESIS | Swiss Centre for Antibiotic Resistance |
| BAL | Bronchoalveolar lavage |
| GM | Galactomannan |
| CFU | Colony-forming unit |
References
- Morens, D.M.; Taubenberger, J.K.; Fauci, A.S. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: Implications for pandemic influenza preparedness. J. Infect. Dis. 2008, 198, 962–970. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Calderon, M.; Gysin, G.; Gujjar, A.; McMaster, A.; King, L.; Comandé, D.; Hunter, E.; Payne, B. Bacterial co-infection and antibiotic stewardship in patients with COVID-19: A systematic review and meta-analysis. BMC Infect. Dis. 2023, 23, 14. [Google Scholar] [CrossRef] [PubMed]
- Langford, B.J.; So, M.; Simeonova, M.; Leung, V.; Lo, J.; Kan, T.; Raybardhan, S.; Sapin, M.E.; Mponponsuo, K.; Farrell, A.; et al. Antimicrobial resistance in patients with COVID-19: A systematic review and meta-analysis. Lancet Microbe 2023, 4, e179–e191. [Google Scholar] [CrossRef]
- Garcia-Vidal, C.; Sanjuan, G.; Moreno-García, E.; Puerta-Alcalde, P.; Garcia-Pouton, N.; Chumbita, M.; Fernandez-Pittol, M.; Pitart, C.; Inciarte, A.; Bodro, M.; et al. Incidence of co-infections and superinfections in hospitalized patients with COVID-19: A retrospective cohort study. Clin. Microbiol. Infect. 2021, 27, 83–88. [Google Scholar] [CrossRef]
- Kubin, C.J.; McConville, T.H.; Dietz, D.; Zucker, J.; May, M.; Nelson, B.; Istorico, E.; Bartram, L.; Small-Saunders, J.; Sobieszczyk, M.E.; et al. Characterization of Bacterial and Fungal Infections in Hospitalized Patients With Coronavirus Disease 2019 and Factors Associated With Health Care-Associated Infections. Open Forum Infect. Dis. 2021, 8, ofab201. [Google Scholar] [CrossRef]
- Hoenigl, M.; Seidel, D.; Sprute, R.; Cunha, C.; Oliverio, M.; Goldman, G.H.; Ibrahim, A.S.; Carvalho, A. COVID-19-associated fungal infections. Nat. Microbiol. 2022, 7, 1127–1140. [Google Scholar] [CrossRef]
- Singh, S.; Verma, N.; Kanaujia, R.; Chakrabarti, A.; Rudramurthy, S.M. Mortality in critically ill patients with coronavirus disease 2019-associated pulmonary aspergillosis: A systematic review and meta-analysis. Mycoses 2021, 64, 1015–1027. [Google Scholar] [CrossRef]
- Søgaard, K.K.; Baettig, V.; Osthoff, M.; Marsch, S.; Leuzinger, K.; Schweitzer, M.; Meier, J.; Bassetti, S.; Bingisser, R.; Nickel, C.H.; et al. Community-acquired and hospital-acquired respiratory tract infection and bloodstream infection in patients hospitalized with COVID-19 pneumonia. J. Intensive Care 2021, 9, 10. [Google Scholar] [CrossRef]
- Thiabaud, A.; Iten, A.; Balmelli, C.; Senn, L.; Troillet, N.; Widmer, A.; Flury, D.; Schreiber, P.W.; Vázquez, M.; Damonti, L.; et al. Cohort profile: SARS-CoV-2/COVID-19 hospitalised patients in Switzerland. Swiss Med. Wkly. 2021, 151, w20475. [Google Scholar] [CrossRef]
- Von Elm, E.; Altman, D.G.; Egger, M.; Pocock, S.J.; Gøtzsche, P.C.; Vandenbroucke, J.P. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: Guidelines for reporting observational studies. Lancet 2007, 370, 1453–1457. [Google Scholar] [CrossRef] [PubMed]
- Bundesamt für Gesundheit, (BAG). Neues Coronavirus (COVID-19). In Verdachts-, Beprobungs- und Meldekriterien vom 01.04.2022. 2022. Available online: https://www.bag.admin.ch/dam/bag/de/dokumente/mt/msys/covid-19-verdachts-beprobungs-meldekriterien.pdf.download.pdf/BAG_Verdachts_Beprobungs_und_Meldekriterien.pdf (accessed on 18 May 2025).
- Koehler, P.; Bassetti, M.; Chakrabarti, A.; Chen, S.C.A.; Colombo, A.L.; Hoenigl, M.; Klimko, N.; Lass-Flörl, C.; Oladele, R.O.; Vinh, D.C.; et al. Defining and managing COVID-19-associated pulmonary aspergillosis: The 2020 ECMM/ISHAM consensus criteria for research and clinical guidance. Lancet Infect. Dis. 2021, 21, e149–e162. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control, (ECDC). Point prevalence survey of healthcare-associated infections and antimicrobial use in European acute care hospitals. In Protocol Version 5.3. 2016. Available online: https://www.ecdc.europa.eu/sites/default/files/media/en/publications/Publications/PPS-HAI-antimicrobial-use-EU-acute-care-hospitals-V5-3.pdf (accessed on 18 May 2025).
- Langford, B.J.; So, M.; Raybardhan, S.; Leung, V.; Soucy, J.R.; Westwood, D.; Daneman, N.; MacFadden, D.R. Antibiotic prescribing in patients with COVID-19: Rapid review and meta-analysis. Clin. Microbiol. Infect. 2021, 27, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Slim, M.A.; Appelman, B.; Peters-Sengers, H.; Dongelmans, D.A.; de Keizer, N.F.; Schade, R.P.; de Boer, M.G.J.; Müller, M.C.A.; Vlaar, A.P.J.; Wiersinga, W.J.; et al. Real-world Evidence of the Effects of Novel Treatments for COVID-19 on Mortality: A Nationwide Comparative Cohort Study of Hospitalized Patients in the First, Second, Third, and Fourth Waves in the Netherlands. Open Forum Infect. Dis. 2022, 9, ofac632. [Google Scholar] [CrossRef]
- University of Bern; Institute for Infectious Diseases. Anresis.ch–Sentinel Surveillance of Antibiotic Resistance in Switzerland. Available online: https://www.anresis.ch/ (accessed on 1 January 2025).
- Westblade, L.F.; Simon, M.S.; Satlin, M.J. Bacterial Coinfections in Coronavirus Disease 2019. Trends Microbiol. 2021, 29, 930–941. [Google Scholar] [CrossRef]
- Jones, R.N. Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin. Infect. Dis. 2010, 51 (Suppl. S1), S81–S87. [Google Scholar] [CrossRef]
- Khan, S.; Hasan, S.S.; Bond, S.E.; Conway, B.R.; Aldeyab, M.A. Antimicrobial consumption in patients with COVID-19: A systematic review and meta-analysis. Expert. Rev. Anti Infect. Ther. 2022, 20, 749–772. [Google Scholar] [CrossRef]
- Friedli, O.; Gasser, M.; Cusini, A.; Fulchini, R.; Vuichard-Gysin, D.; Halder Tobler, R.; Wassilew, N.; Plüss-Suard, C.; Kronenberg, A. Impact of the COVID-19 Pandemic on Inpatient Antibiotic Consumption in Switzerland. Antibiotics 2022, 11, 792. [Google Scholar] [CrossRef]
- Duan, Y.; Wang, J.; Wang, S.; Zhang, R.; Hu, J.; Li, W.; Chen, B. Risk factors, outcomes, and epidemiological and etiological study of hospitalized COVID-19 patients with bacterial co-infection and secondary infections. Eur. J. Clin. Microbiol. Infect. Dis. 2024, 43, 577–586. [Google Scholar] [CrossRef]
- Falcone, M.; Tiseo, G.; Giordano, C.; Leonildi, A.; Menichini, M.; Vecchione, A.; Pistello, M.; Guarracino, F.; Ghiadoni, L.; Forfori, F.; et al. Predictors of hospital-acquired bacterial and fungal superinfections in COVID-19: A prospective observational study. J. Antimicrob. Chemother. 2021, 76, 1078–1084. [Google Scholar] [CrossRef]
- Musuuza, J.S.; Watson, L.; Parmasad, V.; Putman-Buehler, N.; Christensen, L.; Safdar, N. Prevalence and outcomes of co-infection and superinfection with SARS-CoV-2 and other pathogens: A systematic review and meta-analysis. PLoS ONE 2021, 16, e0251170. [Google Scholar] [CrossRef] [PubMed]
- Murray, H.C.; Muleme, M.; Cooper, D.; McNamara, B.J.; Hussain, M.A.; Bartolo, C.; O’Brien, D.P.; Athan, E. Prevalence, risk factors, and outcomes of secondary infections among hospitalized patients with COVID-19 or post-COVID-19 conditions in Victoria, 2020–2023. Int. J. Infect. Dis. 2024, 145, 107078. [Google Scholar] [CrossRef] [PubMed]
- López-Herrero, R.; Sánchez-de Prada, L.; Tamayo-Velasco, A.; Lorenzo-López, M.; Gómez-Pesquera, E.; Sánchez-Quirós, B.; de la Varga-Martínez, O.; Gómez-Sánchez, E.; Resino, S.; Tamayo, E.; et al. Epidemiology of bacterial co-infections and risk factors in COVID-19-hospitalized patients in Spain: A nationwide study. Eur. J. Public Health 2023, 33, 675–681. [Google Scholar] [CrossRef] [PubMed]
- Santus, P.; Danzo, F.; Signorello, J.C.; Rizzo, A.; Gori, A.; Antinori, S.; Gismondo, M.R.; Brambilla, A.M.; Contoli, M.; Rizzardini, G.; et al. Burden and Risk Factors for Coinfections in Patients with a Viral Respiratory Tract Infection. Pathogens 2024, 13, 993. [Google Scholar] [CrossRef]
- Ripa, M.; Galli, L.; Poli, A.; Oltolini, C.; Spagnuolo, V.; Mastrangelo, A.; Muccini, C.; Monti, G.; De Luca, G.; Landoni, G.; et al. Secondary infections in patients hospitalized with COVID-19: Incidence and predictive factors. Clin. Microbiol. Infect. 2021, 27, 451–457. [Google Scholar] [CrossRef]
- Gudiol, C.; Durà-Miralles, X.; Aguilar-Company, J.; Hernández-Jiménez, P.; Martínez-Cutillas, M.; Fernandez-Avilés, F.; Machado, M.; Vázquez, L.; Martín-Dávila, P.; de Castro, N.; et al. Co-infections and superinfections complicating COVID-19 in cancer patients: A multicentre, international study. J. Infect. 2021, 83, 306–313. [Google Scholar] [CrossRef]
- CDC–U.S. Centers For Disease Control And Prevention. Underlying Conditions and the Higher Risk for Severe COVID-19. Available online: https://www.cdc.gov/covid/hcp/clinical-care/underlying-conditions.html (accessed on 20 December 2024).
- Bardi, T.; Pintado, V.; Gomez-Rojo, M.; Escudero-Sanchez, R.; Azzam Lopez, A.; Diez-Remesal, Y.; Martinez Castro, N.; Ruiz-Garbajosa, P.; Pestaña, D. Nosocomial infections associated to COVID-19 in the intensive care unit: Clinical characteristics and outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 495–502. [Google Scholar] [CrossRef]
- Bartoletti, M.; Pascale, R.; Cricca, M.; Rinaldi, M.; Maccaro, A.; Bussini, L.; Fornaro, G.; Tonetti, T.; Pizzilli, G.; Francalanci, E.; et al. Epidemiology of Invasive Pulmonary Aspergillosis Among Intubated Patients With COVID-19: A Prospective Study. Clin. Infect. Dis. 2021, 73, e3606–e3614. [Google Scholar] [CrossRef]
- Fekkar, A.; Lampros, A.; Mayaux, J.; Poignon, C.; Demeret, S.; Constantin, J.M.; Marcelin, A.G.; Monsel, A.; Luyt, C.E.; Blaize, M. Occurrence of Invasive Pulmonary Fungal Infections in Patients with Severe COVID-19 Admitted to the ICU. Am. J. Respir. Crit. Care Med. 2021, 203, 307–317. [Google Scholar] [CrossRef]

| General Information | Total | No COI 1 | Suspected COI 1 | p-Value 2 |
|---|---|---|---|---|
| No. of hospitalizations | 13,265 | 8406 | 4859 | |
| Readmission | 13,265 | <0.001 | ||
| First admission | 7970 (97.4%) | 4159 (92.6%) | ||
| Readmission | 212 (2.6%) | 331 (7.4%) | ||
| Demographic characteristics | ||||
| Age | 13,265 | 69 (56,80) | 73 (62, 82) | <0.001 |
| Male gender | 13,265 | 4623 (55.0%) | 2999 (61.7%) | <0.001 |
| BMI (kg/m2) | 10,723 | 26.9 (23.7, 30.7) | 26.6 (23.5, 30.8) | 0.4 |
| Comorbidities | ||||
| Chronic respiratory disease | 11,027 | 1131 (16.9%) | 950 (22.3%) | <0.001 |
| Asthma | 11,013 | 485 (7.2%) | 315 (7.4%) | 0.9 |
| Diabetes | 11,027 | 1894 (28.4%) | 1361 (32.0%) | <0.001 |
| Hypertension | 11,027 | 4224 (62.4%) | 2741 (64.4%) | 0.052 |
| Cardiovascular disease | 11,027 | 2580 (38.1%) | 1842 (43.3%) | <0.001 |
| Chronic kidney disease | 11,027 | 1338 (19.7%) | 1007 (23.7%) | <0.001 |
| Chronic liver disease | 11,027 | 285 (4.2%) | 199 (4.7%) | 0.3 |
| Chronic neurological impairment | 11,027 | 886 (13.1%) | 664 (15.6%) | <0.001 |
| Dementia | 11,027 | 706 (10.4%) | 494 (11.6%) | 0.040 |
| HIV | 11,027 | 22 (0.3%) | 20 (0.5%) | 0.063 |
| Hematological or immunological disease | 11,010 | 91 (1.3%) | 94 (2.2%) | <0.001 |
| Oncological disease | 11,027 | 904 (13.3%) | 694 (16.3%) | <0.001 |
| Immunosuppressive treatment | 11,010 | 233 (3.4%) | 244 (5.7%) | <0.001 |
| Solid organ transplant | 11,010 | 76 (1.1%) | 54 (1.3%) | 0.13 |
| Risk characteristics | ||||
| Pregnancy | 5642 | 215 (5.7%) | 19 (1.0%) | <0.001 |
| Smoking | 13,246 | 978 (11.7%) | 558 (11.5%) | <0.001 |
| Angiotensin-converting enzyme inhibitor | 13,246 | 1826 (21.8%) | 1226 (25.5%) | <0.001 |
| Charlson Comorbidity Score | 12,993 | 3.0 (1.0, 5.0) | 4.0 (2.0, 6.0) | <0.001 |
| Severity of illness at admission | ||||
| CURB-65 score | 13,265 | <0.001 | ||
| 0–1 | 5942 (71%) | 2469 (51%) | ||
| 2 | 1853 (22%) | 1553 (32%) | ||
| ≥3 | 611 (7.3%) | 837 (17%) | ||
| Complications | ||||
| COVID-19 pneumonia | 9756 | 4626 (86.2%) | 3951 (90.1%) | <0.001 |
| ARDS | 9756 | 475 (8.8%) | 1449 (33.0%) | <0.001 |
| Cardiac and cardiovascular diseases | 11,134 | 767 (12.1%) | 1350 (28.0%) | <0.001 |
| Thrombosis/Embolism | 11,134 | 317 (5.0%) | 528 (11.0%) | <0.001 |
| Neurological complication | 11,134 | 319 (5.0%) | 650 (13.5%) | <0.001 |
| Encephalitis/Encephalopathy | 7802 | 35 (0.8%) | 98 (2.8%) | <0.001 |
| Treatments | ||||
| COVID-19 | ||||
| Corticosteroids | 13,260 | 1657 (19.7%) | 1602 (33.0%) | <0.001 |
| Immunomodulators | 9371 | 30 (0.5%) | 44 (1.2%) | <0.001 |
| Coinfections | ||||
| Antibiotics | 13,261 | 1405 (16.7%) | 4536 (93.4%) | <0.001 |
| Outcomes | ||||
| Length of stay (LOS) in hospital (days) | 13,247 | 7 (4, 12) | 13 (7, 25) | <0.001 |
| Intensive care unit (ICU) stay (no. (%)) | 13,261 | 564 (6.7%) | 1275 (26.2%) | <0.001 |
| ICU LOS (days) | 1837 | 3 (1, 7) | 12 (5, 21) | <0.001 |
| In-hospital death | 13,259 | 800 (9.5%) | 1,149 (23.7%) | <0.001 |
| Discharge destination | 11,399 | <0.001 | ||
| Domicile | 5959 (77.8%) | 2363 (63.1%) | ||
| Another hospital | 250 (3.3%) | 308 (8.2%) | ||
| Rehabilitation clinic | 404 (5.3%) | 419 (11.2%) | ||
| Long-term care facility (LTCF) | 693 (9.1%) | 429 (11.5%) | ||
| Other | 327 (4.3%) | 212 (5.7%) |
| Pathogen | N (%) * |
|---|---|
| Escherichia coli | 179 (14.8%) |
| Staphylococcus aureus | 130 (10.7%) |
| Klebsiella pneumoniae | 74 (6.1%) |
| Pseudomonas aeruginosa | 55 (4.5%) |
| Haemophilus influenzae | 40 (3.3%) |
| Streptococcus pneumoniae | 40 (3.3%) |
| Coagulase-negative staphylococci | 36 (2.9%) |
| Enterobacter cloacae | 35 (2.8%) |
| Enterococcus faecalis | 30 (2.5%) |
| Proteus mirabilis | 28 (2.3%) |
| Pathogen | N = 343 | <3 Days After Hosp., N = 100 1 | 3 to 7 Days After Hosp., N = 79 1 | >7 Days After Hosp., N = 164 1 | p-Value 2 |
|---|---|---|---|---|---|
| Aspergillus fumigatus | 26 | 4 (4.0%) | 6 (7.6%) | 16 (9.8%) | 0.2 |
| Citrobacter spp. | 6 | 1 (1.0%) | 3 (3.8%) | 2 (1.2%) | 0.4 |
| Escherichia coli | 24 | 3 (3.0%) | 3 (3.8%) | 18 (11.0%) | 0.021 |
| Enterobacter cloacae | 16 | 5 (5.0%) | 2 (2.5%) | 9 (5.5%) | 0.6 |
| Haemophilus influenzae | 12 | 6 (6.0%) | 3 (3.8%) | 3 (1.8%) | 0.2 |
| Klebsiella pneumoniae | 29 | 1 (1.0%) | 6 (7.6%) | 22 (13.4%) | 0.002 |
| Pseudomonas aeruginosa | 32 | 10 (10.0%) | 8 (10.1%) | 14 (8.5%) | 0.9 |
| Serratia spp. | 14 | 3 (3.0%) | 3 (3.8%) | 8 (4.9%) | 0.8 |
| Staphylococcus aureus | 87 | 35 (35.0%) | 24 (30.4%) | 28 (17.1%) | 0.003 |
| Streptococcus pneumoniae | 20 | 15 (15.0%) | 3 (3.8%) | 2 (1.2%) | <0.001 |
| Other 3 | 77 | 17 (17.0%) | 18 (22.8%) | 42 (25.6%) | 0.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scanferla, G.; Blöchlinger, A.; Bättig, V.; Buettcher, M.; Cusini, A.; Iten, A.; Keiser, O.; Sommerstein, R.; Sobel, J.; Albrich, W.C. Superinfections in Hospitalized COVID-19 Patients (Super COVID-19): Data from the Multicentric Retrospective CH-SUR Cohort Study in Switzerland. COVID 2025, 5, 86. https://doi.org/10.3390/covid5060086
Scanferla G, Blöchlinger A, Bättig V, Buettcher M, Cusini A, Iten A, Keiser O, Sommerstein R, Sobel J, Albrich WC. Superinfections in Hospitalized COVID-19 Patients (Super COVID-19): Data from the Multicentric Retrospective CH-SUR Cohort Study in Switzerland. COVID. 2025; 5(6):86. https://doi.org/10.3390/covid5060086
Chicago/Turabian StyleScanferla, Giulia, Andrea Blöchlinger, Veronika Bättig, Michael Buettcher, Alexia Cusini, Anne Iten, Olivia Keiser, Rami Sommerstein, Jonathan Sobel, and Werner C. Albrich. 2025. "Superinfections in Hospitalized COVID-19 Patients (Super COVID-19): Data from the Multicentric Retrospective CH-SUR Cohort Study in Switzerland" COVID 5, no. 6: 86. https://doi.org/10.3390/covid5060086
APA StyleScanferla, G., Blöchlinger, A., Bättig, V., Buettcher, M., Cusini, A., Iten, A., Keiser, O., Sommerstein, R., Sobel, J., & Albrich, W. C. (2025). Superinfections in Hospitalized COVID-19 Patients (Super COVID-19): Data from the Multicentric Retrospective CH-SUR Cohort Study in Switzerland. COVID, 5(6), 86. https://doi.org/10.3390/covid5060086






