Abstract
The replication machinery of SARS-CoV-2 is a primary target for therapeutic intervention, and has led to significant progress in antiviral medication discovery. This review consolidates contemporary molecular insights into viral replication and rigorously assesses treatment methods at different phases of viruses’ clinical development. Direct-acting antivirals, such as nucleoside analogs (e.g., remdesivir, molnupiravir) and protease inhibitors (e.g., nirmatrelvir), have shown clinical effectiveness in diminishing morbidity and hospitalization rates. Simultaneously, host-targeted medicines like baricitinib, camostat, and brequinar leverage critical host–virus interactions, providing additional pathways to reduce viral replication while possibly minimizing the development of resistance. Notwithstanding these advancements, constraints in distribution methods, antiviral longevity, and the risk of mutational evasion demand novel strategies. Promising investigational approaches encompass CRISPR-mediated RNA degradation systems, inhalable siRNA-nanoparticle conjugates, and molecular glue degraders that target host and viral proteins. Furthermore, next-generation treatments aimed at underutilized enzyme domains (e.g., NiRAN, ExoN) and host chaperone systems (e.g., TRiC complex) signify a transformative approach in antiviral targeting. The integration of high-throughput phenotypic screening, AI-driven medication repurposing, and systems virology is transforming the antiviral discovery field. An ongoing interdisciplinary endeavor is necessary to convert these findings into versatile, resistance-resistant antiviral strategies that are applicable beyond the present pandemic and in future coronavirus epidemics.
1. Introduction
The global outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in late 2019 has resulted in a pandemic called coronavirus illness 2019 (COVID-19) that has massively affected both the health and economic sectors. On 21 March 2023, an estimated 761 million people were reportedly infected by said virus. Of those who were infected, approximately 6.8 million fatalities were reported [1].
This has led to COVID-19 being one of the most catastrophic global health concerns of the new millennium. In a short span of time, the scientific community made extraordinary progress in understanding the virus and finding countermeasures. These include elucidation of the life cycle of the virus and its viral protein structures in the first year of the global outbreak onset. Over 20 high-resolution structures of the virus’s RNA polymerase complex were reported. This has resulted in in-depth knowledge of the replication mechanism of the virus and methods to inhibit it [2,3,4].
The initial groundwork studies on the molecular basis of SARS-CoV-2 replication have ultimately opened a new research avenue for designing future therapeutic interventions. Based on its viral structure, SARS-CoV-2 is similar to other coronaviruses. It is an enveloped positive-sense RNA virus that has a quite large ~30 kb genomic size, one of the largest of any RNA virus. In comparison, planarian secretory cell nidovirus (PSCNV) has a genome size of 41.1 kb [5], rotaviruses are about 26–30 kb [6], ebolavirus are about ~19 kb [7], and the influenza virus is about ~13.5 kb [8,9].
The viral replication process involves a multi-protein replication/transcription complex that allows the replication of the RNA genome within the infected host cell. The virus interacts with host cellular pathways, which in turn can elude immune defenses. The virus uses host resources for its replication, which leads to dysregulation of the host’s normal host cell functions [8].
In the past five years, antiviral strategies targeting the viral replication cycle have mainly comprised either developed or repurposed drugs, in particular, direct-acting antivirals such as remdesivir and molnupiravir [10] and protease inhibitors (nirmatrelvir in Paxlovid) that indirectly affect viral replication through the inhibition of the viral replicase protein processing [11].
The present review offers a summary of the SARS-CoV-2 replication molecular mechanism and host–cell interactions while assessing the potential impact of this information on the development of future therapeutic strategies. This review examines both wet laboratory experimental findings and computational approaches that have expanded our present understanding of the viral replication mechanism of SARS-CoV-2. In brief, this review includes the molecular mechanism of viral RNA replication, virus–host interactions, and current and emerging antiviral strategies. This paper also looks at future directions in this field by highlighting potential novel targets and possible innovative antiviral strategies. As a whole, this integrative approach aims to synthesize the current advancements in SARS-CoV-2 replication as a guide for future research.
2. Molecular Mechanisms of SARS-CoV-2 Replication
SARS-CoV-2 shares a similar genomic organization and replication method to those of other coronaviruses. Upon entrance in the host cell, the single-stranded positive-sense RNA genome of the virus directly works as a mRNA and has a 5′ cap and 3′ poly(A) tail [8,12,13]. Sixteen non-structural proteins (nsp; nsp1–16) comprise the viral replication and transcription machinery of the virus. These proteins are autocatalytically cleaved by the viral proteases that come from two large polyproteins (pp1a and pp1ab) that are encoded by the 5′ two-thirds of the genome (open reading frames ORF1a and ORF1ab) [8,14].
Meanwhile, the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins serve as auxiliary and structural proteins. These proteins are encoded by the remaining 3′ region of the genome. Furthermore, these proteins are expressed by the sub-genomic mRNAs generated during the infection process (Figure 1) [14,15,16].
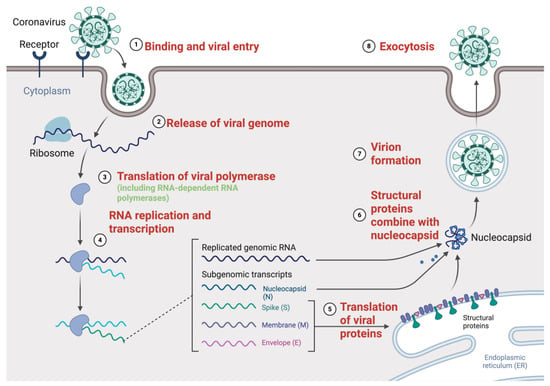
Figure 1.
SARS-CoV-2 replication cycle and key molecular events targeted by antivirals. The SARS-CoV-2 replication cycle starts with (1) binding and viral entry, in which the virus attaches to the host cell receptor (such as ACE2) and is internalized by endocytosis or membrane fusion. (2) The viral genome is then released into the cytoplasm. (3) For translation of viral polyprotein including the RNA-dependent RNA polymerase (RdRp) and other non-structural proteins (nsps), the positive-sense RNA genome is directly employed as mRNA. To generate both full-length genomic RNA and sub-genomic RNAs, these proteins form the replication–transcription complex (RTC), therefore mediating (4) RNA replication and transcription. At the endoplasmic reticulum, the sub-genomic RNA acts as a template for (5) translation of the structural proteins S, E, M, and N. These proteins travel to assembly sites where (6) structural proteins mix with the nucleocapsid, which encapsidates the fresh genomic RNA. Finally, (7) virion assembly occurs and (8) exocytosis releases new virions. Figure created with BioRender.com, adapted from Guo, 2020 [17] and Fehr et al. 2015 [18], licensed under CC BY 4.0.
2.1. Replication Complex and Enzymatic Machinery
The RNA-dependent RNA polymerase (RdRp) complex is a multimeric protein complex that is central to coronavirus replication. The minimum functional components for this multimeric complex are comprised by the nsp12–7–8 super-complex [19,20].
The structure and function of nsp12 are as follows: the protein has a catalytic polymerase subunit that functions with a canonical “right-hand” polymerase domain containing finger, palm and thumb subdomains that allow RNA synthesis [8,21]. In addition, nsp12 has an N-terminal nidovirus-specific RNA-dependent nucleotidyltransferase domain (NiRAN). This plays an important function in viral replication and possibly plays a role in priming or capping [21,22].
The two co-factors, nsp7 and nsp8, form the active polymerase by binding with nsp12. nsp12 has been reported to have little or marginal activity by itself. Two subunits of nsp8 and one nsp7 subunit form a complex with nsp12 to form a functional RdRp holoenzyme. This complex serves as a processivity factor that aids in the gripping of the RNA template and in stabilizing the polymerase during the viral replication process (Figure 2) [19,20].
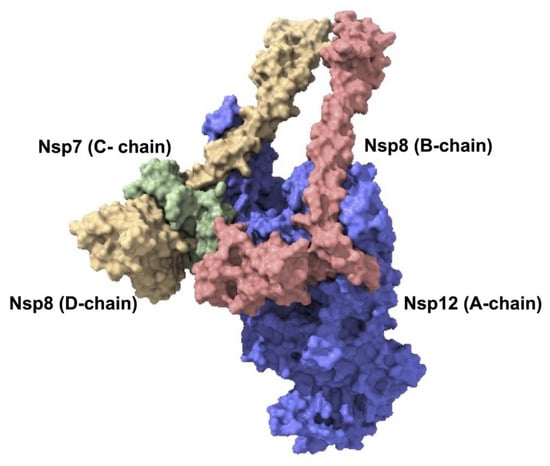
Figure 2.
SARS-CoV-2 viral replication complex. SARS-CoV-2′s core viral replication machinery is made up of the nsp12–nsp7–nsp8 super-complex, which reflects the minimal functional unit needed for RNA synthesis. The nsp12 subunit (A-chain, blue) is the main catalytic polymerase. The nsp7 co-factor (C-chain, depicted in green) binds to nsp12 and stabilizes the system. Acting as co-factors and extensions of the template RNA-binding platform, the nsp8 subunits (B-chain in red and D-chain in yellow) increase the processivity. This structural model was produced using comparative modelling with Robetta; the template structure was PDB ID: 8GWE.
Several other viral nsps play essential functions in genome replication and RNA processing. The replication–transcription complex (RTC) functions by copying the viral RNA. This includes the 5′–3′ helicase/translocase nsp13 that is responsible for the unwinding of the RNA secondary structure and moves along the template during RNA synthesis [23,24]. The nsp14 protein functions as a 3′–5′ exoribonuclease (ExoN) that removes misincorporated nucleotides and thus contributes to increased replication fidelity via the RTC’s proofreading mechanism [25].
This unusual proofreading capability of RNA viruses most likely helps coronaviruses retain their enormous genomes. Moreover, the N7-methyltransferase domain of nsp14 operates in conjunction with nsp10 [26]. nsp16 functions as a 2′-O-methyltransferase with nsp10 as a co-factor. This allows the 5′ end of the viral RNA to establish a cap structure [27]. In turn, this guarantees the efficient translation and stability of viral mRNAs. Other nsps, in particular nsp9 and nsp10, support the RTC through RNA binding and regulation [28].
2.2. Genome Replication Cycle
The multi-step replication of SARS-CoV-2 occurs in the cytoplasm of infected cells [29]. After viral entry and uncoating, the translation of the genomic RNA generates the replicase polyprotein (pp1a/pp1ab) [30]. These are then cleaved to produce nsps. These nsps rapidly form the RTC in the cellular membranes. A characteristic of coronavirus replication is the remodeling of the host intracellular membranes. This generates specific microenvironments for RNA reproduction [31].
Double-membrane vesicles (DMVs) that are derived from the endoplasmic reticulum and work as protected replication organelles are formed upon the induction of SARS-CoV-2 [32,33]. nsp3 and nsp4, viral transmembrane proteins, bind to the endoplasmic reticulum and curate these vesicles. The formed nsp3–nsp4 complex bends the membranes into vesicles that recruit replication proteins [34,35].
The RdRp complex is tethered to the cytosolic faces of DMVs and is concentrated in these compartments [28]. This is due to the N-terminal domain of nsp3, which can directly bind to nsp12 polymerase [8].
The RTC generates a complementary negative-sense RNA from the genomic RNA template within the DMVs [13,36]. This negative strand functions as a template for the synthesis of new positive-sense genomic RNA, a series of sub-genomic RNAs that align with the 3′ regions of the genome, and supports the expression of structural and accessory proteins [12,37]. The polymerases engage in a distinct form of discontinuous transcription during sub-genomic mRNA replication. The RdRp intermittently moves from one portion of the negative-strand template to connect with the leader sequence at the 5′ end. This template-switching mechanism leads to the generation of sub-genomic RNAs that possess a 5′ leader while encoding distinct downstream open reading frames (ORFs) such as S, E, M, and N [37].
Although proofreading provides high fidelity, the virus continues to accumulate mutations over time [38,39]. This, along with RNA recombination events, has resulted in the emergence of various SARS-CoV-2 variants [38]. Following the synthesis of new genomic RNAs, these molecules are encapsulated within virions [40]. The nucleocapsid (N) protein associates with genomic RNA to create a ribonucleoprotein complex [41], which is subsequently enveloped by a lipid membrane that incorporates the S, E, and M proteins at assembly sites within the ER–Golgi intermediate compartment [42]. Progeny virions are transported within vesicles and subsequently released from the cell through exocytosis. An infected cell can release thousands of new virions, which go on to infect neighboring cells and, in an organism, can disseminate to other tissues [40,42].
2.3. Host Factors in Viral Replication
To fulfill its reproduction cycle, SARS-CoV-2 depends on the host proteins and cellular processes. The virus utilizes the host ribosomes for protein synthesis and relies on the host cell′s provision of nucleotides, amino acids, and energy.
Recent proteomic studies have indicated connections between SARS-CoV-2 non-structural proteins and the host proteins. In particular, nsp12 interacts with the host chaperonin complex, often referred to as TRiC (or CCT) [43].
TRiC supports the correct folding or assembly of the polymerase complex. This, in turn, allows reliance on the host for effective replication [44]. The P323L mutation of nsp12 has been shown to influence this interaction. This nsp12 protein variant, alongside the D614G spike mutant, was found to have reduced dependence on the host–phosphatase complex. This indicated a possible adaptive modification for enhanced replication in host human cells [45]. These findings show that SARS-CoV-2 can adapt to its host cellular environment, which can lead to improving its replicative fitness [46]. In such cases, host pathways (including chaperones and phosphatases) might be targeted as host-directed antivirals [47,48]. Such findings underscore how SARS-CoV-2 can adapt to the host cellular environment and potentially improve its replicative fitness. They also point to host pathways (chaperones and phosphatases) that might be targeted by host-directed antivirals [43,45].
3. Drugs Targeting SARS-CoV-2 Replication
The efforts to create therapeutics that inhibit the replication of SARS-CoV-2 have been unprecedented. These drugs encompass direct-acting antivirals that target viral enzymes and host-targeting medications that indirectly impede viral propagation. Here, we discuss four groups of antiviral drugs that disrupt the SARS-CoV-2 replication cycle, along with aspects such as their resistance and efficacy.
3.1. Nucleoside Analogue Polymerase Inhibitors
Targeting the RdRp using nucleoside or nucleotide analogues has been a key tactic to stop viral replication [49]. Remdesivir was the inaugural antiviral medication authorized for COVID-19. It is a mono phosphoramidate prodrug of an adenosine analogue; upon intracellular metabolism to the active remdesivir triphosphate (RTP), it competes with ATP and is integrated into viral RNA by nsp12 [43]. The integration of remdesivir into an elongating RNA strand leads to delayed chain termination, causing RNA synthesis to cease after the addition of many additional nucleotides [4] (Figure 3).
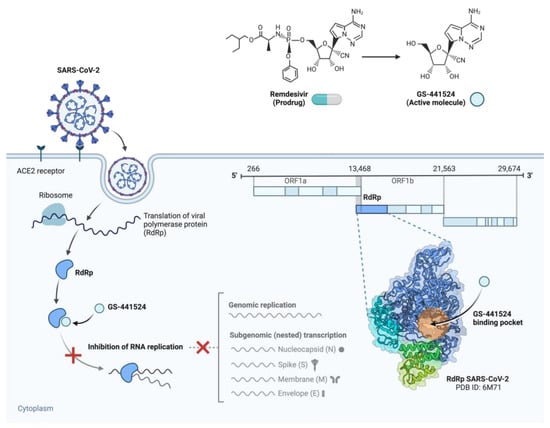
Figure 3.
Action mechanism of remdesivir and its active metabolite GS-441524 against SARS-CoV-2 replication. SARS-CoV-2, upon entering the host cell, uses the host ribosome to convert the RNA-dependent RNA polymerase (RdRp), a main enzyme encoded inside ORF1b. The mono phosphoramidate prodrug remdesivir is metabolized intracellularly to its active nucleoside analogue, GS-441524. RdRp includes this molecule in nascent viral RNA, where it mimics adenosine. Once included, it causes postponed chain termination, therefore stopping more elongation of the RNA strand and thus stopping viral replication. Highlighting the binding pocket of GS-441524, the bottom right inset reveals the RdRp holoenzyme structure (PDB ID: 6M71). Figure created with BioRender.com, adapted from Li, G. & De Clercq, E. (2020) [50], licensed under CC BY 4.0.
Structural studies have shown that remdesivir causes the RdRp to stall by preventing further translocation of the RNA template. In clinical trials, intravenous remdesivir moderately accelerated the recovery of patients who were hospitalized with COVID-19 and continues to be a routine treatment for severe cases [51,52]. Significantly, the virus’s proofreading function (nsp14) does not completely eliminate remdesivir, which enables it to maintain its antiviral effectiveness [53]. However, resistance may develop under selective pressure: in vitro studies have revealed mutations in nsp12 (notably at residues V166 and E802) that provide partial resistance to remdesivir [54]. More importantly, such mutants have not dominated in clinical isolates, possibly due to fitness costs and the relatively short treatment courses that were used.
Another nucleoside analogue that has been approved as an oral antiviral for COVID-19 is molnupiravir [55]. It is a prodrug of β-D-N4-hydroxycytidine that behaves as a mutagenic agent. Unlike remdesivir, molnupiravir does not result in chain termination. Rather, its active form is incorporated into viral RNA in lieu of cytidine or uridine, causing the RdRp to make an erroneous base-pairing (it can pair with guanine or adenine) [56,57]. This leads to the accumulation of mutations in the viral genome, a process known as lethal mutagenesis (Figure 4).
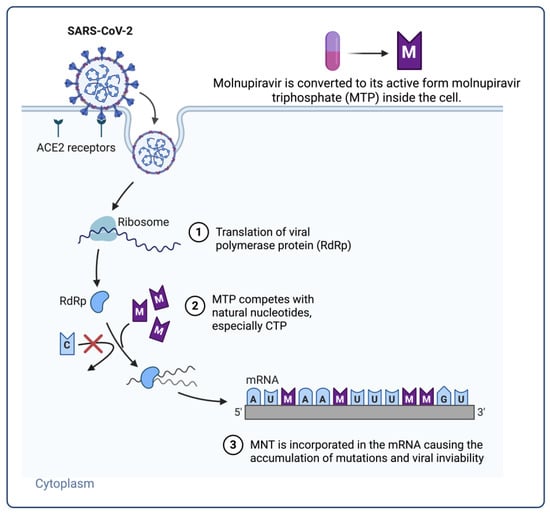
Figure 4.
Action mechanism of mutagenic nucleoside analogues (molnupiravir) in SARS-CoV-2 replication. Viral entry causes the host cell to translate the viral RNA-dependent RNA polymerase (RdRp), which drives RNA synthesis. Competing with natural cytidine triphosphate (CTP), mutagenic nucleoside analogues like the active triphosphate form of molnupiravir (MTP) are misincorporated into viral RNA. Base-pair mismatches and the buildup of mutations follow from this, causing viral inactivation—a process called lethal mutagenesis. This approach enhances molnupiravir’s antiviral effectiveness against several SARS-CoV-2 variants. Figure created with BioRender.com, adapted from Malone, B. & Campbell, E.A. (2021) [58], licensed under CC BY 4.0.
Molnupiravir fundamentally induces errors in the virus, leading to the death of the viral population due to genomic instability [59]. Clinical trials with patients with mild-to-moderate COVID-19 demonstrated that a 5-day oral regimen of molnupiravir markedly decreased the likelihood of hospitalization or mortality compared to placebo [60]. Because molnupiravir targets the conserved polymerase, it is still effective against various SARS-CoV-2 mutations [57,61]. Nonetheless, theoretical apprehensions exist over the potential consequences of its mutagenic action, such as its impact on host DNA or the emergence of novel viral mutations [59]. However, current research suggests that it is harmless and that the viral ExoN proofreading mechanism only partially mitigates its effects [56,59].
Favipiravir (T-705), a purine base analogue, was another nucleoside analogue evaluated early in the pandemic [62,63]. Favipiravir exhibits extensive efficacy against RNA viruses; however, SARS-CoV-2 has demonstrated notable resistance to it [64]. In COVID-19, clinical trials of favipiravir have yielded inconsistent outcomes, and it is not considered a frontline therapy [65,66]. Despite its limited effectiveness when used alone against SARS-CoV-2, ribavirin, a riboside analogue with known activity against some RNA viruses, has been explored in combination regimens [67,68]. The oral derivative of remdesivir′s parent nucleotide (GS-441524), VV116 (deuviridine), is a new nucleoside analogue that has demonstrated potential. The oral bioavailability of VV116 is significantly higher (between 80–90%) [69,70] and, in a clinical trial, it demonstrated non-inferiority to Paxlovid in early COVID-19, making it a potentially useful oral polymerase inhibitor [71].
3.2. Protease Inhibitors
Two proteases that are necessary for breaking down the replicase polyprotein are encoded by SARS-CoV-2: nsp5 major protease and nsp3 papain-like protease. Inhibiting these proteases makes the viral RTC unable to mature [72,73]. Nirmatrelvir is a peptidomimetic inhibitor of the primary protease (3CLpro) that was developed and its combination with the pharmacokinetic enhancer ritonavir (which slows down nirmatrelvir′s metabolism) produces the oral drug Paxlovid [72,74]. Nirmatrelvir indirectly halts RNA replication by inhibiting 3CLpro, as it keeps the polymerase and other nsps from being released from the polyprotein [73]. In the EPIC-HR Phase 2/3 trial, Paxlovid demonstrated remarkable efficacy by decreasing the likelihood of hospitalization or death by ~89% in high-risk outpatients when administered within a few days of symptom onset [72]. For high-risk patients, it has been a game-changer for early intervention. Although mutations in the 3CL-pro substrate-binding pocket can result in resistance to nirmatrelvir (a number of these mutations have been identified in vitro) [75,76], substantial resistance has not yet been extensively disseminated in circulating strains. With the continued usage of Paxlovid, it is crucial to continuously monitor 3CL-pro variations [77].
Additional protease inhibitors have been investigated. The HIV protease inhibitor lopinavir, when tested in combination with ritonavir during early clinical trials, demonstrated no significant benefit for treating COVID-19 [78]. Ensitrelvir, an investigational 3CLpro inhibitor, has demonstrated efficacy in clinical trials conducted in Japan, indicating that protease inhibition represents a validated target for SARS-CoV-2 beyond nirmatrelvir [79]. The papain-like protease (PLpro) represents a potential antiviral target. Inhibitors of PLpro, including GRL-0617, have been identified in preclinical studies. Despite this, none are currently utilized in clinical settings [80].
3.3. Host-Directed Antivirals
One strategy for inhibiting SARS-CoV-2 replication that is currently being explored is host-target antiviral mechanisms. This process targets the host cell processes that are hijacked by the virus during its replication cycle. This entails the repurposing of existing approved drugs that are used beyond their conventional indications. Studies on various compounds that might significantly inhibit SARS-CoV-2 replication and significantly affect therapeutic outcomes via host-directed processes have been reported.
Fluoxetine, a selective serotonin reuptake inhibitor (SSRI), along with GS-441524, has been found to synergistically affect SARS-CoV-2 variants in vitro. Findings show that viral replication was reduced through the targeting of the cellular pathways used by the virus [81]. Compounds that inhibit virus entry into the host cell through the ACE2 receptor and TMPRSS2 protease have been receiving increasing attention. TMPRSS2 inhibitors and ACE2 receptor blockers have limited viral access to host cells. [82,83]. Investigational compounds that inhibit these receptors may enhance the effectiveness of existing antivirals and mitigate viral entry.
The small-molecule inhibitors designed to modulate intracellular signaling pathways that support viral replication have been explored as a route for possible therapeutic intervention. Inhibition of the pathways involved in cellular stress responses or inflammation might indirectly suppress the ability of the virus to replicate efficiently [84].
3.4. Combination Therapies and Drug Synergy
The management of COVID19 has gradually begun to utilize combination techniques, especially in serious cases or to combat resistance. The application of antivirals that target various stages of the replication cycle may yield synergistic effects [85,86]. The observed synergy between remdesivir and nirmatrelvir is an excellent example. In vitro, the combination of these two medications demonstrated synergistic antiviral action, suppressing SARS-CoV-2, including Omicron variant strains, more effectively than either drug alone [87,88,89]. By blocking RNA chain elongation and blocking polyprotein processing, respectively, this combination targets replication through two complementary methods.
Remdesivir has been investigated in conjunction with ribavirin, yielding noteworthy results. Ribavirin, while ineffective as a standalone treatment, functions as a base analogue that promotes mutations in a way that resembles the process of lethal mutagenesis. The combination of remdesivir with ribavirin demonstrated a synergistic effect, resulting in the complete eradication of SARS-CoV-2 in cell cultures at relatively low concentrations [90]. The virus′s capacity to reproduce was overwhelmed by this combination, which drastically increased the polymerase′s mistake rate while also impeding nucleotide addition. This implies a potential therapeutic strategy for “lethal mutagenesis” that involves the combination of a mutagenic agent and a chain terminator [90,91]. Triple combinations that including ribavirin or favipiravir and other antivirals have also been proposed to further boost the efficacy of these drugs and prevent escape mutants [90,92,93].
Molnupiravir has also been evaluated in pharmacological combinations. Molnupiravir synergizes with various other agents, such as nirmatrelvir and specific host-targeting medicines, according to a recent study [87,88,94]. For example, the combination of molnupiravir with the TMPRSS2 inhibitor camostat, which obstructs viral entrance, or with the DHODH inhibitor brequinar, which diminishes the nucleotide pools essential for viral RNA synthesis, resulted in the enhanced suppression of SARS-CoV-2 in lung cell cultures [94,95]. The combination of molnupiravir and nirmatrelvir demonstrated significant synergy, highlighting the advantages of simultaneously targeting polymerase and protease activities [94,96]. In addition, triple therapy using molnupiravir, camostat, and brequinar suppressed viral replication much more strongly in vitro, suggesting that these three medications could be used as a cocktail to treat resistant infections or immunocompromised people who have persistent virus shedding [94,95,97].
Alongside the new antivirals, numerous authorized medications have been modified for to exhibit possible efficacy against SARS-CoV-2. Several of these repurposed candidates were repurposed to focus on host factors, such as the antiparasitic ivermectin or different kinase inhibitors, whereas a select handful exhibited direct or indirect antiviral properties. An illustrative instance is the antidepressant fluoxetine and the antifungal itraconazole. These medicines, each with unique primary targets (serotonin reuptake and fungal sterol synthesis, respectively), were shown to augment the antiviral efficacy of remdesivir in cell culture [98,99]. The combination of fluoxetine with remdesivir or itraconazole with remdesivir diminished infectious viral generation by over 90% compared to remdesivir administered alone [100]. The precise antiviral contributions of fluoxetine and itraconazole are still being investigated, but fluoxetine may have lysosomotropic effects that impede viral egress, while itraconazole may disrupt cholesterol trafficking for viral replication. The use of safe, off-patent medications to increase the effectiveness of direct antivirals is nevertheless made possible by these findings [101].
In addition to antivirals that directly inhibit replication, immunomodulators have been utilized as adjunctive therapies in COVID-19 treatment to manage hyperinflammation. Although not classified as antivirals, medications such as the corticosteroid dexamethasone and the JAK inhibitor baricitinib (which received emergency use authorization in conjunction with remdesivir) contribute to improved outcomes by alleviating the harmful inflammatory response to antivirals [100,102]. Interestingly, baricitinib may also have minor antiviral effects due to blocking clathrin-mediated endocytosis, which is necessary for viral entry. This makes it a host-targeted treatment that can be used in conjunction with direct antivirals [100,103,104].
A comprehensive survey of antiviral therapies that directly or indirectly target SARS-CoV-2 replication is summarized in Table 1. The list includes molecules and their molecular targets, such as the RNA-dependent RNA polymerase (nsp12), main protease (3CLpro), papain-like protease (PLpro), and host cellular factors. Moreover, host-targeting chemicals that change cellular pathways for viral replication or entrance were included. Combination therapies that enhance viral efficacy offer lower resistance probability. Overall, this emphasizes the need for multi-target strategies in COVID-19 control and underscores the current available treatments.

Table 1.
Summary of antiviral therapeutics targeting SARS-CoV-2.
3.5. Antiviral Resistance Considerations
The rapid development of antiviral medication raises questions about resistance. SARS-CoV-2 is an excellent example of an RNA virus’s high rate of mutation, even though its proofreading capacity slows its rate of change [91,105]. Clinical resistance to the main antivirals is not yet a major concern. Patients treated with remdesivir have been observed to occasionally shed variants containing nsp12 mutations, and in vitro studies indicate that the virus can acquire these alterations [106,107]. Molnupiravir′s mechanism complicates the virus′s ability to develop resistance, as any single mutation it may acquire is equally probably harmful as it is beneficial. This is because the drug drives the virus into an evolutionary dead end via an error catastrophe [108,109]. Based on substantial experience with HIV and HCV therapy, resistance to protease inhibitors is a serious problem. With the continued usage of Paxlovid, it will be essential to continuously monitor its usage, and it will also be essential to continuously monitor 3CL-pro sequences in clinical isolates [110]. Because the virus needs to acquire numerous independent mutations at the same time to evade multiple medications, combination therapy dramatically reduces the possibility of resistance. This notion serves as the foundation of current approaches to COVID-19 treatments and future pandemic preparedness [111,112].
5. Conclusions and Future Directions
The COVID-19 pandemic has advanced our understanding of coronavirus replication and renewed interest in antiviral drugs and strategies, which are reviewed in this paper (Table 3). SARS-CoV-2 replication involves complicated viral protein activities and dynamic host–virus interactions. Over the past few years, researchers have discovered the polymerase complex’s atomic structure, its RNA manufacturing mechanism, and how the SARS-CoV-2 virus alters cellular membranes and avoids immune detection. This comprehension has directly impacted the development of targeted therapeutics. RNA polymerase inhibitors like remdesivir and molnupiravir and protease inhibitors like nirmatrelvir were developed to limit viral polyprotein processing. These interventions have shown their effectiveness in reducing disease severity and saving lives.

Table 3.
Summary of reviewed recent antiviral strategies and their current status.
Despite significant progress in SARS-CoV-2 antiviral research, substantial gaps persist, offering avenues for additional exploration and innovation. The demand for comprehensive, resistance-resistant therapeutics that efficiently combat diverse coronaviruses and their emerging variants constitutes a significant challenge and underscores the importance of pan-coronaviral targeting approaches [85]. Innovative protease inhibitors in preclinical stages and non-nucleoside RdRp inhibitors signify two increasingly significant therapeutic strategies [152]. Host-targeted medicines, including autophagy modulators and TMPRSS2 inhibitors, bolster defensive systems by diminishing viral replication and thwarting resistance [153,154]. RNA-based therapies, including siRNAs and antisense oligonucleotides (ASOs), when employed with sophisticated nanoparticle delivery systems, may provide targeted and localized treatment for pulmonary disorders [155,156]. The amalgamation of host-directed and viral medicines may augment therapeutic efficacy and inhibit the development of resistance [85]. The antiviral pipeline is being transformed by sophisticated drug discovery methodologies, such as high-throughput phenotypic screening, artificial intelligence-facilitated repurposing, and the CRISPR-based exploration of host–virus interactions [157]. A worldwide project is underway to improve readiness for future zoonotic coronavirus epidemics, as the SARS-CoV-2 pandemic has markedly heightened interest in treatments with pan-coronaviral capabilities. Converting these substantial discoveries into clinically viable medicines necessitates ongoing interdisciplinary collaboration [158].
Current therapies, such as early administration and intravenous remdesivir delivery, are limited by the virus’s development and require caution. While not yet a clinical issue for SARS-CoV-2, antiviral resistance remains a concern. The advent of variants with changed replication or immune evasion features highlights the need to expand our antiviral therapies. Researchers are exploring next-generation antivirals. These include cutting-edge tactics like CRISPR-based antivirals, host-targeted treatments that may provide broad protection against several coronaviruses, and inhibitors of several viral enzymes (helicases, exonucleases, etc.). AI- and computer-based approaches speed up drug discovery.
There are encouraging possibilities for future therapies across various study domains. Low-toxicity host-targeted antivirals have been developed using RNA-based therapies, specifically siRNA/Cas13 inhalable nanoparticles and inhibitors, that are aimed at less-redundant viral enzymes such as the NiRAN and ExoN domains [159,160,161]. The existing research is inadequate in applying artificial intelligence to forecasting resistance patterns and to guide the formulation of tailored combination therapies [85,162]. Moreover, the application of molecular glues and degradable technologies that target both host and viral proteins may facilitate the advancement of particular antiviral pharmaceuticals. Future pandemic preparedness could be significantly enhanced through the development of broad-spectrum antivirals that target conserved patterns across coronaviruses [163,164].
To summarize, the development of successful therapeutic strategies for COVID-19 depends on an understanding of the molecular mechanisms underlying SARS-CoV-2 replication. To contain the pandemic and prepare for future outbreaks, researchers are discovering novel viral inhibitory targets and techniques. Addressing SARS-CoV-2 and other deadly viruses requires a synergistic approach that combines molecular virology, pharmacology, and new technology. The insights and antivirals developed during the COVID-19 pandemic will shape future viral disease management.
Author Contributions
Conceptualization: B.J.J.S. and I.L.F.; writing: original draft preparation: B.J.J.S. and I.L.F.; writing review and editing: B.J.J.S. and I.L.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
B.J.J.S. would like to acknowledge the support of the Ministry of Education, Culture, Sports, Science and Technology (MEXT).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, A.; Preiss, A.J.; Xiao, X.; Brannock, M.D.; Alexander, G.C.; Chew, R.F.; Davis, H.; Fitzgerald, M.; Hill, E.; Kelly, E.P.; et al. Effect of nirmatrelvir/ritonavir (Paxlovid) on hospitalization among adults with COVID-19: An electronic health record-based target trial emulation from N3C. PLoS Med. 2025, 22, e1004493. [Google Scholar] [CrossRef]
- Faisal, H.M.N.; Katti, K.S.; Katti, D.R. Differences in Interactions Within Viral Replication Complexes of SARS-CoV-2 (COVID-19) and SARS-CoV Coronaviruses Control RNA Replication Ability. JOM (1989) 2021, 73, 1684–1695. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Mao, C.; Luan, X.; Shen, D.-D.; Shen, Q.; Su, H.; Wang, X.; Zhou, F.; Zhao, W.; Gao, M.; et al. Structural Basis for Inhibition of the RNA-Dependent RNA Polymerase from SARS-CoV-2 by Remdesivir. Science 2020, 368, 1499–1504. [Google Scholar] [CrossRef]
- Saberi, A.; Gulyaeva, A.A.; Brubacher, J.L.; Newmark, P.A.; Gorbalenya, A.E. A Planarian Nidovirus Expands the Limits of RNA Genome Size. PLoS Pathog. 2018, 14, e1007314. [Google Scholar] [CrossRef] [PubMed]
- Krell, P.J. An Introduction to Viruses of Invertebrates. In Encyclopedia of Virology; Elsevier: Amsterdam, The Netherlands, 2021; pp. 699–723. ISBN 978-0-12-814516-6. [Google Scholar]
- Bettini, A.; Lapa, D.; Garbuglia, A.R. Diagnostics of Ebola Virus. Front. Public. Health 2023, 11, 1123024. [Google Scholar] [CrossRef]
- Hillen, H.S. Structure and Function of SARS-CoV-2 Polymerase. Curr. Opin. Virol. 2021, 48, 82–90. [Google Scholar] [CrossRef]
- Terrier, O.; Si-Tahar, M.; Ducatez, M.; Chevalier, C.; Pizzorno, A.; Le Goffic, R.; Crépin, T.; Simon, G.; Naffakh, N. Influenza Viruses and Coronaviruses: Knowns, Unknowns, and Common Research Challenges. PLoS Pathog. 2021, 17, e1010106. [Google Scholar] [CrossRef]
- Singh, A.K.; Singh, A.; Singh, R.; Misra, A. Molnupiravir in COVID-19: A Systematic Review of Literature. Diabetes Metab. Syndr. Clin. Res. Rev. 2021, 15, 102329. [Google Scholar] [CrossRef]
- Qiu, Y.; Wen, H.; Wang, H.; Sun, W.; Li, G.; Li, S.; Wang, Y.; Zhai, J.; Zhan, Y.; Su, Y.; et al. Real-World Effectiveness and Safety of Nirmatrelvir-Ritonavir (Paxlovid)-Treated for COVID-19 Patients with Onset of More than 5 Days: A Retrospective Cohort Study. Front. Pharmacol. 2024, 15, 1401658. [Google Scholar] [CrossRef]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 Genome, Structure, Evolution, Pathogenesis and Therapies: Structural Genomics Approach. Biochim. Et. Biophys. Acta (BBA)—Mol. Basis Disease 2020, 1866, 165878. [Google Scholar] [CrossRef] [PubMed]
- Brant, A.C.; Tian, W.; Majerciak, V.; Yang, W.; Zheng, Z.-M. SARS-CoV-2: From Its Discovery to Genome Structure, Transcription, and Replication. Cell Biosci. 2021, 11, 136. [Google Scholar] [CrossRef] [PubMed]
- Polatoğlu, I.; Oncu-Oner, T.; Dalman, I.; Ozdogan, S. COVID-19 in Early 2023: Structure, Replication Mechanism, Variants of SARS-CoV-2, Diagnostic Tests, and Vaccine & Drug Development Studies. MedComm 2023, 4, e228. [Google Scholar] [CrossRef] [PubMed]
- Baggen, J.; Vanstreels, E.; Jansen, S.; Daelemans, D. Cellular Host Factors for SARS-CoV-2 Infection. Nat. Microbiol. 2021, 6, 1219–1232. [Google Scholar] [CrossRef]
- Yan, W.; Zheng, Y.; Zeng, X.; He, B.; Cheng, W. Structural Biology of SARS-CoV-2: Open the Door for Novel Therapies. Sig. Transduct. Target. Ther. 2022, 7, 26. [Google Scholar] [CrossRef]
- Guo, Y.-R.; Cao, Q.-D.; Hong, Z.-S.; Tan, Y.-Y.; Chen, S.-D.; Jin, H.-J.; Tan, K.-S.; Wang, D.-Y.; Yan, Y. The Origin, Transmission and Clinical Therapies on Coronavirus Disease 2019 (COVID-19) Outbreak—An Update on the Status. Mil. Med. Res. 2020, 7, 11. [Google Scholar] [CrossRef]
- Fehr, A.R.; Perlman, S. Coronaviruses: An Overview of Their Replication and Pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar] [CrossRef]
- Subong, B.J.J.; Ozawa, T. Bio-Chemoinformatics-Driven Analysis of Nsp7 and Nsp8 Mutations and Their Effects on Viral Replication Protein Complex Stability. CIMB 2024, 46, 2598–2619. [Google Scholar] [CrossRef]
- Reshamwala, S.M.S.; Likhite, V.; Degani, M.S.; Deb, S.S.; Noronha, S.B. Mutations in SARS-CoV-2 Nsp7 and Nsp8 Proteins and Their Predicted Impact on Replication/Transcription Complex Structure. J. Med. Virol. 2021, 93, 4616–4619. [Google Scholar] [CrossRef]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-Dependent RNA Polymerase from COVID-19 Virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef]
- Slanina, H.; Madhugiri, R.; Bylapudi, G.; Schultheiß, K.; Karl, N.; Gulyaeva, A.; Gorbalenya, A.E.; Linne, U.; Ziebuhr, J. Coronavirus Replication–Transcription Complex: Vital and Selective NMPylation of a Conserved Site in Nsp9 by the NiRAN-RdRp Subunit. Proc. Natl. Acad. Sci. USA 2021, 118, e2022310118. [Google Scholar] [CrossRef] [PubMed]
- Mickolajczyk, K.J.; Shelton, P.M.M.; Grasso, M.; Cao, X.; Warrington, S.E.; Aher, A.; Liu, S.; Kapoor, T.M. Force-Dependent Stimulation of RNA Unwinding by SARS-CoV-2 Nsp13 Helicase. Biophys. J. 2021, 120, 1020–1030. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Q.; Malone, B.; Llewellyn, E.; Pechersky, Y.; Maruthi, K.; Eng, E.T.; Perry, J.K.; Campbell, E.A.; Shaw, D.E.; et al. Ensemble Cryo-EM Reveals Conformational States of the Nsp13 Helicase in the SARS-CoV-2 Helicase Replication–Transcription Complex. Nat. Struct. Mol. Biol. 2022, 29, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Chinthapatla, R.; Sotoudegan, M.; Srivastava, P.; Anderson, T.K.; Moustafa, I.M.; Passow, K.T.; Kennelly, S.A.; Moorthy, R.; Dulin, D.; Feng, J.Y.; et al. Interfering with Nucleotide Excision by the Coronavirus 3′-to-5′ Exoribonuclease. Nucleic Acids Res. 2023, 51, 315–336. [Google Scholar] [CrossRef]
- Niu, X.; Kong, F.; Hou, Y.J.; Wang, Q. Crucial Mutation in the Exoribonuclease Domain of Nsp14 of PEDV Leads to High Genetic Instability during Viral Replication. Cell Biosci. 2021, 11, 106. [Google Scholar] [CrossRef]
- Bouvet, M.; Lugari, A.; Posthuma, C.C.; Zevenhoven, J.C.; Bernard, S.; Betzi, S.; Imbert, I.; Canard, B.; Guillemot, J.-C.; Lécine, P.; et al. Coronavirus Nsp10, a Critical Co-Factor for Activation of Multiple Replicative Enzymes. J. Biol. Chem. 2014, 289, 25783–25796. [Google Scholar] [CrossRef]
- Yang, H.; Rao, Z. Structural Biology of SARS-CoV-2 and Implications for Therapeutic Development. Nat. Rev. Microbiol. 2021, 19, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.; Cortese, M.; Winter, S.L.; Wachsmuth-Melm, M.; Neufeldt, C.J.; Cerikan, B.; Stanifer, M.L.; Boulant, S.; Bartenschlager, R.; Chlanda, P. SARS-CoV-2 Structure and Replication Characterized by in Situ Cryo-Electron Tomography. Nat. Commun. 2020, 11, 5885. [Google Scholar] [CrossRef]
- Malone, B.; Urakova, N.; Snijder, E.J.; Campbell, E.A. Structures and Functions of Coronavirus Replication–Transcription Complexes and Their Relevance for SARS-CoV-2 Drug Design. Nat. Rev. Mol. Cell Biol. 2022, 23, 21–39. [Google Scholar] [CrossRef]
- Wong, L.H.; Edgar, J.R.; Martello, A.; Ferguson, B.J.; Eden, E.R. Exploiting Connections for Viral Replication. Front. Cell Dev. Biol. 2021, 9, 640456. [Google Scholar] [CrossRef]
- Roingeard, P.; Eymieux, S.; Burlaud-Gaillard, J.; Hourioux, C.; Patient, R.; Blanchard, E. The Double-Membrane Vesicle (DMV): A Virus-Induced Organelle Dedicated to the Replication of SARS-CoV-2 and Other Positive-Sense Single-Stranded RNA Viruses. Cell. Mol. Life Sci. 2022, 79, 425. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, T.; Zhong, L.; Zhang, W.; Zhang, Y.; Yu, X.; Yuan, S.; Ni, T. Molecular Architecture of Coronavirus Double-Membrane Vesicle Pore Complex. Nature 2024, 633, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.M.; Chen, Y.-J.; Cho, W.J.; Tai, A.W.; Tsai, B. Reticulons Promote Formation of ER-Derived Double-Membrane Vesicles That Facilitate SARS-CoV-2 Replication. J. Cell Biol. 2023, 222, e202203060. [Google Scholar] [CrossRef] [PubMed]
- Hagemeijer, M.C.; Monastyrska, I.; Griffith, J.; Van Der Sluijs, P.; Voortman, J.; Van Bergen En Henegouwen, P.M.; Vonk, A.M.; Rottier, P.J.M.; Reggiori, F.; De Haan, C.A.M. Membrane Rearrangements Mediated by Coronavirus Nonstructural Proteins 3 and 4. Virology 2014, 458–459, 125–135. [Google Scholar] [CrossRef]
- Alexandersen, S.; Chamings, A.; Bhatta, T.R. SARS-CoV-2 Genomic and Subgenomic RNAs in Diagnostic Samples Are Not an Indicator of Active Replication. Nat. Commun. 2020, 11, 6059. [Google Scholar] [CrossRef]
- Telwatte, S.; Martin, H.A.; Marczak, R.; Fozouni, P.; Vallejo-Gracia, A.; Kumar, G.R.; Murray, V.; Lee, S.; Ott, M.; Wong, J.K.; et al. Novel RT-ddPCR Assays for Measuring the Levels of Subgenomic and Genomic SARS-CoV-2 Transcripts. Methods 2022, 201, 15–25. [Google Scholar] [CrossRef]
- Markov, P.V.; Ghafari, M.; Beer, M.; Lythgoe, K.; Simmonds, P.; Stilianakis, N.I.; Katzourakis, A. The Evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 361–379. [Google Scholar] [CrossRef]
- Mack, A.H.; Menzies, G.; Southgate, A.; Jones, D.D.; Connor, T.R. A Proofreading Mutation with an Allosteric Effect Allows a Cluster of SARS-CoV-2 Viruses to Rapidly Evolve. Mol. Biol. Evol. 2023, 40, msad209. [Google Scholar] [CrossRef]
- Prydz, K.; Saraste, J. The Life Cycle and Enigmatic Egress of Coronaviruses. Mol. Microbiol. 2022, 117, 1308–1316. [Google Scholar] [CrossRef]
- Perdikari, T.M.; Murthy, A.C.; Ryan, V.H.; Watters, S.; Naik, M.T.; Fawzi, N.L. SARS-CoV-2 Nucleocapsid Protein Phase-separates with RNA and with Human hnRNPs. EMBO J. 2020, 39, e106478. [Google Scholar] [CrossRef]
- Pearson, G.J.; Mears, H.V.; Broncel, M.; Snijders, A.P.; Bauer, D.L.V.; Carlton, J.G. ER-Export and ARFRP1/AP-1–Dependent Delivery of SARS-CoV-2 Envelope to Lysosomes Controls Late Stages of Viral Replication. Sci. Adv. 2024, 10, eadl5012. [Google Scholar] [CrossRef]
- Gordon, D.E.; Hiatt, J.; Bouhaddou, M.; Rezelj, V.V.; Ulferts, S.; Braberg, H.; Jureka, A.S.; Obernier, K.; Guo, J.Z.; Batra, J.; et al. Comparative Host-Coronavirus Protein Interaction Networks Reveal Pan-Viral Disease Mechanisms. Science 2020, 370, eabe9403. [Google Scholar] [CrossRef]
- Smith, T.; Wang, S.; Kwon, Y.; Reid, A.A.; Robison, R.; Shen, P.; Willardson, B. Folding of the SARS-CoV-2 RNA Polymerase by the Cytosolic Chaperonin CCT. FASEB J. 2022, 36. [Google Scholar] [CrossRef]
- Alruwaili, M.; Armstrong, S.; Prince, T.; Erdmann, M.; Matthews, D.A.; Davidson, A.; Aljabr, W.; Hiscox, J.A. SARS-CoV-2 NSP12 Associates with the TRiC Complex and the P323L Substitution Is a Host Adaption. bioRxiv 2023. [Google Scholar]
- Liu, Q.; Zhao, S.; Hou, Y.; Ye, S.; Sha, T.; Su, Y.; Zhao, W.; Bao, Y.; Xue, Y.; Chen, H. Ongoing Natural Selection Drives the Evolution of SARS-CoV-2 Genomes. MedRxiv 2020. [Google Scholar]
- Shi, J.; Du, T.; Wang, J.; Tang, C.; Lei, M.; Yu, W.; Yang, Y.; Ma, Y.; Huang, P.; Chen, H.; et al. Aryl Hydrocarbon Receptor Is a Proviral Host Factor and a Candidate Pan-SARS-CoV-2 Therapeutic Target. Sci. Adv. 2023, 9, eadf0211. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Pu, Y.; Yuan, S.; Pache, L.; Churas, C.; Weston, S.; Riva, L.; Simons, L.M.; Cisneros, W.; Clausen, T.; et al. Global siRNA Screen Reveals Critical Human Host Factors of SARS-CoV-2 Multicycle Replication. bioRxiv 2024. [Google Scholar]
- Jockusch, S.; Tao, C.; Li, X.; Anderson, T.K.; Chien, M.; Kumar, S.; Russo, J.J.; Kirchdoerfer, R.N.; Ju, J. A Library of Nucleotide Analogues Terminate RNA Synthesis Catalyzed by Polymerases of Coronaviruses That Cause SARS and COVID-19. Antivir. Res. 2020, 180, 104857. [Google Scholar] [CrossRef]
- Li, G.; De Clercq, E. Therapeutic Options for the 2019 Novel Coronavirus (2019-nCoV). Nat. Rev. Drug Discov. 2020, 19, 149–150. [Google Scholar] [CrossRef]
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the Treatment of Covid-19—Final Report. N. Engl. J. Med. 2020, 383, 1813–1826. [Google Scholar] [CrossRef]
- Olender, S.A.; Perez, K.K.; Go, A.S.; Balani, B.; Price-Haywood, E.G.; Shah, N.S.; Wang, S.; Walunas, T.L.; Swaminathan, S.; Slim, J.; et al. Remdesivir for Severe Coronavirus Disease 2019 (COVID-19) Versus a Cohort Receiving Standard of Care. Clin. Infect. Dis. 2021, 73, e4166–e4174. [Google Scholar] [CrossRef] [PubMed]
- Ogando, N.S.; Zevenhoven-Dobbe, J.C.; Van Der Meer, Y.; Bredenbeek, P.J.; Posthuma, C.C.; Snijder, E.J. The Enzymatic Activity of the Nsp14 Exoribonuclease Is Critical for Replication of MERS-CoV and SARS-CoV-2. J. Virol. 2020, 94, e01246-20. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.-Y.; Lahser, F.; Warren, C.; He, X.; Murray, E.; Wang, D. The Impact of SARS-CoV-2 Nsp14 Proofreading on Nucleoside Antiviral Activity: Insights from Genetic and Pharmacological Investigations. bioRxiv 2024. [Google Scholar]
- Lee, C.-C.; Hsieh, C.-C.; Ko, W.-C. Molnupiravir—A Novel Oral Anti-SARS-CoV-2 Agent. Antibiotics 2021, 10, 1294. [Google Scholar] [CrossRef]
- Kabinger, F.; Stiller, C.; Schmitzová, J.; Dienemann, C.; Kokic, G.; Hillen, H.S.; Höbartner, C.; Cramer, P. Mechanism of Molnupiravir-Induced SARS-CoV-2 Mutagenesis. Nat. Struct. Mol. Biol. 2021, 28, 740–746. [Google Scholar] [CrossRef]
- Tian, L.; Pang, Z.; Li, M.; Lou, F.; An, X.; Zhu, S.; Song, L.; Tong, Y.; Fan, H.; Fan, J. Molnupiravir and Its Antiviral Activity Against COVID-19. Front. Immunol. 2022, 13, 855496. [Google Scholar] [CrossRef]
- Malone, B.; Campbell, E.A. Molnupiravir: Coding for Catastrophe. Nat. Struct. Mol. Biol. 2021, 28, 706–708. [Google Scholar] [CrossRef]
- Masyeni, S.; Iqhrammullah, M.; Frediansyah, A.; Nainu, F.; Tallei, T.; Emran, T.B.; Ophinni, Y.; Dhama, K.; Harapan, H. Molnupiravir: A Lethal Mutagenic Drug against Rapidly Mutating Severe Acute Respiratory Syndrome Coronavirus 2—A Narrative Review. J. Med. Virol. 2022, 94, 3006–3016. [Google Scholar] [CrossRef]
- Caraco, Y.; Crofoot, G.E.; Moncada, P.A.; Galustyan, A.N.; Musungaie, D.B.; Payne, B.; Kovalchuk, E.; Gonzalez, A.; Brown, M.L.; Williams-Diaz, A.; et al. Phase 2/3 Trial of Molnupiravir for Treatment of Covid-19 in Nonhospitalized Adults. NEJM Evid. 2022, 1. [Google Scholar] [CrossRef]
- Strizki, J.M.; Gaspar, J.M.; Howe, J.A.; Hutchins, B.; Mohri, H.; Nair, M.S.; Kinek, K.C.; McKenna, P.; Goh, S.L.; Murgolo, N. Molnupiravir Maintains Antiviral Activity against SARS-CoV-2 Variants and Exhibits a High Barrier to the Development of Resistance. Antimicrob. Agents Chemother. 2024, 68, e00953-23. [Google Scholar] [CrossRef]
- Erdem, H.A.; Korkmaz Ekren, P.; Çağlayan, D.; Işikgöz Taşbakan, M.; Yamazhan, T.; Taşbakan, M.S.; Sayiner, A.; Gökengi̇n, D. Treatment of SARS-CoV-2 Pneumonia with Favipiravir: Early Results from the Ege University Cohort, Turkey. Turk. J. Med. Sci. 2021, 51, 912–920. [Google Scholar] [CrossRef] [PubMed]
- Ghasemnejad-Berenji, M.; Pashapour, S. Favipiravir and COVID-19: A Simplified Summary. Drug Res. 2021, 71, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Manabe, T.; Kambayashi, D.; Akatsu, H.; Kudo, K. Favipiravir for the Treatment of Patients with COVID-19: A Systematic Review and Meta-Analysis. BMC Infect. Dis. 2021, 21, 489. [Google Scholar] [CrossRef]
- Juul, S.; Nielsen, E.E.; Feinberg, J.; Siddiqui, F.; Jørgensen, C.K.; Barot, E.; Holgersson, J.; Nielsen, N.; Bentzer, P.; Veroniki, A.A.; et al. Interventions for Treatment of COVID-19: Second Edition of a Living Systematic Review with Meta-Analyses and Trial Sequential Analyses (The LIVING Project). PLoS ONE 2021, 16, e0248132. [Google Scholar] [CrossRef]
- Shrestha, D.B.; Budhathoki, P.; Khadka, S.; Shah, P.B.; Pokharel, N.; Rashmi, P. Favipiravir versus Other Antiviral or Standard of Care for COVID-19 Treatment: A Rapid Systematic Review and Meta-Analysis. Virol. J. 2020, 17, 141. [Google Scholar] [CrossRef]
- Tong, S.; Su, Y.; Yu, Y.; Wu, C.; Chen, J.; Wang, S.; Jiang, J. Ribavirin Therapy for Severe COVID-19: A Retrospective Cohort Study. Int. J. Antimicrob. Agents 2020, 56, 106114. [Google Scholar] [CrossRef] [PubMed]
- Messina, E.; Danise, A.; Ferrari, G.; Andolina, A.; Chiurlo, M.; Razanakolona, M.; Barakat, M.; Israel, R.J.; Castagna, A. Ribavirin Aerosol in the Treatment of SARS-CoV-2: A Case Series. Infect. Dis. Ther. 2021, 10, 2791–2804. [Google Scholar] [CrossRef]
- Qian, H.; Wang, Y.; Zhang, M.; Xie, Y.; Wu, Q.; Liang, L.; Cao, Y.; Duan, H.; Tian, G.; Ma, J.; et al. Safety, Tolerability, and Pharmacokinetics of VV116, an Oral Nucleoside Analog against SARS-CoV-2, in Chinese Healthy Subjects. Acta Pharmacol. Sin. 2022, 43, 3130–3138. [Google Scholar] [CrossRef]
- Xiao, N.; Huang, X.; Kang, X.; Zang, W.; Li, B.; Kiselev, S. The Safety and Efficacy of Oral Antiviral Drug VV116 for Treatment of COVID-19: A Systematic Review. Medicine 2023, 102, e34105. [Google Scholar] [CrossRef]
- McCarthy, M.W. VV116 as a Potential Treatment for COVID-19. Expert Opin. Pharmacother. 2023, 24, 675–678. [Google Scholar] [CrossRef]
- Hammond, J.; Leister-Tebbe, H.; Gardner, A.; Abreu, P.; Bao, W.; Wisemandle, W.; Baniecki, M.; Hendrick, V.M.; Damle, B.; Simón-Campos, A.; et al. Oral Nirmatrelvir for High-Risk, Nonhospitalized Adults with Covid-19. N. Engl. J. Med. 2022, 386, 1397–1408. [Google Scholar] [CrossRef]
- Marzi, M.; Vakil, M.K.; Bahmanyar, M.; Zarenezhad, E. Paxlovid: Mechanism of Action, Synthesis, and In Silico Study. BioMed Res. Int. 2022, 2022, 7341493. [Google Scholar] [CrossRef] [PubMed]
- Marzolini, C.; Kuritzkes, D.R.; Marra, F.; Boyle, A.; Gibbons, S.; Flexner, C.; Pozniak, A.; Boffito, M.; Waters, L.; Burger, D.; et al. Recommendations for the Management of Drug–Drug Interactions Between the COVID-19 Antiviral Nirmatrelvir/Ritonavir (Paxlovid) and Comedications. Clin. Pharma Ther. 2022, 112, 1191–1200. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Gammeltoft, K.A.; Ryberg, L.A.; Pham, L.V.; Fahnøe, U.; Binderup, A.; Hernandez, C.R.D.; Offersgaard, A.; Fernandez-Antunez, C.; Peters, G.H.J.; et al. Nirmatrelvir Resistant SARS-CoV-2 Variants with High Fitness in Vitro. Sci. Adv. 2022, 51, eadd7197. [Google Scholar] [CrossRef] [PubMed]
- Iketani, S.; Mohri, H.; Culbertson, B.; Hong, S.J.; Duan, Y.; Luck, M.I.; Annavajhala, M.K.; Guo, Y.; Sheng, Z.; Uhlemann, A.-C.; et al. Multiple Pathways for SARS-CoV-2 Resistance to Nirmatrelvir. Nature 2022, 613, 558–564. [Google Scholar] [CrossRef]
- Costacurta, F.; Dodaro, A.; Bante, D.; Schöppe, H.; Peng, J.-Y.; Sprenger, B.; He, X.; Moghadasi, S.A.; Egger, L.M.; Fleischmann, J.; et al. A Comprehensive Study of SARS-CoV-2 Main Protease (Mpro) Inhibitor-Resistant Mutants Selected in a VSV-Based System. PLoS Pathog. 2024, 20, e1012522. [Google Scholar] [CrossRef]
- Cao, B.; Wang, Y.; Wen, D.; Liu, W.; Wang, J.; Fan, G.; Ruan, L.; Song, B.; Cai, Y.; Wei, M.; et al. A Trial of Lopinavir–Ritonavir in Adults Hospitalized with Severe Covid-19. N. Engl. J. Med. 2020, 382, 1787–1799. [Google Scholar] [CrossRef]
- Kuroda, T.; Nobori, H.; Fukao, K.; Baba, K.; Matsumoto, K.; Yoshida, S.; Tanaka, Y.; Watari, R.; Oka, R.; Kasai, Y.; et al. Efficacy Comparison of 3CL Protease Inhibitors Ensitrelvir and Nirmatrelvir against SARS-CoV-2 in Vitro and in Vivo. J. Antimicrob. Chemother. 2023, 78, 946–952. [Google Scholar] [CrossRef]
- Garnsey, M.R.; Robinson, M.C.; Nguyen, L.T.; Cardin, R.; Tillotson, J.; Mashalidis, E.; Yu, A.; Aschenbrenner, L.; Balesano, A.; Behzadi, A.; et al. Discovery of SARS-CoV-2 Papain-like Protease (PLpro ) Inhibitors with Efficacy in a Murine Infection Model. Sci. Adv. 2024, 10, eado4288. [Google Scholar] [CrossRef]
- Brunotte, L.; Zheng, S.; Mecate-Zambrano, A.; Tang, J.; Ludwig, S.; Rescher, U.; Schloer, S. Combination Therapy with Fluoxetine and the Nucleoside Analog GS-441524 Exerts Synergistic Antiviral Effects against Different SARS-CoV-2 Variants In Vitro. Pharmaceutics 2021, 13, 1400. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Liu, M.; Wang, C.; Xu, W.; Lan, Q.; Feng, S.; Qi, F.; Bao, L.; Du, L.; Liu, S.; et al. Inhibition of SARS-CoV-2 (Previously 2019-nCoV) Infection by a Highly Potent Pan-Coronavirus Fusion Inhibitor Targeting Its Spike Protein That Harbors a High Capacity to Mediate Membrane Fusion. Cell Res. 2020, 30, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.; Chen, X.; Wu, J.; Duan, X.; Men, K. Small Molecules in the Treatment of COVID-19. Signal Transduct. Target. Ther. 2022, 7, 387. [Google Scholar] [CrossRef]
- Batool, S.; Chokkakula, S.; Jeong, J.H.; Baek, Y.H.; Song, M.-S. SARS-CoV-2 Drug Resistance and Therapeutic Approaches. Heliyon 2025, 11, e41980. [Google Scholar] [CrossRef]
- Rahmah, L.; Abarikwu, S.O.; Arero, A.G.; Essouma, M.; Jibril, A.T.; Fal, A.; Flisiak, R.; Makuku, R.; Marquez, L.; Mohamed, K.; et al. Oral Antiviral Treatments for COVID-19: Opportunities and Challenges. Pharmacol. Rep. 2022, 74, 1255–1278. [Google Scholar] [CrossRef]
- Gidari, A.; Sabbatini, S.; Schiaroli, E.; Bastianelli, S.; Pierucci, S.; Busti, C.; Saraca, L.M.; Capogrossi, L.; Pasticci, M.B.; Francisci, D. Synergistic Activity of Remdesivir–Nirmatrelvir Combination on a SARS-CoV-2 In Vitro Model and a Case Report. Viruses 2023, 15, 1577. [Google Scholar] [CrossRef]
- Gidari, A.; Sabbatini, S.; Schiaroli, E.; Bastianelli, S.; Pierucci, S.; Busti, C.; Comez, L.; Libera, V.; Macchiarulo, A.; Paciaroni, A.; et al. The Combination of Molnupiravir with Nirmatrelvir or GC376 Has a Synergic Role in the Inhibition of SARS-CoV-2 Replication In Vitro. Microorganisms 2022, 10, 1475. [Google Scholar] [CrossRef] [PubMed]
- Woodall, M.; Ellis, S.; Zhang, S.; Kembou-Ringert, J.; Kite, K.-A.; Buggiotti, L.; Jacobs, A.I.; Agyeman, A.A.; Masonou, T.; Palor, M.; et al. Efficient in Vitro Assay for Evaluating Drug Efficacy and Synergy against Emerging SARS-CoV-2 Strains. Antimicrob. Agents Chemother. 2025, 69, e01233-24. [Google Scholar] [CrossRef]
- García-Crespo, C.; De Ávila, A.I.; Gallego, I.; Soria, M.E.; Durán-Pastor, A.; Somovilla, P.; Martínez-González, B.; Muñoz-Flores, J.; Mínguez, P.; Salar-Vidal, L.; et al. Synergism between Remdesivir and Ribavirin Leads to SARS-CoV-2 Extinction in Cell Culture. Br. J. Pharmacol. 2024, 181, 2636–2654. [Google Scholar] [CrossRef]
- Robson, F.; Khan, K.S.; Le, T.K.; Paris, C.; Demirbag, S.; Barfuss, P.; Rocchi, P.; Ng, W.-L. Coronavirus RNA Proofreading: Molecular Basis and Therapeutic Targeting. Mol. Cell 2020, 79, 710–727. [Google Scholar] [CrossRef]
- Do, T.N.D.; Abdelnabi, R.; Boda, B.; Constant, S.; Neyts, J.; Jochmans, D. The Triple Combination of Remdesivir (GS-441524), Molnupiravir and Ribavirin Is Highly Efficient in Inhibiting Coronavirus Replication in Human Nasal Airway Epithelial Cell Cultures and in a Hamster Infection Model. Antivir. Res. 2024, 231, 105994. [Google Scholar] [CrossRef] [PubMed]
- Mayor, J.; Engler, O.; Rothenberger, S. Antiviral Efficacy of Ribavirin and Favipiravir against Hantaan Virus. Microorganisms 2021, 9, 1306. [Google Scholar] [CrossRef]
- Wagoner, J.; Herring, S.; Hsiang, T.-Y.; Ianevski, A.; Biering, S.B.; Xu, S.; Hoffmann, M.; Pöhlmann, S.; Gale, M.; Aittokallio, T.; et al. Combinations of Host- and Virus-Targeting Antiviral Drugs Confer Synergistic Suppression of SARS-CoV-2. Microbiol. Spectr. 2022, 10, e03331-22. [Google Scholar] [CrossRef]
- White, J.M.; Schiffer, J.T.; Bender Ignacio, R.A.; Xu, S.; Kainov, D.; Ianevski, A.; Aittokallio, T.; Frieman, M.; Olinger, G.G.; Polyak, S.J. Drug Combinations as a First Line of Defense against Coronaviruses and Other Emerging Viruses. mBio 2021, 12, e03347-21. [Google Scholar] [CrossRef]
- Jeong, J.H.; Chokkakula, S.; Min, S.C.; Kim, B.K.; Choi, W.-S.; Oh, S.; Yun, Y.S.; Kang, D.H.; Lee, O.-J.; Kim, E.-G.; et al. Combination Therapy with Nirmatrelvir and Molnupiravir Improves the Survival of SARS-CoV-2 Infected Mice. Antivir. Res. 2022, 208, 105430. [Google Scholar] [CrossRef]
- Mahendran, T.R.; Cynthia, B.; Thevendran, R.; Maheswaran, S. Prospects of Innovative Therapeutics in Combating the COVID-19 Pandemic. Mol. Biotechnol. 2024, 67, 2598–2606. [Google Scholar] [CrossRef]
- Abdulaziz, L.; Elhadi, E.; Abdallah, E.A.; Alnoor, F.A.; Yousef, B.A. Antiviral Activity of Approved Antibacterial, Antifungal, Antiprotozoal and Anthelmintic Drugs: Chances for Drug Repurposing for Antiviral Drug Discovery. JEP 2022, 14, 97–115. [Google Scholar] [CrossRef] [PubMed]
- Hamid, A.; Mäser, P.; Mahmoud, A.B. Drug Repurposing in the Chemotherapy of Infectious Diseases. Molecules 2024, 29, 635. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, B.; Thakur, S.S. Remdesivir and Its Combination With Repurposed Drugs as COVID-19 Therapeutics. Front. Immunol. 2022, 13, 830990. [Google Scholar] [CrossRef]
- Mahdi, M.; Hermán, L.; Réthelyi, J.M.; Bálint, B.L. Potential Role of the Antidepressants Fluoxetine and Fluvoxamine in the Treatment of COVID-19. IJMS 2022, 23, 3812. [Google Scholar] [CrossRef]
- Al-Hajeri, H.; Baroun, F.; Abutiban, F.; Al-Mutairi, M.; Ali, Y.; Alawadhi, A.; Albasri, A.; Aldei, A.; AlEnizi, A.; Alhadhood, N.; et al. Therapeutic Role of Immunomodulators during the COVID-19 Pandemic– a Narrative Review. Postgrad. Med. 2022, 134, 160–179. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, S.C.J.; Tse, C.L.Y.; Burry, L.; Dresser, L.D. Baricitinib: A Review of Pharmacology, Safety, and Emerging Clinical Experience in COVID-19. Pharmacotherapy 2020, 40, 843–856. [Google Scholar] [CrossRef]
- Frediansyah, A.; Tiwari, R.; Sharun, K.; Dhama, K.; Harapan, H. Antivirals for COVID-19: A Critical Review. Clin. Epidemiol. Glob. Health 2021, 9, 90–98. [Google Scholar] [CrossRef]
- Villa, T.G.; Abril, A.G.; Sánchez, S.; De Miguel, T.; Sánchez-Pérez, A. Animal and Human RNA Viruses: Genetic Variability and Ability to Overcome Vaccines. Arch. Microbiol. 2021, 203, 443–464. [Google Scholar] [CrossRef]
- Checkmahomed, L.; Carbonneau, J.; Du Pont, V.; Riola, N.C.; Perry, J.K.; Li, J.; Paré, B.; Simpson, S.M.; Smith, M.A.; Porter, D.P.; et al. In Vitro Selection of Remdesivir-Resistant SARS-CoV-2 Demonstrates High Barrier to Resistance. Antimicrob. Agents Chemother. 2022, 66, e00198-22. [Google Scholar] [CrossRef] [PubMed]
- Szemiel, A.M.; Merits, A.; Orton, R.J.; MacLean, O.A.; Pinto, R.M.; Wickenhagen, A.; Lieber, G.; Turnbull, M.L.; Wang, S.; Furnon, W.; et al. In Vitro Selection of Remdesivir Resistance Suggests Evolutionary Predictability of SARS-CoV-2. PLoS Pathog. 2021, 17, e1009929. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, T.; Hisner, R.; Donovan-Banfield, I.; Hartman, H.; Løchen, A.; Peacock, T.P.; Ruis, C. A Molnupiravir-Associated Mutational Signature in Global SARS-CoV-2 Genomes. Nature 2023, 623, 594–600. [Google Scholar] [CrossRef]
- Zhao, Y.; He, G.; Huang, W. A Novel Model of Molnupiravir against SARS-CoV-2 Replication: Accumulated RNA Mutations to Induce Error Catastrophe. Sig Transduct. Target. Ther. 2021, 6, 410. [Google Scholar] [CrossRef]
- Lee, J.T.; Yang, Q.; Gribenko, A.; Perrin, B.S.; Zhu, Y.; Cardin, R.; Liberator, P.A.; Anderson, A.S.; Hao, L. Genetic Surveillance of SARS-CoV-2 Mpro Reveals High Sequence and Structural Conservation Prior to the Introduction of Protease Inhibitor Paxlovid. mBio 2022, 13, e00869-22. [Google Scholar] [CrossRef]
- Peluso, M.J.; Deeks, S.G. Mechanisms of Long COVID and the Path toward Therapeutics. Cell 2024, 187, 5500–5529. [Google Scholar] [CrossRef]
- Lupașcu (Moisi), R.E.; Ilie, M.I.; Velescu, B.Ș.; Udeanu, D.I.; Sultana, C.; Ruță, S.; Arsene, A.L. COVID-19-Current Therapeutical Approaches and Future Perspectives. Processes 2022, 10, 1053. [Google Scholar] [CrossRef]
- Chala, B.; Tilaye, T.; Waktole, G. Re-Emerging COVID-19: Controversy of Its Zoonotic Origin, Risks of Severity of Reinfection and Management. IJGM 2023, 16, 4307–4319. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Zheng, M.; Gao, Y.; Lin, S.; Zhang, X.; Chen, C.; Zhu, H.; Sun, W.; Zhang, Y. Overview of Host-Directed Antiviral Targets for Future Research and Drug Development. Acta Pharm. Sin. B 2025, 15, 1723–1751. [Google Scholar] [CrossRef]
- Schuller, M.; Zarganes-Tzitzikas, T.; Bennett, J.; De Cesco, S.; Fearon, D.; Von Delft, F.; Fedorov, O.; Brennan, P.E.; Ahel, I. Discovery and Development Strategies for SARS-CoV-2 NSP3 Macrodomain Inhibitors. Pathogens 2023, 12, 324. [Google Scholar] [CrossRef]
- Petushkova, A.I.; Zamyatnin, A.A. Papain-Like Proteases as Coronaviral Drug Targets: Current Inhibitors, Opportunities, and Limitations. Pharmaceuticals 2020, 13, 277. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, M.; Damalanka, V.C.; Tartell, M.A.; Chung, D.H.; Lourenço, A.L.; Pwee, D.; Mayer Bridwell, A.E.; Hoffmann, M.; Voss, J.; Karmakar, P.; et al. A Novel Class of TMPRSS2 Inhibitors Potently Block SARS-CoV-2 and MERS-CoV Viral Entry and Protect Human Epithelial Lung Cells. Proc. Natl. Acad. Sci. USA 2021, 118, e2108728118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yu, W. Heat Shock Proteins and Viral Infection. Front. Immunol. 2022, 13, 947789. [Google Scholar] [CrossRef]
- Wickramaratne, A.C.; Wickner, S.; Kravats, A.N. Hsp90, a Team Player in Protein Quality Control and the Stress Response in Bacteria. Microbiol. Mol. Biol. Rev. 2024, 88, e00176-22. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, T.; Song, M.; Jin, M.; Liu, S.; Guo, K.; Zhang, Y. Rab1b-GBF1-ARFs Mediated Intracellular Trafficking Is Required for Classical Swine Fever Virus Replication in Swine Umbilical Vein Endothelial Cells. Vet. Microbiol. 2020, 246, 108743. [Google Scholar] [CrossRef]
- Chikhoune, L.; Poggi, C.; Moreau, J.; Dubucquoi, S.; Hachulla, E.; Collet, A.; Launay, D. JAK Inhibitors (JAKi): Mechanisms of Action and Perspectives in Systemic and Autoimmune Diseases. La. Rev. De Médecine Interne 2025, 46, 89–106. [Google Scholar] [CrossRef]
- Sodeifian, F.; Nikfarjam, M.; Kian, N.; Mohamed, K.; Rezaei, N. The Role of Type I Interferon in the Treatment of COVID-19. J. Med. Virol. 2022, 94, 63–81. [Google Scholar] [CrossRef] [PubMed]
- Savan, R.; Gale, M. Innate Immunity and Interferon in SARS-CoV-2 Infection Outcome. Immunity 2023, 56, 1443–1450. [Google Scholar] [CrossRef] [PubMed]
- Floresta, G.; Zagni, C.; Gentile, D.; Patamia, V.; Rescifina, A. Artificial Intelligence Technologies for COVID-19 De Novo Drug Design. IJMS 2022, 23, 3261. [Google Scholar] [CrossRef]
- Chodera, J.; Lee, A.A.; London, N.; Von Delft, F. Crowdsourcing Drug Discovery for Pandemics. Nat. Chem. 2020, 12, 581. [Google Scholar] [CrossRef]
- Faisal, S.; Badshah, S.L.; Kubra, B.; Sharaf, M.; Emwas, A.-H.; Jaremko, M.; Abdalla, M. Computational Study of SARS-CoV-2 RNA Dependent RNA Polymerase Allosteric Site Inhibition. Molecules 2021, 27, 223. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Yang, S.; He, S.; Li, F. AI Drug Discovery Tools and Analysis Technology: New Methods Aid in Studying the Compatibility of Traditional Chinese Medicine. Pharmacol. Res. Mod. Chin. Med. 2025, 14, 100566. [Google Scholar] [CrossRef]
- Maghsoudi, S.; Taghavi Shahraki, B.; Rameh, F.; Nazarabi, M.; Fatahi, Y.; Akhavan, O.; Rabiee, M.; Mostafavi, E.; Lima, E.C.; Saeb, M.R.; et al. A Review on Computer-aided Chemogenomics and Drug Repositioning for Rational COVID-19 Drug Discovery. Chem. Biol. Drug Des. 2022, 100, 699–721. [Google Scholar] [CrossRef]
- Mottaqi, M.S.; Mohammadipanah, F.; Sajedi, H. Contribution of Machine Learning Approaches in Response to SARS-CoV-2 Infection. Inform. Med. Unlocked 2021, 23, 100526. [Google Scholar] [CrossRef]
- Rajput, A.; Thakur, A.; Mukhopadhyay, A.; Kamboj, S.; Rastogi, A.; Gautam, S.; Jassal, H.; Kumar, M. Prediction of Repurposed Drugs for Coronaviruses Using Artificial Intelligence and Machine Learning. Comput. Struct. Biotechnol. J. 2021, 19, 3133–3148. [Google Scholar] [CrossRef]
- Huchting, J. Targeting Viral Genome Synthesis as Broad-Spectrum Approach against RNA Virus Infections. Antivir. Chem. Chemother. 2020, 28, 204020662097678. [Google Scholar] [CrossRef]
- Tolksdorf, B.; Heinze, J.; Niemeyer, D.; Röhrs, V.; Berg, J.; Drosten, C.; Kurreck, J. Development of a Highly Stable, Active Small Interfering RNA with Broad Activity against SARS-CoV Viruses. Antivir. Res. 2024, 226, 105879. [Google Scholar] [CrossRef] [PubMed]
- Abbott, T.R.; Dhamdhere, G.; Liu, Y.; Lin, X.; Goudy, L.; Zeng, L.; Chemparathy, A.; Chmura, S.; Heaton, N.S.; Debs, R.; et al. Development of CRISPR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza. Cell 2020, 181, 865–876.e12. [Google Scholar] [CrossRef]
- Li, T.; Yang, Y.; Qi, H.; Cui, W.; Zhang, L.; Fu, X.; He, X.; Liu, M.; Li, P.; Yu, T. CRISPR/Cas9 Therapeutics: Progress and Prospects. Signal Transduct. Target. Ther. 2023, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Najafi, S.; Tan, S.C.; Aghamiri, S.; Raee, P.; Ebrahimi, Z.; Jahromi, Z.K.; Rahmati, Y.; Sadri Nahand, J.; Piroozmand, A.; Jajarmi, V.; et al. Therapeutic Potentials of CRISPR-Cas Genome Editing Technology in Human Viral Infections. Biomed. Pharmacother. 2022, 148, 112743. [Google Scholar] [CrossRef] [PubMed]
- Zhong, G.; Chang, X.; Xie, W.; Zhou, X. Targeted Protein Degradation: Advances in Drug Discovery and Clinical Practice. Signal Transduct. Target. Ther. 2024, 9, 308. [Google Scholar] [CrossRef]
- Kubryń, N.; Fijałkowski, Ł.; Nowaczyk, J.; Jamil, A.; Nowaczyk, A. PROTAC Technology as a New Tool for Modern Pharmacotherapy. Molecules 2025, 30, 2123. [Google Scholar] [CrossRef]
- Liu, Z.; Hu, M.; Yang, Y.; Du, C.; Zhou, H.; Liu, C.; Chen, Y.; Fan, L.; Ma, H.; Gong, Y.; et al. An Overview of PROTACs: A Promising Drug Discovery Paradigm. Mol. Biomed. 2022, 3, 46. [Google Scholar] [CrossRef]
- Reboud-Ravaux, M.; El Amri, C. COVID-19 Therapies: Protease Inhibitions and Novel Degrader Strategies. Front. Drug Discov. 2022, 2, 892057. [Google Scholar] [CrossRef]
- Low, Z.Y.; Zabidi, N.Z.; Yip, A.J.W.; Puniyamurti, A.; Chow, V.T.K.; Lal, S.K. SARS-CoV-2 Non-Structural Proteins and Their Roles in Host Immune Evasion. Viruses 2022, 14, 1991. [Google Scholar] [CrossRef]
- Alugubelli, Y.R.; Xiao, J.; Khatua, K.; Kumar, S.; Sun, L.; Ma, Y.; Ma, X.R.; Vulupala, V.R.; Atla, S.; Blankenship, L.R.; et al. Discovery of First-in-Class PROTAC Degraders of SARS-CoV-2 Main Protease. J. Med. Chem. 2024, 67, 6495–6507. [Google Scholar] [CrossRef]
- Pan, B.; Mountford, S.J.; Kiso, M.; Anderson, D.E.; Papadakis, G.; Jarman, K.E.; Tilmanis, D.R.; Maher, B.; Tran, T.; Shortt, J.; et al. Targeted Protein Degraders of SARS-CoV-2 Mpro Are More Active than Enzymatic Inhibition Alone with Activity against Nirmatrelvir Resistant Virus. Commun. Med. 2025, 5, 140. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhao, J.; Zhong, K.; Tong, A.; Jia, D. Targeted Protein Degradation: Mechanisms, Strategies and Application. Signal Transduct. Target. Ther. 2022, 7, 113. [Google Scholar] [CrossRef] [PubMed]
- Von Delft, A.; Hall, M.D.; Kwong, A.D.; Purcell, L.A.; Saikatendu, K.S.; Schmitz, U.; Tallarico, J.A.; Lee, A.A. Accelerating Antiviral Drug Discovery: Lessons from COVID-19. Nat. Rev. Drug Discov. 2023, 22, 585–603. [Google Scholar] [CrossRef]
- Schreiber, A.; Ludwig, S. Host-Targeted Antivirals against SARS-CoV-2 in Clinical Development—Prospect or Disappointment? Antivir. Res. 2025, 235, 106101. [Google Scholar] [CrossRef]
- Seley-Radtke, K.L.; Thames, J.E.; Waters, C.D. Broad Spectrum Antiviral Nucleosides—Our Best Hope for the Future. In Annual Reports in Medicinal Chemistry; Elsevier: Amsterdam, The Netherlands, 2021; Volume 57, pp. 109–132. ISBN 978-0-323-91511-3. [Google Scholar]
- Kirchdoerfer, R.N.; Ward, A.B. Structure of the SARS-CoV Nsp12 Polymerase Bound to Nsp7 and Nsp8 Co-Factors. Nat. Commun. 2019, 10, 2342. [Google Scholar] [CrossRef]
- Xiong, R.; Zhang, L.; Li, S.; Sun, Y.; Ding, M.; Wang, Y.; Zhao, Y.; Wu, Y.; Shang, W.; Jiang, X.; et al. Novel and Potent Inhibitors Targeting DHODH Are Broad-Spectrum Antivirals against RNA Viruses Including Newly-Emerged Coronavirus SARS-CoV-2. Protein Cell 2020, 11, 723–739. [Google Scholar] [CrossRef]
- Sievers, B.L.; Cheng, M.T.K.; Csiba, K.; Meng, B.; Gupta, R.K. SARS-CoV-2 and Innate Immunity: The Good, the Bad, and the “Goldilocks”. Cell Mol. Immunol. 2023, 21, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Italiya, J.; Bhavsar, T.; Černý, J. Assessment and Strategy Development for SARS-CoV-2 Screening in Wildlife: A Review. Vet. World 2023, 16, 1193–1200. [Google Scholar] [CrossRef] [PubMed]
- Karim, M.; Lo, C.-W.; Einav, S. Preparing for the next Viral Threat with Broad-Spectrum Antivirals. J. Clin. Investig. 2023, 133, e170236. [Google Scholar] [CrossRef]
- Barghash, R.F.; Gemmati, D.; Awad, A.M.; Elbakry, M.M.M.; Tisato, V.; Awad, K.; Singh, A.V. Navigating the COVID-19 Therapeutic Landscape: Unveiling Novel Perspectives on FDA-Approved Medications, Vaccination Targets, and Emerging Novel Strategies. Molecules 2024, 29, 5564. [Google Scholar] [CrossRef]
- Aligolighasemabadi, F.; Bakinowska, E.; Kiełbowski, K.; Sadeghdoust, M.; Coombs, K.M.; Mehrbod, P.; Ghavami, S. Autophagy and Respiratory Viruses: Mechanisms, Viral Exploitation, and Therapeutic Insights. Cells 2025, 14, 418. [Google Scholar] [CrossRef] [PubMed]
- Farkaš, B.; Minneci, M.; Misevicius, M.; Rozas, I. A Tale of Two Proteases: MPro and TMPRSS2 as Targets for COVID-19 Therapies. Pharmaceuticals 2023, 16, 834. [Google Scholar] [CrossRef] [PubMed]
- Tani, H. Recent Advances and Prospects in RNA Drug Development. IJMS 2024, 25, 12284. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Hilles, A.R.; Almurisi, S.H.; Bhatia, A.; Mahmood, S. Micro and Nano-Carriers-Based Pulmonary Drug Delivery System: Their Current Updates, Challenges, and Limitations—A Review. JCIS Open 2023, 12, 100095. [Google Scholar] [CrossRef]
- Rehman, A.U.; Li, M.; Wu, B.; Ali, Y.; Rasheed, S.; Shaheen, S.; Liu, X.; Luo, R.; Zhang, J. Role of Artificial Intelligence in Revolutionizing Drug Discovery. Fundam. Res. 2024, 5, 1273–1287. [Google Scholar] [CrossRef]
- Li, G.; Hilgenfeld, R.; Whitley, R.; De Clercq, E. Therapeutic Strategies for COVID-19: Progress and Lessons Learned. Nat. Rev. Drug Discov. 2023, 22, 449–475. [Google Scholar] [CrossRef]
- Paunovska, K.; Loughrey, D.; Dahlman, J.E. Drug Delivery Systems for RNA Therapeutics. Nat. Rev. Genet. 2022, 23, 265–280. [Google Scholar] [CrossRef]
- De Jesús-González, L.A.; León-Juárez, M.; Lira-Hernández, F.I.; Rivas-Santiago, B.; Velázquez-Cervantes, M.A.; Méndez-Delgado, I.M.; Macías-Guerrero, D.I.; Hernández-Castillo, J.; Hernández-Rodríguez, X.; Calderón-Sandate, D.N.; et al. Advances and Challenges in Antiviral Development for Respiratory Viruses. Pathogens 2024, 14, 20. [Google Scholar] [CrossRef]
- Huang, Y.; Guo, X.; Wu, Y.; Chen, X.; Feng, L.; Xie, N.; Shen, G. Nanotechnology’s Frontier in Combatting Infectious and Inflammatory Diseases: Prevention and Treatment. Signal Transduct. Target. Ther. 2024, 9, 34. [Google Scholar] [CrossRef]
- Fu, C.; Chen, Q. The Future of Pharmaceuticals: Artificial Intelligence in Drug Discovery and Development. J. Pharm. Anal. 2025, 101248. [Google Scholar] [CrossRef]
- Chakravarty, A.; Yang, P.L. Targeted Protein Degradation as an Antiviral Approach. Antivir. Res. 2023, 210, 105480. [Google Scholar] [CrossRef] [PubMed]
- Chitalia, V.C.; Munawar, A.H. A Painful Lesson from the COVID-19 Pandemic: The Need for Broad-Spectrum, Host-Directed Antivirals. J. Transl. Med. 2020, 18, 390. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).