1. Introduction
The human immune system is a marvel of biological defense, orchestrated by an intricate web of molecules and cells which are designed to identify and eliminate invading pathogens. Key players in this complex immune response in humans are the human leukocyte antigens (HLAs), also known as the major histocompatibility complex (MHC). These proteins are responsible for capturing fragments of pathogens and presenting them to T cells, enabling the adaptive immune system to recognize and mount targeted defenses. The HLA system, characterized by its remarkable polymorphism, offers a vast array of allelic variations, allowing the immune system to engage with a diverse range of microbial peptides. This diversity, in turn, contributes to an individual’s unique immune responses and susceptibility or resistance to infections [
1,
2,
3].
HLA genes can significantly impact the outcome of viral infections. Notably, certain HLA alleles have been linked to susceptibility or resistance to viral pathogens such as HIV, hepatitis C, and influenza [
4,
5,
6]. These associations underscore the profound influence of HLA genetics on immune responses and disease outcomes. HLA polymorphisms result in differences in epitope binding affinity and the T cell repertoire developed during thymic selection, thus affecting the immune system’s ability to recognize and respond to viral threats [
7,
8].
The advent of SARS-CoV-2 in late 2019, leading to the global COVID-19 pandemic, has posed unprecedented challenges to public health systems worldwide. While the majority of individuals infected with SARS-CoV-2 experience mild to moderate symptoms or remain asymptomatic, a subset of patients develops severe—and, in some cases, fatal—pneumonia. Notably, the clinical outcomes of COVID-19 cannot be fully explained by demographics and pre-existing health conditions alone, suggesting that genetic factors may play a substantial role in determining the severity of the disease [
9].
Recent studies have begun to unravel the relationships between HLA alleles and the severity of COVID-19 infection. These investigations have associated specific HLA haplotypes and alleles with either protection against or increased risk of severe COVID-19, highlighting the significant influence of HLA genetics on the disease’s outcomes [
8,
10,
11]. The proposed mechanism underlying this association lies in the differential binding and presentation of viral epitopes by distinct HLA alleles, which can modulate the efficacy of the host’s immune response.
However, it is important to recognize that the relationship between HLA alleles and COVID-19 severity exhibits notable variations across different populations. The Arab world, with a total population of approximately 450 million people [
12], shares significant genetic material due to historical migration patterns and common ancestry [
13]. Saudi Arabia, with a population of about 35.5 million as of 2022 [
14], represents a significant portion of this genetic pool. Despite this large population base, there has been limited research examining HLA profiles and COVID-19 outcomes in Arab populations, with most existing studies focusing on Western populations. While substantial research has been conducted in certain populations, such as Italian or American patients, racially diverse non-Western groups such as Saudi Arabians remain relatively understudied. Considering the genetic diversity in Saudi Arabia and the limited COVID-19 genetic research in this population, there is a compelling need for population-specific investigations. Such studies can offer crucial insights into the intricacies of COVID-19 immunity under diverse genetic backgrounds and reveal unique facets regarding the epidemiology of the disease [
15,
16,
17].
This study seeks to contribute to the growing body of knowledge on the subject through investigating whether viral mutations in successive SARS-CoV-2 waves have led to escape from common HLA alleles in the Saudi Arabian population, with a primary focus on HLA class I alleles. We aim to explore the binding affinities of SARS-CoV-2 epitope peptides for select common HLA class I alleles which are prevalent in Saudis. Through assessing potential differences in binding affinities across recently dominant SARS-CoV-2 strains, we aim to shed light on the evolution of the virus and its impact on the immune system.
The significance of this research extends beyond its immediate context. It has the potential to uncover patterns of viral evolution and elucidate the mechanisms of immune evasion. The insights gained from this research may not only inform our understanding of T cell immunity in the natural course of infection, but also contribute to the design of more effective vaccines tailored to populations with specific HLA profiles.
2. Materials and Methods
This study analyzed HLA allele frequencies in a cohort of 45,457 potential stem cell donors registered at the King Faisal Specialist Hospital and Research Centre in Riyadh, Saudi Arabia [
18]. This donor population had existing HLA genotyping data available. HLA class I (HLA-A, HLA-B, HLA-C) and class II (HLA-DRB1, HLA-DQB1, HLA-DPB1) allele frequencies were determined for the cohort based on next-generation sequencing of exons 2 and 3, as previously described [
19].
Based on the HLA allele frequencies in the Saudi Arabian cohort, the three most common HLA class I alleles were selected for analysis: HLA-A_02:01, HLA-B_51:01, and HLA-C*06:02. These alleles had frequencies of 18.5%, 14.1%, and 16.1% in the population, respectively.
Viral epitope peptides with predicted or experimentally validated HLA binding for the alleles of interest were identified from the Immune Epitope Database (IEDB) [
20]. Epitopes located in the spike (S) protein were prioritized, as this is the major target of neutralizing antibodies and T cell responses [
21]. Seven epitopes were selected for each allele, based on the strength of predicted affinity and coverage of key regions (e.g., the receptor binding domain).
The binding affinities between the selected HLA class I alleles and corresponding SARS-CoV-2 epitope peptides were analyzed using the NetMHCpan 4.1 server [
22]. NetMHCpan uses artificial neural networks to predict peptide–HLA binding based on training using affinity and mass spectrometry data. For each allele, seven epitope peptides were selected based on their predicted binding affinity scores. Epitopes were categorized as strong binders (scores < 0.5), weak binders (scores between 0.5 and 2.0), or non-binders (scores > 2.0).
Four globally dominant SARS-CoV-2 strains were selected for comparison: Alpha, Epsilon, Gamma, and Omicron. The predicted binding affinity for each epitope–HLA allele pair was determined across the four strains.
One-way ANOVA was performed to assess the differences in binding affinity scores among SARS-CoV-2 strains for each HLA allele. Additionally, one-sample t-tests were conducted to compare the mean binding affinity scores of the 7 selected epitopes to a theoretical mean of 0 for each strain–HLA allele combination. Pairwise chi-square tests were used to compare the distribution of epitope binding categories (strong binders, weak binders, and non-binders) between each pair of SARS-CoV-2 strains for each HLA allele. Statistical significance was set at p < 0.05.
3. Results
3.1. Overall Binding Affinity Analysis
This study analyzed binding affinities between three common HLA class I alleles in the Saudi Arabian population (HLA-A02:01, HLA-C06:02, and HLA-B*51:01) and viral epitopes from four major SARS-CoV-2 strains (Alpha, Epsilon, Gamma, and Omicron). The binding affinities were predicted using the NetMHCpan 4.1 server, with seven epitope peptides selected for each allele based on their frequency in the spike protein. HLA binding was predicted according to the Immune Epitope Database.
3.2. HLA-A02:01 Analysis
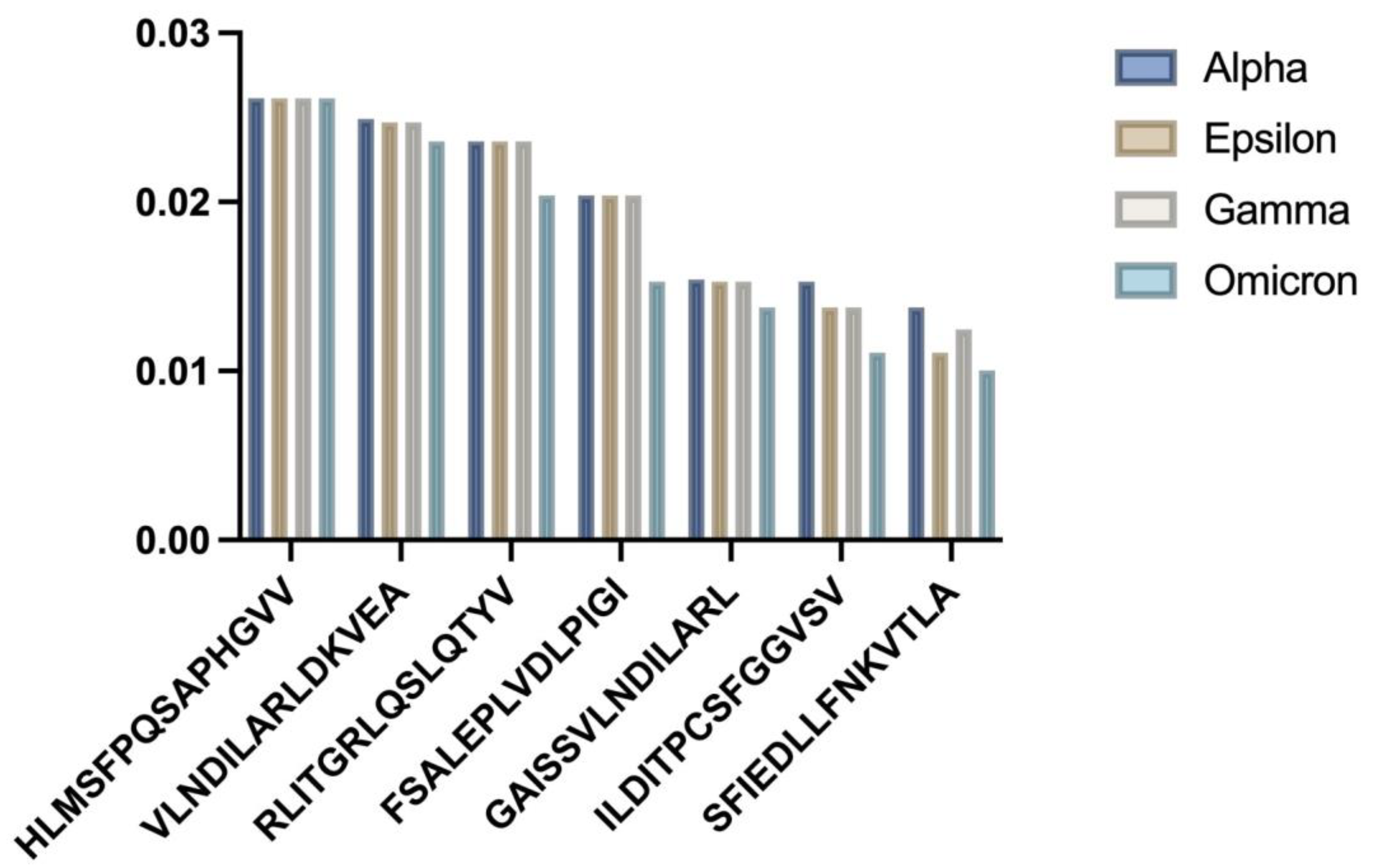
Analysis of the frequent allele HLA-A02:01 showed statistically significant differences in binding affinities for the seven selected epitopes across the SARS-CoV-2 strains (
Figure 1).
Figure 1 HLA-A02:01 binding affinity analysis:
- (a)
Bar chart showing comparative binding affinities across SARS-CoV-2 variants;
- (b)
Statistical distribution of binding patterns.
The one-way ANOVA results revealed significant differences among the strains (
p = 0.009954). The Alpha variant epitopes generally showed the strongest predicted binding, which weakened for later strains such as Omicron. One-sample
t-tests comparing the mean binding affinity scores showed significant differences for all strains (
Table 1).
3.3. HLA-C06:02 Analysis
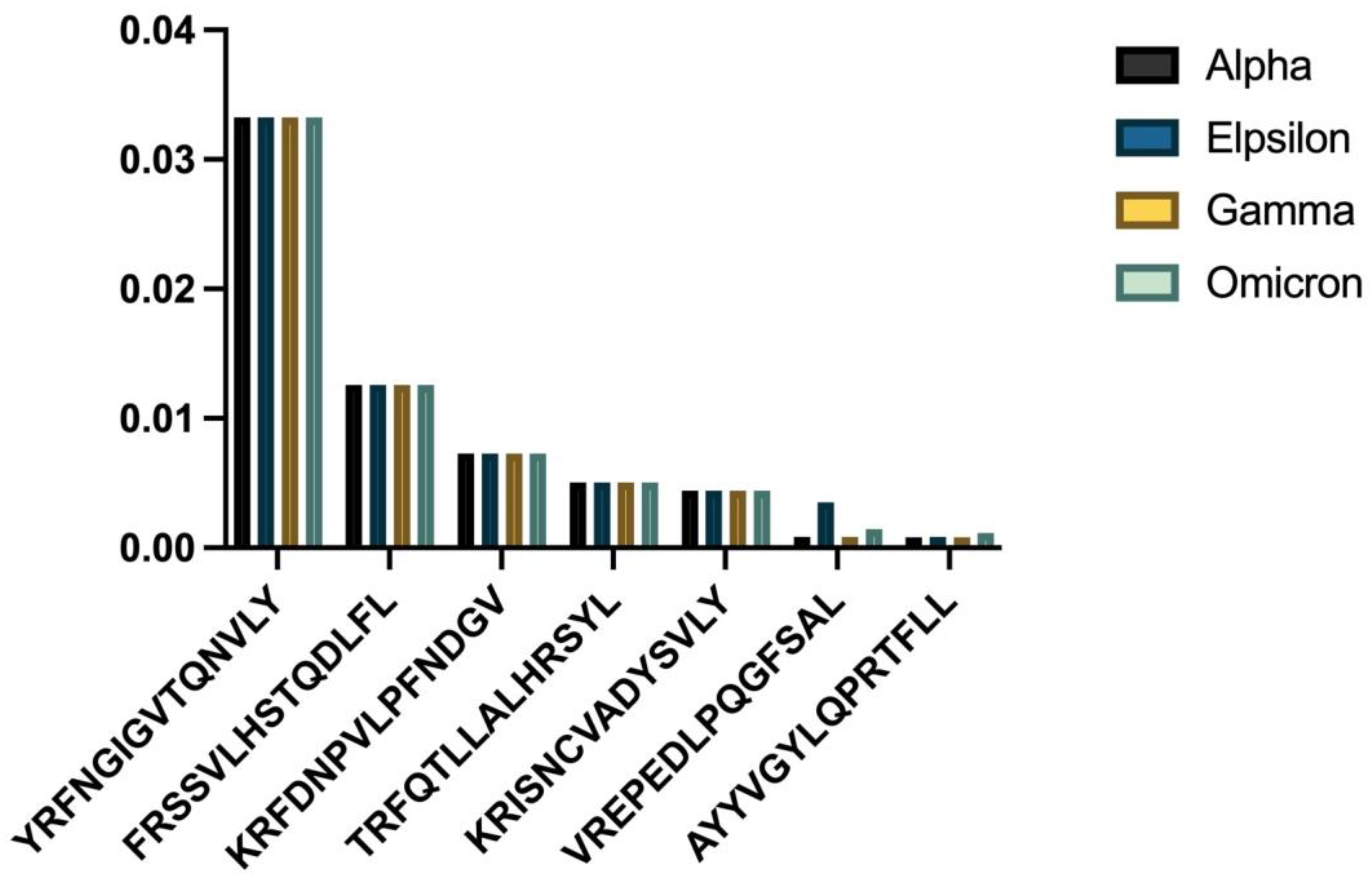
HLA-C06:02 demonstrated a similar trend of declining binding affinities in newer viral strains (
Figure 2).
- (a)
Comparative binding strengths across variants;
- (b)
Trend analysis showing temporal changes.
The one-way ANOVA results indicated significant differences among the strains (
p = 0.025474), with the Omicron variant showing the weakest HLA-C06:02 binding for many epitopes. One-sample
t-tests revealed significant differences in mean binding affinity scores for all strains (
Table 2).
3.4. HLA-B51:01 Analysis
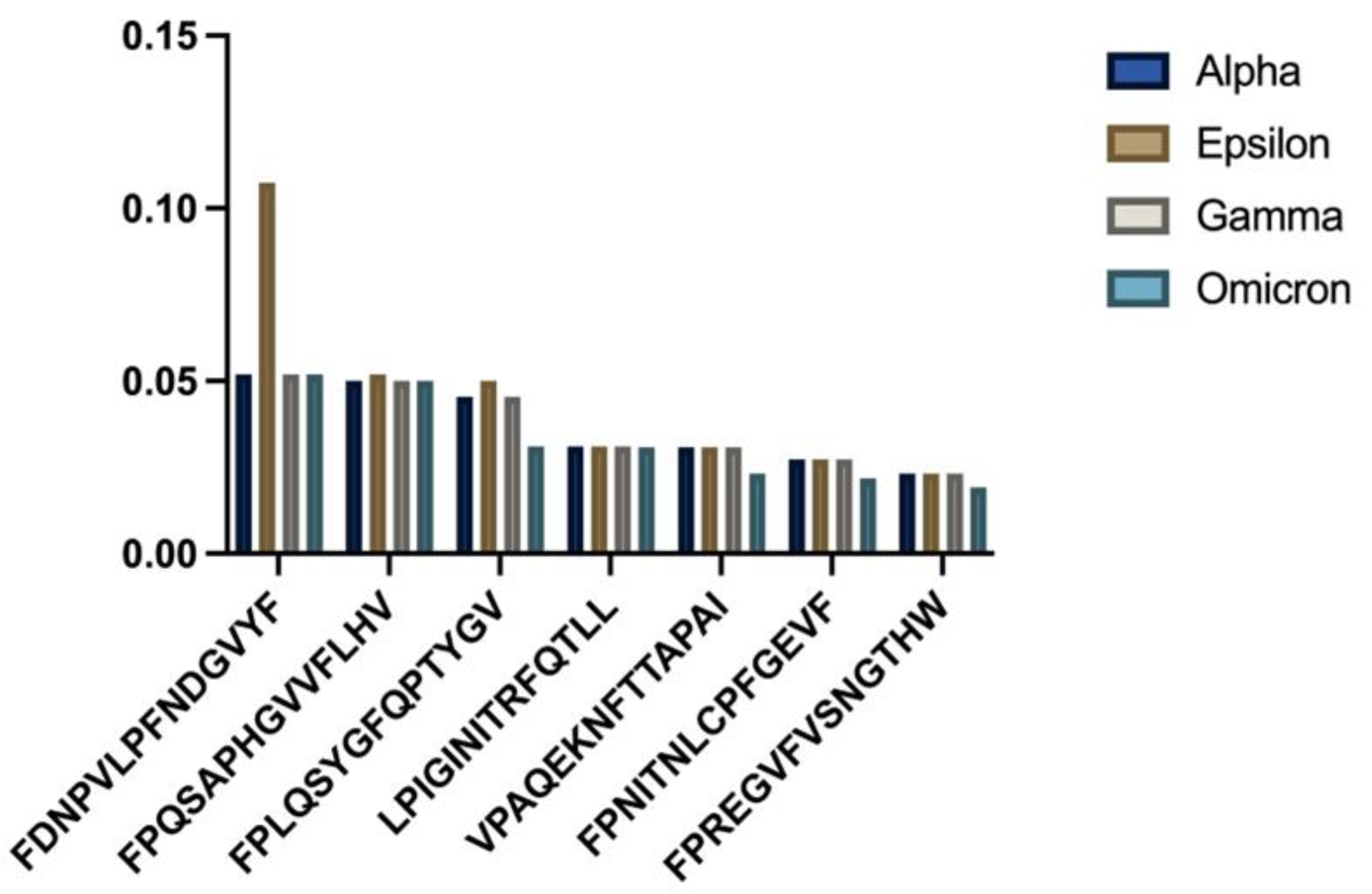
HLA-B51:01 presented different patterns when compared to the other alleles (
Figure 3).
- (a)
Consistency analysis across variants;
- (b)
Statistical comparison of binding affinities.
The one-way ANOVA results revealed non-significant
p-values for all strains (
p = 0.967107), and one-sample
t-tests showed no significant differences in mean binding affinity scores (
Table 3).
3.5. Comparative Analysis Across Variants
Pairwise chi-square analyses were performed to compare the distribution of epitope binding categories between the SARS-CoV-2 strains (
Table 4). Significant differences were observed between the Alpha and Epsilon strains, as well as Epsilon and Gamma strains, for HLA-B*51:01 (
p = 0.030197).
4. Discussion
This study investigated whether mutations in successive variants of SARS-CoV-2 have led to viral escape from common HLA class I alleles in the Saudi Arabian population. The results provide evidence for the significant differences in binding affinities among SARS-CoV-2 strains for HLA-A02:01 and HLA-C06:02, but not for HLA-B51:01, as demonstrated via the one-way ANOVA. However, the pairwise chi-square analyses did not reveal significant differences in the distribution of specific peptides presented by HLA-A02:01 and HLA-C*06:02 across the strains.
The significant differences in binding affinities among strains for HLA-A02:01 and HLA-C06:02 suggest that SARS-CoV-2 mutations may impact the overall strength of epitope binding to these alleles, which could potentially affect the efficiency of antigen presentation and subsequent T cell responses. These findings align with previous studies showing that SARS-CoV-2 mutations confer resistance to CD8+ T cell responses; particularly in the receptor-binding domain of the spike protein, where most analyzed epitopes were located [
23,
24]. Our results provide novel evidence that this viral escape is occurring in the context of common HLA alleles in Saudis. Population-specific analyses are critical, as HLA binding is dependent on the unique peptide-binding pockets of alleles that vary in frequency across people of different ethnicities.
Escape from dominant HLA alleles could have important consequences for SARS-CoV-2 immunity at the population level. HLA-A02:01 and HLA-C06:02 are predicted to present a significant proportion of viral epitopes to CD8+ T cells in Saudis, based on their gene frequencies. Thus, mutations reducing binding to these alleles may diminish viral control through compromising antigen presentation to cytotoxic T cells. This evasion of cellular immunity likely contributes to the increased transmissibility and ability to re-infect previously exposed individuals observed in newer SARS-CoV-2 variants.
Moreover, understanding HLA escape mutations provides insights that could optimize COVID-19 vaccines for the Saudi population. For example, the use of ancestral Wuhan or Alpha spike immunogens may lead to relatively ineffective T cell responses against newer viral strains, and vaccine design efforts should consider incorporating spike protein mutations that restore HLA binding lost due to viral evolution [
25]. Functional analysis of how the identified mutations affect antigen processing and presentation is also warranted.
However, the lack of significant differences in the distribution of specific peptides presented by HLA-A02:01 and HLA-C06:02 across the strains indicates that the mutations may not substantially alter the repertoire of epitopes presented by these alleles. This underscores the importance of considering both the strength of epitope binding and the diversity of the presented peptides when assessing viral escape from HLA-mediated immunity. Interestingly, HLA-B51:01 showed no significant differences in binding affinities across the analyzed SARS-CoV-2 variants, suggesting this allele may be less susceptible to viral escape mutations. This stability could be attributed to several potential mechanisms. First, the peptide-binding groove of HLA-B51:01 may recognize conserved regions of the spike protein that are under functional constraint and therefore less prone to mutation. Second, the specific anchor residues preferred by HLA-B51:01 might be maintained across variants due to the structural requirements of the spike protein. From a clinical perspective, the stable binding profile of HLA-B51:01 could potentially confer more consistent T cell responses against emerging variants in individuals carrying this allele. This finding suggests that targeting epitopes presented by HLA-B51:01 might be particularly valuable for vaccine design, as such epitopes may provide broader cross-variant protection. Future studies should investigate whether Saudi individuals with HLA-B51:01 demonstrate more consistent immune responses or clinical outcomes across different SARS-CoV-2 variants compared to those with other HLA alleles.
For HLA-B51:01, the absence of significant differences in both binding affinities and peptide distribution suggests that this allele may be less affected by SARS-CoV-2 mutations, potentially indicating a lower level of immune pressure on the virus. However, the pairwise chi-square analyses revealed some differences in the distribution of epitope binding categories between certain SARS-CoV-2 strains for HLA-B51:01, specifically between the Alpha and Epsilon strains and between the Epsilon and Gamma strains. This suggests that the impact of viral mutations on HLA binding may vary across different alleles and highlights the importance of considering allele-specific effects when assessing immune escape.
Overall, this study provided evidence for the ongoing interplay between SARS-CoV-2 evolution and human immunity. Tracking HLA escape will be vital for containing COVID-19 in diverse populations globally. Integrating HLA genetics into epidemiologic, therapeutic, and vaccine research remains an important frontier to comprehensively understand and combat this pandemic.
5. Limitations and Future Directions
We acknowledge that our study relies primarily on computational predictions using NetMHCpan, and experimental validation through in vitro binding assays or T cell activation experiments would strengthen our findings. Future work should include HLA binding assays with synthesized peptides from various SARS-CoV-2 variants to confirm the predicted differences in binding affinities. Additionally, T cell activation assays using peripheral blood mononuclear cells from HLA-typed Saudi individuals would help validate the functional impact of these binding differences on immune responses. Such experimental validation would provide crucial evidence on whether the computationally predicted changes in HLA binding translate to altered T cell recognition and activation.
While this study provides novel insights into the impacts of SARS-CoV-2 mutations on HLA-mediated immune responses in the Saudi Arabian population, it is important to acknowledge its limitations. One key limitation is the reliance on computational predictions of binding affinities, which may not fully capture the complexity of in vivo antigen presentation. As such, experimental validation of the predicted impacts of mutations on HLA binding and T cell recognition is necessary to confirm these findings. In vitro assays using HLA-typed cells and patient-derived T cells could provide valuable functional data to complement the computational analyses.
A key limitation of our study is the absence of clinical data correlating our binding predictions with actual disease outcomes. Future research should aim to integrate our computational findings with clinical metrics including disease severity, viral load, and immune response parameters from Saudi patients with confirmed SARS-CoV-2 infections. A prospective study design collecting samples from HLA-typed patients across the spectrum of COVID-19 severity would allow us to determine whether the predicted binding differences correlate with clinical outcomes. Such data would provide insights into the real-world implications of viral escape from HLA-mediated immunity and could help identify protective or risk-associated HLA alleles in this population.
Another limitation is the lack of clinical data, preventing direct assessment of the impacts of the observed differences in binding affinities on T cell responses and disease outcomes. Future studies should aim to integrate binding affinity data with clinical measures of disease severity, viral load, and immune responses in infected individuals. Such analyses would enable a more comprehensive understanding of the functional consequences of SARS-CoV-2 mutations on HLA-mediated immunity and their potential implications for disease progression and transmission.
Furthermore, this study focused on a limited set of common HLA class I alleles in the Saudi Arabian population. Extending the analysis to a broader range of HLA alleles, including class II alleles, would provide a more comprehensive picture of the impacts of SARS-CoV-2 mutations on HLA-mediated immunity across diverse genetic backgrounds. Class II HLA molecules present viral epitopes to CD4+ T cells, which play crucial roles in orchestrating adaptive immune responses and providing help to B cells and CD8+ T cells. Investigating the effects of SARS-CoV-2 mutations on class II HLA binding and presentation could reveal additional insights into viral immune escape mechanisms and better inform vaccine design strategies.
Future studies should also include a broader range of HLA alleles, including class II alleles, which would provide a more comprehensive picture of the impacts of SARS-CoV-2 mutations on HLA-mediated immunity across diverse genetic backgrounds. Our study focused exclusively on HLA class I alleles and their interaction with CD8+ T cell epitopes. However, CD4+ T cells, which recognize antigens presented by HLA class II molecules, play crucial roles in orchestrating adaptive immune responses, providing help to B cells, and supporting CD8+ T cell function. Future studies should extend this analysis to include common HLA class II alleles in the Saudi population to provide a more comprehensive picture of T cell immunity against SARS-CoV-2. Additionally, while we analyzed several important SARS-CoV-2 variants (Alpha, Epsilon, Gamma, and Omicron), our study did not include all major variants, notably Delta. A more comprehensive analysis including all major variants would provide a more complete evolutionary perspective on viral escape from HLA-mediated immunity. These limitations should be considered when interpreting our findings and addressing them represents an important direction for future research.
Future studies should also consider the potential impacts of SARS-CoV-2 mutations on other aspects of the immune response, such as antibody recognition and neutralization. Integrating data on HLA binding escape with information on antibody epitopes and neutralizing antibody responses could provide a more holistic view of how viral evolution shapes immune evasion and protection.
Additionally, longitudinal studies that track HLA binding affinities and immune responses over time in infected individuals or vaccinated cohorts could shed light on the dynamics of viral escape and the durability of HLA-mediated immunity. Such studies could also help to identify key epitopes or HLA alleles that are associated with better clinical outcomes or more robust and long-lasting immune protection.
Finally, expanding the study to include more diverse populations and HLA allele frequencies would enable a better understanding of the generalizability of these findings and the potential for population-specific immune escape patterns. Collaborative efforts to share data and integrate findings across different study cohorts could accelerate progress in this area, informing global vaccine and immunotherapy development efforts.
In conclusion, while this study provides valuable insights into the impacts of SARS-CoV-2 mutations on HLA-mediated immunity in the Saudi Arabian population, further research is needed to validate these findings experimentally, expand the scope of the analysis, and translate the results into actionable strategies for vaccine design and pandemic control. Addressing these limitations and pursuing the identified future directions will be crucial for advancing our understanding of SARS-CoV-2 immune escape and developing more effective interventions to combat this ongoing global health threat.
6. Conclusions
This study analyzed binding affinities between common HLA class I alleles in Saudi Arabia (HLA-A02:01, HLA-C06:02, and HLA-B51:01) and epitopes from major SARS-CoV-2 strains (Alpha, Epsilon, Gamma, and Omicron). The results demonstrated significant differences in binding affinities among the strains for HLA-A02:01 and HLA-C*06:02, as evidenced by the nested one-way ANOVA, suggesting that viral evolution may impact the overall strength of epitope binding to these alleles; however, pairwise chi-square analyses did not reveal significant differences in the distribution of specific peptides presented by these alleles across the strains.
In contrast, HLA-B*51:01 did not show evidence of significant differences in either binding affinity or peptide distribution among the strains, indicating that this allele may be less affected by SARS-CoV-2 mutations.
These findings provide novel insights into the complex interplay between SARS-CoV-2 evolution and HLA-mediated immune responses in the Saudi Arabian population. While mutations may impact the overall binding affinity of epitopes to certain HLA alleles, the specific peptides presented by these alleles may remain relatively conserved. This highlights the importance of considering both the strength of epitope binding and the diversity of presented peptides when assessing viral escape from HLA-mediated immunity.
Mapping HLA escape mutations will be important for tracking viral evolution and understanding population-level immunity. The insights gained from this study can guide vaccine design efforts through informing the selection of epitopes that are less susceptible to escape mutations and more likely to elicit effective T cell responses across diverse HLA profiles.
Further research is warranted in order to experimentally validate the predicted impacts of mutations on HLA binding and investigate the functional consequences of the observed differences in binding affinities on T cell responses and clinical outcomes. Additionally, extending the analysis to include HLA class II alleles and integrating clinical data could provide a more comprehensive understanding of the impacts of SARS-CoV-2 mutations on immune responses in the Saudi Arabian population.
Overall, this study highlighted the ongoing interplay between SARS-CoV-2 and human HLA genetics, emphasizing the importance of population-specific analyses in deciphering the complex dynamics of viral evolution and host immunity. The findings contribute to the growing body of knowledge on SARS-CoV-2 immune escape and underscore the need to integrate HLA genetics into epidemiologic, therapeutic, and vaccine research, in order to develop effective interventions that account for the genetic diversity of global populations and are resilient to viral escape mechanisms.