Gene-Level Analyses of Novel Olfactory-Related Signal from Severe SARS-CoV-2 GWAS Reveal Association with Disease Mortality
Abstract
- Key Messages
1. Introduction
2. Materials & Methods
2.1. Study Population
2.2. Patients and Public Involvement
2.3. Genotype Quality Control
2.4. Genome-Wide Association Analysis
2.5. Optimally Adjusted SNP-Set Kernel Association Test
2.6. Functional Prediction & eQTL
3. Results
3.1. Genome-Wide Significance
3.2. SNP-Set (Sequence) Kernel Association Test
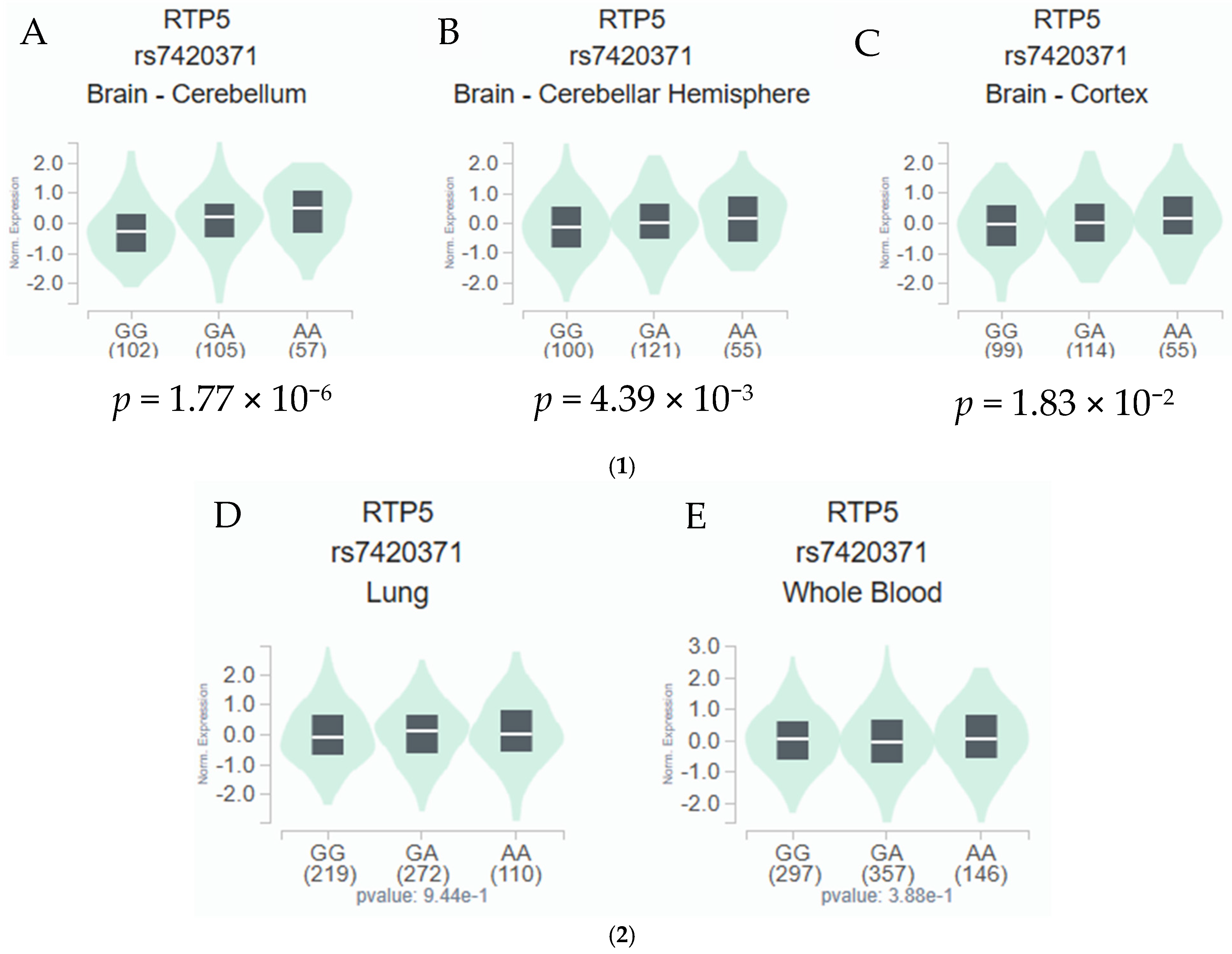
3.3. eQTL and Functional Prediction
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Biancolella, M.; Colona, V.L.; Mehrian-Shai, R.; Watt, J.L.; Luzzatto, L.; Novelli, G.; Reichardt, J.K.V. COVID-19 2022 update: Transition of the pandemic to the endemic phase. Hum Genom. 2022, 16, 19. [Google Scholar] [CrossRef] [PubMed]
- Alkhalil, E.; Jarad, B.; Saleh, T.; Aldarwish, W.; Joujeh, D. BioNTech/Pfizer (BNT162b2) COVID-19 mRNA vaccine: Manufacturing, immunogenicity, efficacy and safety. Prospect. Pharm. Sci. 2025, 23, 31–46. [Google Scholar] [CrossRef]
- World Health Organization. WHO (COVID-19) Dashboard. 2022. Available online: https://covid19.who.int (accessed on 14 February 2023).
- Esakandari, H.; Nabi-Afjadi, M.; Fakkari-Afjadi, J.; Farahmandian, N.; Miresmaeili, S.-M.; Bahreini, E. A comprehensive review of COVID-19 characteristics. Biol. Proced. Online 2020, 22, 19. [Google Scholar] [CrossRef] [PubMed]
- Severe COVID-19 GWAS Group; Ellinghaus, D.; Degenhardt, F.; Bujanda, L.; Buti, M.; Albillos, A.; Invernizzi, P.; Fernández, J.; Prati, D.; Baselli, G.; et al. Genomewide Association Study of Severe COVID-19 with Respiratory Failure. N. Engl. J. Med. 2020, 383, 1522–1534. [Google Scholar] [CrossRef]
- Degenhardt, F.; Ellinghaus, D.; Juzenas, S.; Lerga-Jaso, J.; Wendorff, M.; Maya-Miles, D.; Uellendahl-Werth, F.; ElAbd, H.; Rühlemann, M.C.; Arora, J.; et al. Detailed stratified GWAS analysis for severe COVID-19 in four European populations. Hum. Mol. Genet. 2022, 31, 3945–3966. [Google Scholar] [CrossRef]
- COVID-19 Host Genetics Initiative. Mapping the human genetic architecture of COVID-19. Nature 2021, 600, 472–477. [Google Scholar] [CrossRef]
- Lee, S.; Emond, M.J.; Bamshad, M.J.; Barnes, K.C.; Rieder, M.J.; Nickerson, D.A.; Christiani, D.C.; Wurfel, M.M.; Lin, X. Optimal Unified Approach for Rare-Variant Association Testing with Application to Small-Sample Case-Control Whole-Exome Sequencing Studies. Am. J. Hum. Genet. 2012, 91, 224–237. [Google Scholar] [CrossRef]
- Rubinacci, S.; Ribeiro, D.M.; Hofmeister, R.J.; Delaneau, O. Efficient phasing and imputation of low-coverage sequencing data using large reference panels. Nat. Genet. 2021, 53, 120–126. [Google Scholar] [CrossRef]
- Durinck, S.; Moreau, Y.; Kasprzyk, A.; Davis, S.; De Moor, B.; Brazma, A.; Huber, W. BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 2005, 21, 3439–3440. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 9 December 2025).
- SAS Institute Inc. SAS/STAT® 15.3 User’s Guide; SAS Institute Inc.: Cary, NC, USA, 2023. [Google Scholar]
- Genomes Project Consortium; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [PubMed]
- The GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Boyle, A.P.; Hong, E.L.; Hariharan, M.; Cheng, Y.; Schaub, M.A.; Kasowski, M.; Karczewski, K.J.; Park, J.; Hitz, B.C.; Weng, S.; et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012, 22, 1790–1797. [Google Scholar] [CrossRef] [PubMed]
- Ward, L.D.; Kellis, M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012, 40, D930–D934. [Google Scholar] [CrossRef]
- Luo, Y.; Hitz, B.C.; Gabdank, I.; A Hilton, J.; Kagda, M.S.; Lam, B.; Myers, Z.; Sud, P.; Jou, J.; Lin, K.; et al. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 2020, 48, 882–889. [Google Scholar] [CrossRef]
- Boughton, A.P.; Welch, R.P.; Flickinger, M.; VandeHaar, P.; Taliun, D.; Abecasis, G.R.; Boehnke, M. LocusZoom.js: Interactive and embeddable visualization of genetic association study results. Bioinformatics 2021, 37, 3017–3018. [Google Scholar] [CrossRef]
- Zhou, H.; Arapoglou, T.; Li, X.; Li, Z.; Zheng, X.; Moore, J.; Asok, A.; Kumar, S.; E Blue, E.; Buyske, S.; et al. FAVOR: Functional annotation of variants online resource and annotator for variation across the human genome. Nucleic Acids Res. 2022, 51, 1300–1311. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 51, 1–33. [Google Scholar] [CrossRef]
- Claus, L.E.; Leland, E.M.; Tai, K.Y.; Schlosser, R.J.; Kamath, V.; Lane, A.P.; Rowan, N.R. Olfactory Loss and Beyond: A Practical Review of Chemosensory Dysfunction. J. Am. Board Fam. Med. 2022, 35, 406–419. [Google Scholar] [CrossRef]
- Mastrangelo, A.; Bonato, M.; Cinque, P. Smell and taste disorders in COVID-19: From pathogenesis to clinical features and outcomes. Neurosci. Lett. 2021, 748, 135694. [Google Scholar] [CrossRef] [PubMed]
- Elkazzaz, M.; Ahmed, A.; Abo-Amer, Y.E.-E.; Hydara, T.; Haikal, A.; El Razek, D.N.A.; Eltayb, W.A.; Wang, X.; Karpiński, T.M.; Hamza, D.; et al. In Silico Discovery of GPCRs and GnRHRs as Novel Binding Receptors of SARS-CoV-2 Spike Protein Could Explain Neuroendocrine Disorders in COVID-19. Vaccines 2022, 10, 1500. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- COVID-19 Host Genetics Initiative. A first update on mapping the human genetic architecture of COVID-19. Nature 2022, 608, E1–E10. [Google Scholar] [CrossRef]
- Huang, X.; Guan, W.; Xiang, B.; Wang, W.; Xie, Y.; Zheng, J. MUC5B regulates goblet cell differentiation and reduces inflammation in a murine COPD model. Respir. Res. 2022, 23, 11. [Google Scholar] [CrossRef]
- Khademul, A.B.; Kamran Khan, A.A. Lung transcriptome of a COVID-19 patient and systems biology predictions suggest impaired surfactant production which may be druggable by surfactant therapy. Sci. Rep. 2020, 10, 19395. [Google Scholar] [CrossRef]
- van Moorsel, C.H.M.; van der Vis, J.J.; Duckworth, A.; Scotton, C.J.; Benschop, C.; Ellinghaus, D.; Ruven, H.J.T.; Quanjel, M.J.R.; Grutters, J.C. The MUC5B Promoter Polymorphism Associates with Severe COVID-19 in the European Population. Front. Med. 2021, 8, 668024. [Google Scholar] [CrossRef]
- Pietzner, M.; Chua, R.L.; Wheeler, E.; Jechow, K.; Willett, J.D.S.; Radbruch, H.; Trump, S.; Heidecker, B.; Zeberg, H.; Heppner, F.L.; et al. ELF5 is a potential respiratory epithelial cell-specific risk gene for severe COVID-19. Nat. Commun. 2022, 13, 4484. [Google Scholar] [CrossRef]

| Covariates | Number of Patients (%) | 30 Day Deaths (%) | 60 Day Deaths (%) | Median Survival Time (Days) a |
|---|---|---|---|---|
| Total | 370 | 79 (21.4) | 86 (23.2) | 16.0 |
| Age (Years) | ||||
| <50 | 86 (23.2) | 8 (10.1) | 9 (10.5) | 16.5 |
| 50 | 284 (76.8) | 71 (89.9) | 77 (89.5) | 16.0 |
| Race & Ethnicity | ||||
| Non-Hispanic White | 245 (66.2) | 62 (78.5) | 67 (77.9) | 15.0 |
| Hispanic | 125 (33.8) | 17 (21.5) | 19 (22.1) | 18.0 |
| Sex | ||||
| Male | 231 (62.4) | 58 (73.4) | 63 (73.3) | 16.0 |
| Female | 139 (37.6) | 21 (26.6) | 23 (26.7) | 15.0 |
| Smoking Status | ||||
| Never | 211 (57.0) | 35 (44.3) | 38 (44.2) | 15.0 |
| Former | 127 (34.3) | 31 (39.2) | 35 (40.7) | 17.0 |
| Current | 23 (6.2) | 7 (8.9) | 7 (8.1) | 18.0 |
| BMI (kg/m2) | ||||
| 25 | 84 (22.7) | 19 (24.1) | 23 (26.7) | 16.5 |
| >25 | 286 (77.3) | 60 (75.9) | 63 (73.3) | 15.5 |
| ICU Status | ||||
| Floor | 140 (37.8) | 12 (15.2) | 13 (15.1) | 7.0 |
| ICU | 230 (62.2) | 67 (84.8) | 73 (84.9) | 23.0 |
| COVID Surge | ||||
| Wave 1 b | 203 (54.9) | 40 (50.6) | 41 (47.7) | 19.0 |
| Wave 2 c | 167 (45.1) | 39 (49.4) | 45 (52.3) | 13.0 |
| MGH COVID Genotypes (n = 370) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Mortality | SNPs a | Chromosome | Allele | MAF | Gene | FDR b | Bonferroni c | OR (95% CI) d | SE |
| 30-day | rs7420371 | 2 | G>A | 0.3926 | RTP5 | 0.017 | 0.031 | 2.32 (1.59, 3.39) | 0.19 |
| 60-day | rs7420371 | 2 | G>A | 0.3926 | RTP5 | 0.027 | 0.050 | 2.06 (1.43, 2.97) | 0.19 |
| Mortality Outcomes p-Value | ||
|---|---|---|
| Top GWAS Signal & HGI Signals | 30-Day Mortality b,c | 60-Day Mortality b,c |
| RTP5 d | 5.90 × 10−5 | 6.17 × 10−5 |
| SFTPD | 2.04 × 10−5 | 3.85 × 10−5 |
| MUC5B | 1.42 × 10−5 | 3.46 × 10−5 |
| ELF5 | 2.41 × 10−6 | 1.10 × 10−5 |
| FBRSL1 | 1.76 × 10−5 | 2.23 × 10−5 |
| SLC22A31 | 1.69 × 10−5 | 1.55 × 10−5 |
| TMPRSS2 | 2.17 × 10−5 | 3.31 × 10−5 |
| NR1H2 | 0.0001 | 0.0002 |
| THBS3 | 0.0013 | 0.0006 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.C.; Wang, X.; Lu, Y.; Dey, R.; Liu, Y.; Giacona, F.; Abe, E.A.; White, E.; Su, L.; Wei, Q.; et al. Gene-Level Analyses of Novel Olfactory-Related Signal from Severe SARS-CoV-2 GWAS Reveal Association with Disease Mortality. COVID 2025, 5, 206. https://doi.org/10.3390/covid5120206
Zhao YC, Wang X, Lu Y, Dey R, Liu Y, Giacona F, Abe EA, White E, Su L, Wei Q, et al. Gene-Level Analyses of Novel Olfactory-Related Signal from Severe SARS-CoV-2 GWAS Reveal Association with Disease Mortality. COVID. 2025; 5(12):206. https://doi.org/10.3390/covid5120206
Chicago/Turabian StyleZhao, Yu Chen, Xinan Wang, Yujia Lu, Rounak Dey, Yuchen Liu, Francesca Giacona, Elizabeth A. Abe, Emma White, Li Su, Qingyi Wei, and et al. 2025. "Gene-Level Analyses of Novel Olfactory-Related Signal from Severe SARS-CoV-2 GWAS Reveal Association with Disease Mortality" COVID 5, no. 12: 206. https://doi.org/10.3390/covid5120206
APA StyleZhao, Y. C., Wang, X., Lu, Y., Dey, R., Liu, Y., Giacona, F., Abe, E. A., White, E., Su, L., Wei, Q., Lin, X., Mucci, L. A., Alladina, J., & Christiani, D. C. (2025). Gene-Level Analyses of Novel Olfactory-Related Signal from Severe SARS-CoV-2 GWAS Reveal Association with Disease Mortality. COVID, 5(12), 206. https://doi.org/10.3390/covid5120206






