Nasopharyngeal Proteomic Profiles from Patients Hospitalized Due to COVID-19 in Manaus, Amazonas, Brazil
Abstract
1. Introduction
2. Methods
2.1. Patient Profiles and Sample Collection
2.2. Nasal Swab Sample Preparation, LC-MS-MS Processing and Analysis
3. Results
3.1. Biochemical Blood Markers of Patients Hospitalized
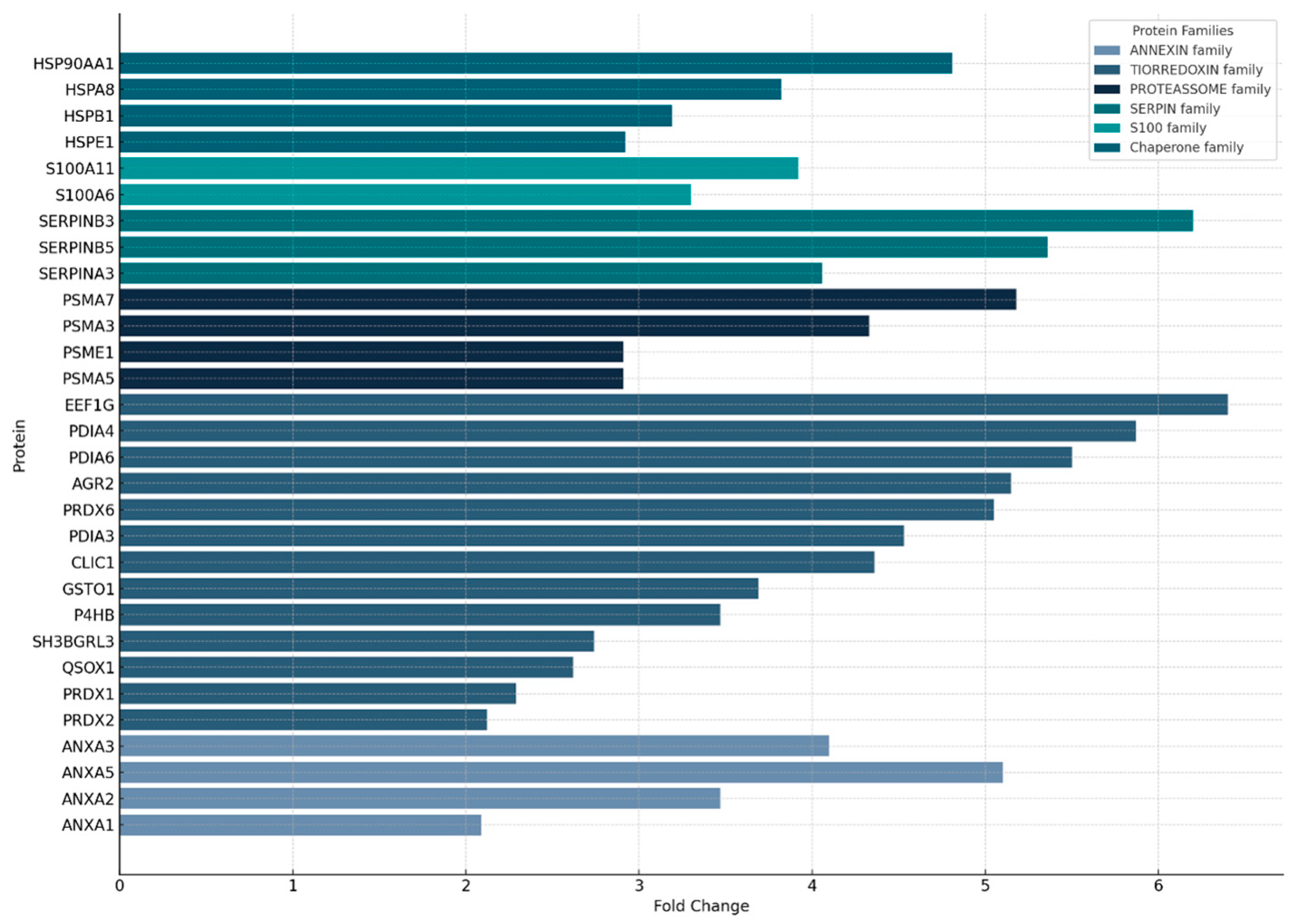
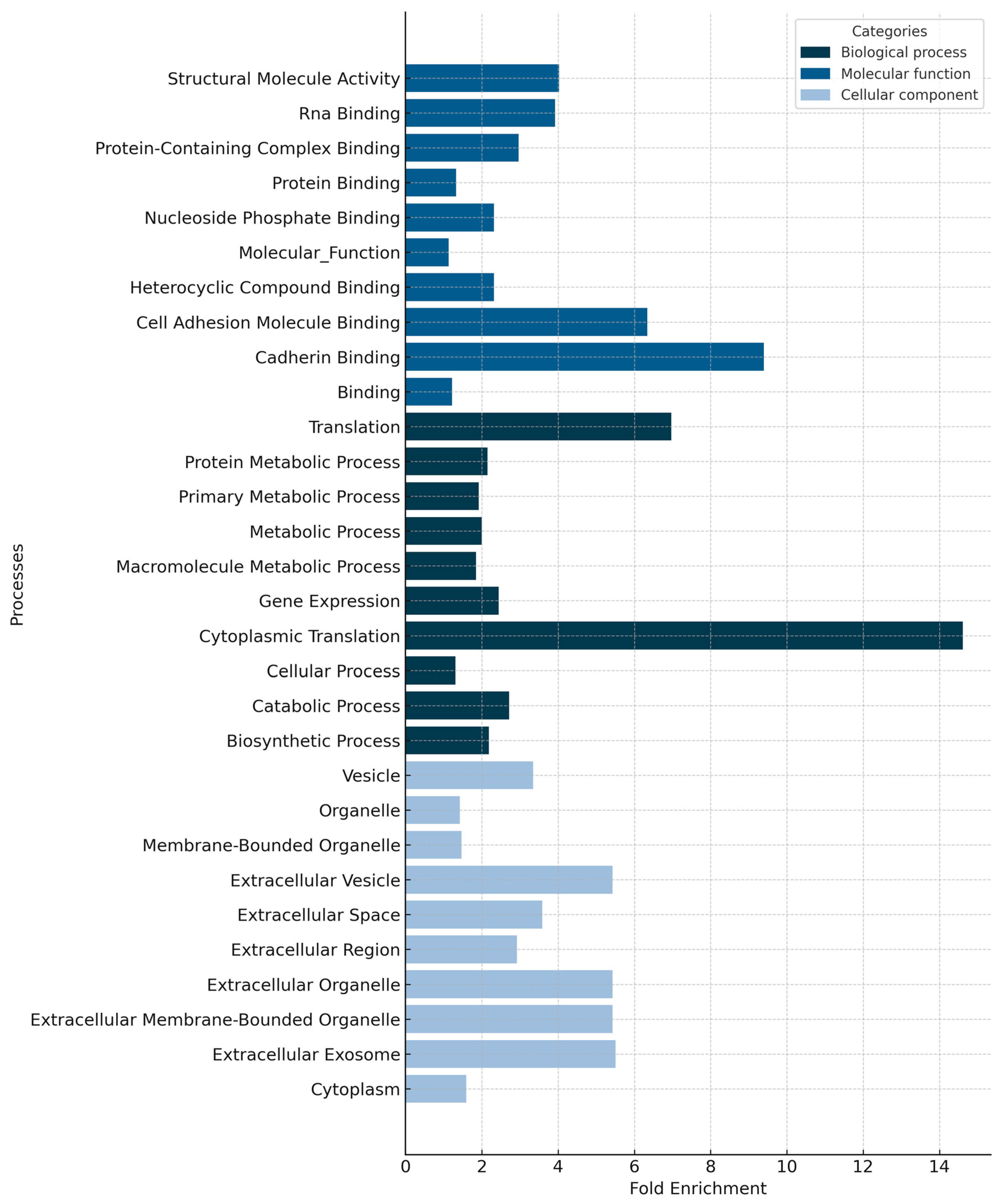
3.2. General Proteomic Profile of Nasopharyngeal Smear Samples
4. Discussion
4.1. The General Proteomic Profile of Nasopharyngeal Swabs
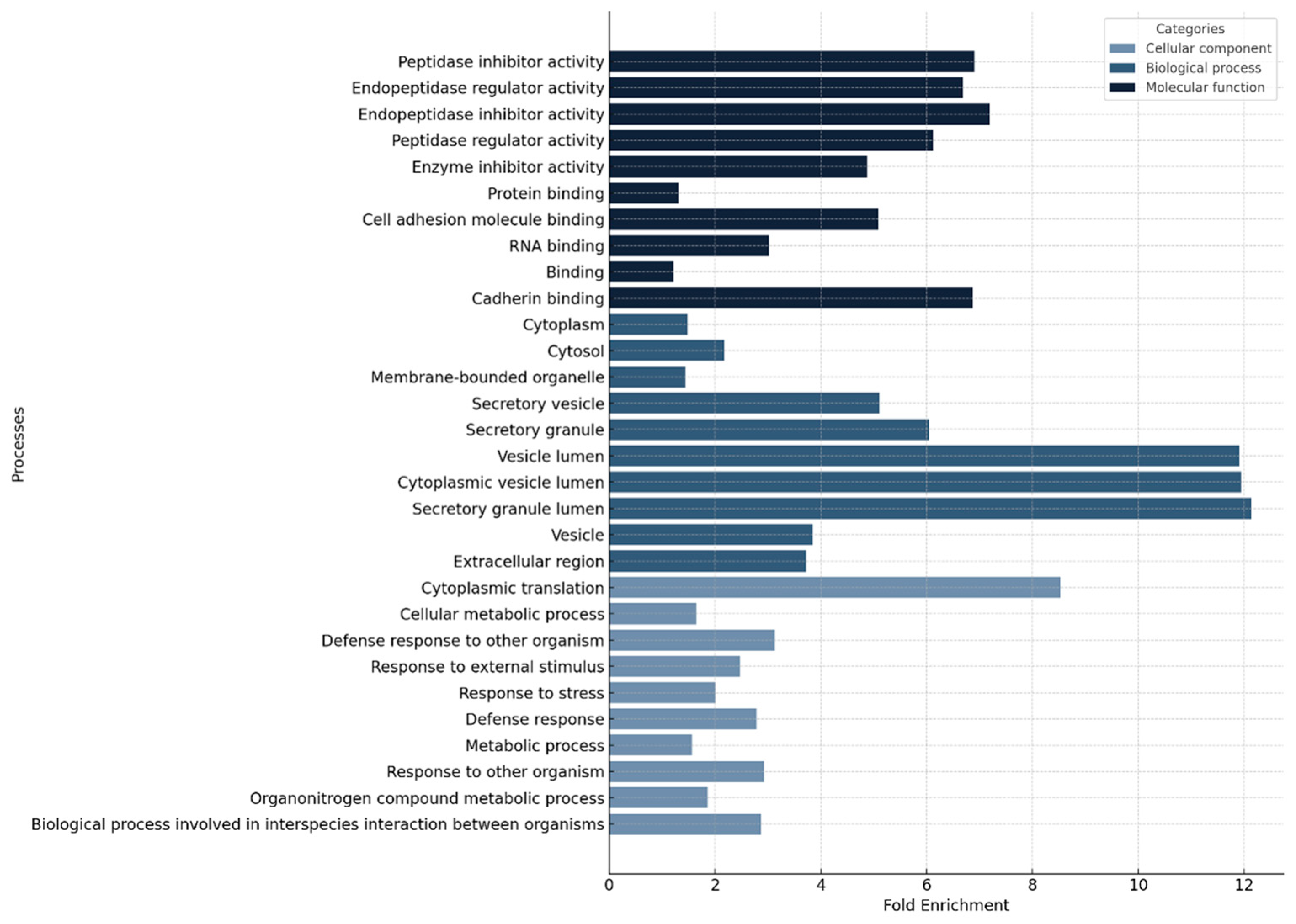
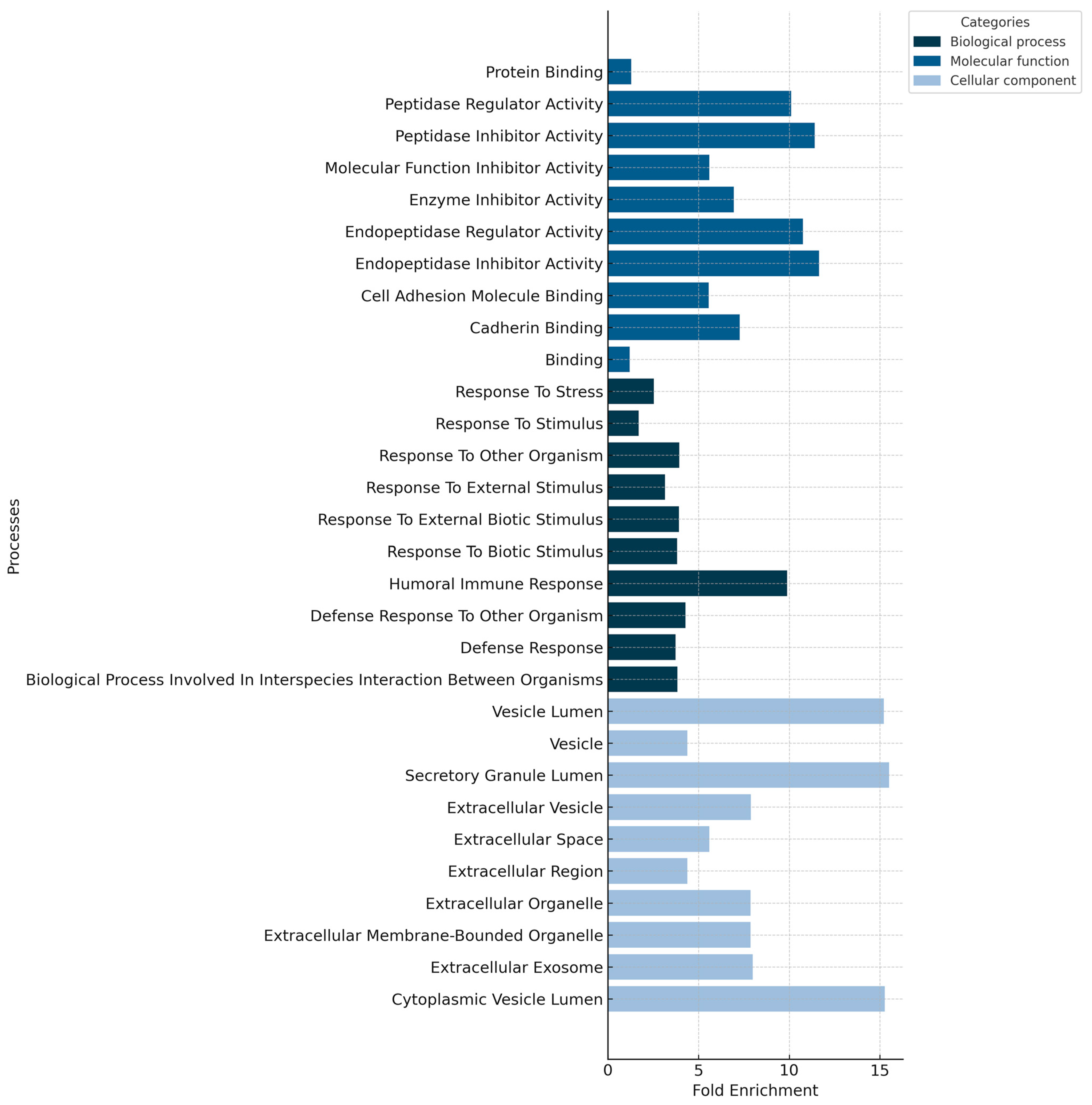
4.2. The Proteomic Profiles of Nasopharyngeal Swabs Associated with Fatalities
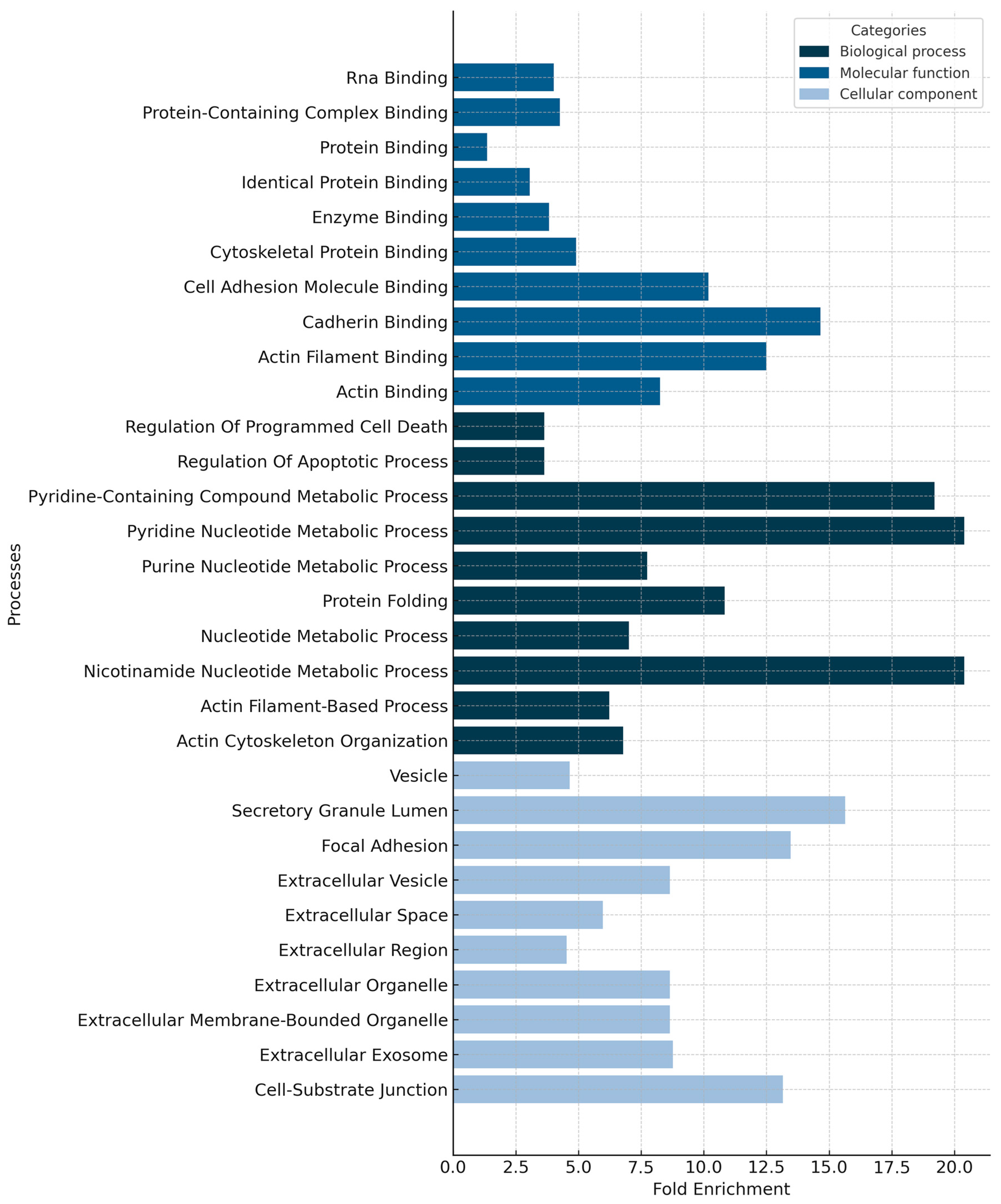
4.3. The Proteomic Profiles of Nasopharyngeal Swabs Associated with Survival
4.4. Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. COVID-19 Cases | WHO COVID-19 Dashboard. Available online: https://data.who.int/dashboards/covid19/cases (accessed on 9 September 2025).
- Zeiser, F.A.; Donida, B.; da Costa, C.A.; Ramos, G.d.O.; Scherer, J.N.; Barcellos, N.T.; Alegretti, A.P.; Ikeda, M.L.R.; Müller, A.P.W.C.; Bohn, H.C.; et al. First and Second COVID-19 Waves in Brazil: A Cross-Sectional Study of Patients’ Characteristics Related to Hospitalization and in-Hospital Mortality. Lancet Reg. Health Am. 2021, 6, 100107. [Google Scholar] [CrossRef]
- Barreto, I.C.d.H.C.; Costa Filho, R.V.; Ramos, R.F.; de Oliveira, L.G.; Martins, N.R.A.V.; Cavalcante, F.V.; Andrade, L.O.M.d.; Santos, L.M.P. Health Collapse in Manaus: The Burden of Not Adhering to Non-Pharmacological Measures to Reduce the Transmission of COVID-19. Saúde Debate 2021, 45, 1126–1139. [Google Scholar] [CrossRef]
- Moura, E.C.; Cortez-Escalante, J.; Cavalcante, F.V.; Barreto, I.C.d.H.C.; Sanchez, M.N.; Santos, L.M.P. COVID-19: Temporal Evolution and Immunization in the Three Epidemiological Waves, Brazil, 2020–2022. Rev. Saúde Pública 2022, 56, 105. [Google Scholar] [CrossRef]
- Schimke, L.F.; Marques, A.H.C.; Baiocchi, G.C.; de Souza Prado, C.A.; Fonseca, D.L.M.; Freire, P.P.; Rodrigues Plaça, D.; Salerno Filgueiras, I.; Coelho Salgado, R.; Jansen-Marques, G.; et al. Severe COVID-19 Shares a Common Neutrophil Activation Signature with Other Acute Inflammatory States. Cells 2022, 11, 847. [Google Scholar] [CrossRef]
- Bojkova, D.; Klann, K.; Koch, B.; Widera, M.; Krause, D.; Ciesek, S.; Cinatl, J.; Münch, C. Proteomics of SARS-CoV-2-Infected Host Cells Reveals Therapy Targets. Nature 2020, 583, 469–472. [Google Scholar] [CrossRef]
- Zeng, H.; Chen, D.; Yan, J.; Yang, Q.; Han, Q.; Li, S.; Cheng, L. Proteomic Characteristics of Bronchoalveolar Lavage Fluid in Critical COVID-19 Patients. FEBS J. 2021, 288, 5190–5200. [Google Scholar] [CrossRef]
- Chatterjee, S.; Zaia, J. Proteomics-Based Mass Spectrometry Profiling of SARS-CoV-2 Infection from Human Nasopharyngeal Samples. Mass Spectrom. Rev. 2022, 43, 193–229. [Google Scholar] [CrossRef] [PubMed]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for Micro-Purification, Enrichment, Pre-Fractionation and Storage of Peptides for Proteomics Using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef]
- Hallal, P.C.; Hartwig, F.P.; Horta, B.L.; Silveira, M.F.; Struchiner, C.J.; Vidaletti, L.P.; Neumann, N.A.; Pellanda, L.C.; Dellagostin, O.A.; Burattini, M.N.; et al. SARS-CoV-2 Antibody Prevalence in Brazil: Results from Two Successive Nationwide Serological Household Surveys. Lancet Glob. Health 2020, 8, e1390–e1398. [Google Scholar] [CrossRef] [PubMed]
- Fundação de Vigilância em Saúde do Amazonas (FVS-AM). Boletim Diário de COVID-19 no Amazonas, 31 May 2020; Fundação de Vigilância em Saúde do Amazonas: Manaus, Brazil, 2020; p. 5.
- Naveca, F.G.; Nascimento, V.; de Souza, V.C.; Corado, A.d.L.; Nascimento, F.; Silva, G.; Costa, Á.; Duarte, D.; Pessoa, K.; Mejía, M.; et al. COVID-19 in Amazonas, Brazil, Was Driven by the Persistence of Endemic Lineages and P.1 Emergence. Nat. Med. 2021, 27, 1230–1238. [Google Scholar] [CrossRef]
- He, D.; Lin, L.; Artzy-Randrup, Y.; Demirhan, H.; Cowling, B.J.; Stone, L. Resolving the Enigma of Iquitos and Manaus: A Modeling Analysis of Multiple COVID-19 Epidemic Waves in Two Amazonian Cities. Proc. Natl. Acad. Sci. USA 2023, 120, e2211422120. [Google Scholar] [CrossRef]
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Reduction and functional exhaustion of t cells in patients with coronavirus disease 2019 (COVID-19). Front. Immunol. 2020, 11, 544639. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A Retrospective Cohort Study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J. HLH Across Speciality Collaboration, UK COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Zheng, H.-Y.; Zhang, M.; Yang, C.-X.; Zhang, N.; Wang, X.-C.; Yang, X.-P.; Dong, X.-Q.; Zheng, Y.-T. Elevated exhaustion levels and reduced functional diversity of t cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol. Immunol. 2020, 17, 541–543. [Google Scholar] [CrossRef]
- Rock, J.R.; Randell, S.H.; Hogan, B.L.M. Airway basal stem cells: A perspective on their roles in epithelial homeostasis and remodeling. Dis. Model. Mech. 2010, 3, 545–556. [Google Scholar] [CrossRef]
- Gelzo, M.; Scialò, F.; Cacciapuoti, S.; Pinchera, B.; De Rosa, A.; Cernera, G.; Comegna, M.; Tripodi, L.; Schiano Moriello, N.; Mormile, M.; et al. Inducible nitric oxide synthase (iNOS): Why a different production in COVID-19 patients of the two waves? Viruses 2022, 14, 534. [Google Scholar] [CrossRef] [PubMed]
- Bankar, R.; Suvarna, K.; Ghantasala, S.; Banerjee, A.; Biswas, D.; Choudhury, M.; Palanivel, V.; Salkar, A.; Verma, A.; Singh, A. Proteomic investigation reveals dominant alterations of neutrophil degranulation and mRNA translation pathways in patients with COVID-19. iScience 2021, 24, 102135. [Google Scholar] [CrossRef]
- Banu, S.; Nagaraj, R.; Idris, M.M. A proteomic perspective and involvement of cytokines in SARS-CoV-2 infection. PLoS ONE 2023, 18, e0279998. [Google Scholar] [CrossRef] [PubMed]
- Kloc, M.; Uosef, A.; Wosik, J.; Kubiak, J.Z.; Ghobrial, R.M. Virus interactions with the actin cytoskeleton—What we know and do not know about SARS-CoV-2. Arch. Virol. 2022, 167, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Morrison, C.B.; Edwards, C.E.; Shaffer, K.M.; Araba, K.C.; Wykoff, J.A.; Williams, D.R.; Asakura, T.; Dang, H.; Morton, L.C.; Gilmore, R.C.; et al. SARS-CoV-2 infection of airway cells causes intense viral and cell shedding, two spreading mechanisms affected by IL-13. Proc. Natl. Acad. Sci. USA 2022, 119, e2119680119. [Google Scholar] [CrossRef]
- Lu, W.; Liu, X.; Wang, T.; Liu, F.; Zhu, A.; Lin, Y.; Luo, J.; Ye, F.; He, J.; Zhao, J.; et al. Elevated MUC1 and MUC5AC mucin protein levels in airway mucus of critical Ill COVID-19 patients. J. Med. Virol. 2021, 93, 582–584. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Na, H.G.; Choi, Y.S.; Bae, C.H.; Song, S.-Y.; Kim, Y.-D. SARS-CoV-2 Induces Expression of cytokine and MUC5AC/5B in human nasal epithelial cell through ACE 2 receptor. BioMed Res. Int. 2022, 2022, 2743046. [Google Scholar] [CrossRef]
- Yang, X.; Chi, H.; Wu, M.; Wang, Z.; Lang, Q.; Han, Q.; Wang, X.; Liu, X.; Li, Y.; Wang, X.; et al. Discovery and characterization of SARS-CoV-2 reactive and neutralizing antibodies from humanized CAMouseHG mice through rapid hybridoma screening and high-throughput single-cell V(D)J sequencing. Front. Immunol. 2022, 13, 992787. [Google Scholar] [CrossRef]
- Pankov, R.; Yamada, K.M. Fibronectin at a glance. J. Cell Sci. 2002, 115, 3861–3863. [Google Scholar] [CrossRef] [PubMed]
- Lemańska-Perek, A.; Krzyżanowska-Gołąb, D.; Dragan, B.; Tyszko, M.; Adamik, B. Fibronectin as a marker of disease severity in critically Ill COVID-19 patients. Cells 2022, 11, 1566. [Google Scholar] [CrossRef] [PubMed]
- Ciccosanti, F.; Antonioli, M.; Sacchi, A.; Notari, S.; Farina, A.; Beccacece, A.; Fusto, M.; Vergori, A.; D’Offizi, G.; Taglietti, F.; et al. Proteomic analysis identifies a signature of disease severity in the plasma of COVID-19 pneumonia patients associated to neutrophil, platelet and complement activation. Clin. Proteom. 2022, 19, 38. [Google Scholar] [CrossRef]
- Messner, C.B.; Demichev, V.; Wendisch, D.; Michalick, L.; White, M.; Freiwald, A.; Textoris-Taube, K.; Vernardis, S.I.; Egger, A.-S.; Kreidl, M.; et al. Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection. Cell Syst. 2020, 11, 11–24.e4. [Google Scholar] [CrossRef] [PubMed]
- Ermert, D.; Blom, A.M. C4b-Binding Protein: The good, the bad and the deadly. novel functions of an old friend. Immunol. Lett. 2016, 169, 82–92. [Google Scholar] [CrossRef]
- Mohammed, R.N.; Tamjidifar, R.; Rahman, H.S.; Adili, A.; Ghoreishizadeh, S.; Saeedi, H.; Thangavelu, L.; Shomali, N.; Aslaminabad, R.; Marofi, F.; et al. A comprehensive review about immune responses and exhaustion during coronavirus disease (COVID-19). Cell Commun. Signal. 2022, 20, 79. [Google Scholar] [CrossRef] [PubMed]
- Hahne, H.; Pachl, F.; Ruprecht, B.; Maier, S.K.; Klaeger, S.; Helm, D.; Médard, G.; Wilm, M.; Lemeer, S.; Kuster, B. DMSO enhances electrospray response, boosting sensitivity of proteomic experiments. Nat. Methods 2013, 10, 989–991. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.D.M.; Lima, D.B.; Fischer, J.S.G.; Clasen, M.A.; Kurt, L.U.; Camillo-Andrade, A.C.; Monteiro, L.C.; de Aquino, P.F.; Neves-Ferreira, A.G.C.; Valente, R.H.; et al. Simple, efficient and thorough shotgun proteomic analysis with PatternLab V. Nat. Protoc. 2022, 17, 1553–1578. [Google Scholar] [CrossRef]
- Carvalho, P.C.; Fischer, J.S.G.; Xu, T.; Cociorva, D.; Balbuena, T.S.; Valente, R.H.; Perales, J.; Yates, J.R.; Barbosa, V.C. Search Engine Processor: Filtering and organizing PSMs. Proteomics 2012, 12, 944–949. [Google Scholar] [CrossRef]
- Barboza, R.; Cociorva, D.; Xu, T.; Barbosa, V.C.; Perales, J.; Valente, R.H.; França, F.M.G.; Yates, J.R.; Carvalho, P.C. Can the false-discovery rate be misleading? Proteomics 2011, 11, 4105–4108. [Google Scholar] [CrossRef] [PubMed]
- Yates, J.R.; Park, S.K.R.; Delahunty, C.M.; Xu, T.; Savas, J.N.; Cociorva, D.; Carvalho, P.C. Toward objective evaluation of proteomic algorithms. Nat. Methods 2012, 9, 455–456. [Google Scholar] [CrossRef]
- Carvalho, P.C.; Lima, D.B.; Leprevost, F.V.; Santos, M.D.M.; Fischer, J.S.G.; Aquino, P.F.; Moresco, J.J.; Yates, J.R.; Barbosa, V.C. Integrated analysis of shotgun proteomic data with patternlab for proteomics 4.0. Nat. Protoc. 2016, 11, 102–117. [Google Scholar] [CrossRef]
- Carvalho, P.C.; Yates, J.R.; Barbosa, V.C. Improving the TFold test for differential shotgun proteomics. Bioinformatics 2012, 28, 1652–1654. [Google Scholar] [CrossRef]
- Zybailov, B.; Mosley, A.L.; Sardiu, M.E.; Coleman, M.K.; Florens, L.; Washburn, M.P. Statistical analysis of membrane proteome expression changes in saccharomyces cerevisiae. J. Proteome Res. 2006, 5, 2339–2347. [Google Scholar] [CrossRef]
- Mi, H.; Poudel, S.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. PANTHER version 10: Expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016, 44, D336–D342. [Google Scholar] [CrossRef]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017, 45, D183–D189. [Google Scholar] [CrossRef] [PubMed]

| Locus | Gene | Description | Fold Change |
|---|---|---|---|
| P80188 | LCN2 | Neutrophil gelatinase-associated lipocalin | 2.01 |
| P06753 | TPM3 | Tropomyosin alpha-3 chain | 2.02 |
| P61626 | LYZ | Lysozyme C | 2.05 |
| Q96C19 | EFHD2 | EF-hand domain-containing protein D2 | 2.08 |
| P04083 | ANXA1 | Annexin A1 | 2.09 |
| P32119 | PRDX2 | Peroxiredoxin-2 | 2.12 |
| P13796 | LCP1 | Plastin-2 | 2.14 |
| P52566 | ARHGDIB | Rho GDP-dissociation inhibitor 2 | 2.15 |
| P06733 | ENO1 | Alpha-enolase | 2.18 |
| P47756 | CAPZB | F-actin-capping protein subunit beta | 2.21 |
| P06396 | GSN | Gelsolin | 2.22 |
| P05387 | RPLP2 | Large ribosomal subunit protein P2 | 2.22 |
| P28799 | GRN | Progranulin | 2.25 |
| P37802 | TAGLN2 | Transgelin-2 | 2.28 |
| Q06830 | PRDX1 | Peroxiredoxin-1 | 2.29 |
| P13987 | CD59 | CD59 glycoprotein | 2.33 |
| P52565 | ARHGDIA | Rho GDP-dissociation inhibitor 1 | 2.34 |
| P80303 | NUCB2 | Nucleobindin-2 | 2.35 |
| P37837 | TALDO1 | Transaldolase | 2.36 |
| P07737 | PFN1 | Profilin-1 | 2.44 |
| P04040 | CAT | Catalase | 2.46 |
| Q13510 | ASAH1 | Acid ceramidase | 2.47 |
| P04075 | ALDOA | Fructose-bisphosphate aldolase A | 2.52 |
| P62937 | PPIA | Peptidyl-prolyl cis-trans isomerase A | 2.52 |
| Q9UGM3 | DMBT1 | Scavenger receptor cysteine-rich domain-containing protein DMBT1 | 2.55 |
| P14618 | PKM | Pyruvate kinase PKM | 2.59 |
| P20700 | LMNB1 | Lamin-B1 | 2.61 |
| O00391 | QSOX1 | Sulfhydryl oxidase 1 | 2.62 |
| P31944 | CASP14 | Caspase-14 | 2.62 |
| O43707 | ACTN4 | Alpha-actinin-4 | 2.63 |
| P09960 | LTA4H | Leukotriene A-4 hydrolase | 2.64 |
| P26583 | HMGB2 | High mobility group protein B2 | 2.65 |
| P18669 | PGAM1 | Phosphoglycerate mutase 1 | 2.66 |
| P20061 | TCN1 | Transcobalamin-1 | 2.71 |
| P06744 | GPI | Glucose-6-phosphate isomerase | 2.72 |
| O15143 | ARPC1B | Actin-related protein 2/3 complex subunit 1B | 2.74 |
| Q9H299 | SH3BGRL3 | SH3 domain-binding glutamic acid-rich-like protein 3 | 2.74 |
| P09758 | TACSTD2 | Tumor-associated calcium signal transducer 2 | 2.76 |
| P17931 | LGALS3 | Galectin-3 | 2.78 |
| P60660 | MYL6 | Myosin light polypeptide 6 | 2.82 |
| P03973 | SLPI | Antileukoproteinase | 2.85 |
| P28066 | PSMA5 | Proteasome subunit alpha type-5 | 2.91 |
| Q06323 | PSME1 | Proteasome activator complex subunit 1 | 2.91 |
| P61604 | HSPE1 | 10 kDa heat shock protein, mitochondrial | 2.92 |
| P16401 | H1-5 | Histone H1.5 | 2.92 |
| P29401 | TKT | Transketolase | 3.04 |
| P07998 | RNASE1 | Ribonuclease pancreatic | 3.08 |
| P26038 | MSN | Moesin | 3.09 |
| P04632 | CAPNS1 | Calpain small subunit 1 | 3.11 |
| P04406 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 3.11 |
| P23528 | CFL1 | Cofilin-1 | 3.14 |
| P40121 | CAPG | Macrophage-capping protein | 3.17 |
| P19957 | PI3 | Elafin | 3.18 |
| P04792 | HSPB1 | Heat shock protein beta-1 | 3.19 |
| P10909 | CLU | Clusterin | 3.19 |
| P12273 | PIP | Prolactin-inducible protein | 3.21 |
| P63104 | YWHAZ | 14-3-3 protein zeta/delta | 3.23 |
| P40926 | MDH2 | Malate dehydrogenase, mitochondrial | 3.26 |
| P06703 | S100A6 | Protein S100-A6 | 3.30 |
| P07339 | CTSD | Cathepsin D | 3.35 |
| P00352 | ALDH1A1 | Aldehyde dehydrogenase 1A1 | 3.36 |
| P20810 | CAST | Calpastatin | 3.36 |
| Q09666 | AHNAK | Neuroblast differentiation-associated protein AHNAK | 3.39 |
| P23284 | PPIB | Peptidyl-prolyl cis-trans isomerase B | 3.41 |
| O14745 | NHERF1 | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | 3.42 |
| P07355 | ANXA2 | Annexin A2 | 3.47 |
| P07237 | P4HB | Protein disulfide-isomerase | 3.47 |
| P11413 | G6PD | Glucose-6-phosphate 1-dehydrogenase | 3.48 |
| O15144 | ARPC2 | Actin-related protein 2/3 complex subunit 2 | 3.50 |
| P61158 | ACTR3 | Actin-related protein 3 | 3.50 |
| P59998 | ARPC4 | Actin-related protein 2/3 complex subunit 4 | 3.57 |
| Q14508 | WFDC2 | WAP four-disulfide core domain protein 2 | 3.62 |
| Q01518 | CAP1 | Adenylyl cyclase-associated protein 1 | 3.62 |
| P07858 | CTSB | Cathepsin B | 3.63 |
| P00338 | LDHA | L-lactate dehydrogenase A chain | 3.65 |
| P52209 | PGD | 6-phosphogluconate dehydrogenase, decarboxylating | 3.66 |
| P80723 | BASP1 | Brain acid soluble protein 1 | 3.66 |
| P78417 | GSTO1 | Glutathione S-transferase omega-1 | 3.69 |
| P14314 | PRKCSH | Glucosidase 2 subunit beta | 3.72 |
| P15311 | EZR | Ezrin | 3.73 |
| P61981 | YWHAG | 14-3-3 protein gamma | 3.76 |
| P61916 | NPC2 | NPC intracellular cholesterol transporter 2 | 3.76 |
| P35579 | MYH9 | Myosin-9 | 3.81 |
| P27797 | CALR | Calreticulin | 3.81 |
| P11142 | HSPA8 | Heat shock cognate 71 kDa protein | 3.82 |
| Q8TDL5 | BPIFB1 | BPI fold-containing family B member 1 | 3.83 |
| P60174 | TPI1 | Triosephosphate isomerase | 3.84 |
| P07602 | PSAP | Prosaposin | 3.86 |
| P31949 | S100A11 | Protein S100-A11 | 3.92 |
| Q99497 | PARK7 | Parkinson disease protein 7 | 3.99 |
| P07195 | LDHB | L-lactate dehydrogenase B chain | 4.00 |
| P01011 | SERPINA3 | Alpha-1-antichymotrypsin | 4.06 |
| P12429 | ANXA3 | Annexin A3 | 4.10 |
| P50395 | GDI2 | Rab GDP dissociation inhibitor beta | 4.32 |
| P25788 | PSMA3 | Proteasome subunit alpha type-3 | 4.33 |
| O00299 | CLIC1 | Chloride intracellular channel protein 1 | 4.36 |
| P31946 | YWHAB | 14-3-3 protein beta/alpha | 4.37 |
| Q96DA0 | ZG16B | Pancreatic adenocarcinoma up-regulated factor | 4.43 |
| P00558 | PGK1 | Phosphoglycerate kinase 1 | 4.47 |
| P30101 | PDIA3 | Protein disulfide-isomerase A3 | 4.53 |
| P47929 | LGALS7 | Galectin-7 | 4.60 |
| P43490 | NAMPT | Nicotinamide phosphoribosyltransferase | 4.61 |
| P55072 | VCP | Transitional endoplasmic reticulum ATPase | 4.62 |
| Q6P5S2 | LEG1 | Protein LEG1 homolog | 4.75 |
| P62258 | YWHAE | 14-3-3 protein epsilon | 4.79 |
| P07900 | HSP90AA1 | Heat shock protein HSP 90-alpha | 4.81 |
| O75083 | WDR1 | WD repeat-containing protein 1 | 4.82 |
| P61978 | HNRNPK | Heterogeneous nuclear ribonucleoprotein K | 4.83 |
| P51149 | RAB7A | Ras-related protein Rab-7a | 4.87 |
| P22626 | HNRNPA2B1 | Heterogeneous nuclear ribonucleoproteins A2/B1 | 4.87 |
| P02545 | LMNA | Prelamin-A/C | 4.88 |
| P08238 | HSP90AB1 | Heat shock protein HSP 90-beta | 4.93 |
| P30041 | PRDX6 | Peroxiredoxin-6 | 5.05 |
| P08758 | ANXA5 | Annexin A5 | 5.10 |
| O95994 | AGR2 | Anterior gradient protein 2 homolog | 5.15 |
| O14818 | PSMA7 | Proteasome subunit alpha type-7 | 5.18 |
| P36952 | SERPINB5 | Serpin B5 | 5.36 |
| Q15084 | PDIA6 | Protein disulfide-isomerase A6 | 5.50 |
| P52907 | CAPZA1 | F-actin-capping protein subunit alpha-1 | 5.57 |
| P25705 | ATP5F1A | ATP synthase subunit alpha, mitochondrial | 5.57 |
| P08133 | ANXA6 | Annexin A6 | 5.64 |
| P46940 | IQGAP1 | Ras GTPase-activating-like protein IQGAP1 | 5.70 |
| P23396 | RPS3 | Small ribosomal subunit protein uS3 | 5.85 |
| P13667 | PDIA4 | Protein disulfide-isomerase A4 | 5.87 |
| P29508 | SERPINB3 | Serpin B3 | 6.20 |
| P26641 | EEF1G | Elongation factor 1-gamma | 6.40 |
| P98088 | MUC5AC | Mucin-5AC | 6.48 |
| P18206 | VCL | Vinculin | 6.57 |
| P27824 | CANX | Calnexin | 7.21 |
| P13639 | EEF2 | Elongation factor 2 | 7.24 |
| Q13813 | SPTAN1 | Spectrin alpha chain, non-erythrocytic 1 | 7.54 |
| Q14764 | MVP | Major vault protein | 8.05 |
| Q9NP55 | BPIFA1 | BPI fold-containing family A member 1 | 8.32 |
| Q00610 | CLTC | Clathrin heavy chain 1 | 9.54 |
| P14625 | HSP90B1 | Endoplasmin | 9.93 |
| Locus | Replicate Count | Description | Gene |
|---|---|---|---|
| P02533 | 4 | Keratin, type I cytoskeletal 14 | KRT14 |
| P35241 | 4 | Radixin | RDX |
| P00505 | 4 | Aspartate aminotransferase, mitochondrial | GOT2 |
| Q9BR76 | 4 | Coronin-1B | CORO1B |
| P47755 | 4 | F-actin-capping protein subunit alpha-2 | CAPZA2 |
| P46926 | 4 | Glucosamine-6-phosphate isomerase 1 | GNPDA1 |
| Q92499 | 4 | ATP-dependent RNA helicase DDX1 | DDX1 |
| Q16222 | 4 | UDP-N-acetylhexosamine pyrophosphorylase | UAP1 |
| O60716 | 4 | Catenin delta-1 | CTNND1 |
| P47897 | 4 | Glutamine-tRNA ligase | QARS1 |
| P36405 | 4 | ADP-ribosylation factor-like protein 3 | ARL3 |
| Q9UHL4 | 4 | Dipeptidyl peptidase 2 | DPP7 |
| P14866 | 4 | Heterogeneous nuclear ribonucleoprotein L | HNRNPL |
| Q04446 | 4 | 1,4-alpha-glucan-branching enzyme | GBE1 |
| P61313 | 4 | Large ribosomal subunit protein eL15 | RPL15 |
| P12268 | 4 | Inosine-5′-monophosphate dehydrogenase 2 | IMPDH2 |
| Q16698 | 4 | 2,4-dienoyl-CoA reductase [(3E)-enoyl-CoA-producing], mitochondrial | DECR1 |
| O43747 | 4 | AP-1 complex subunit gamma-1 | AP1G1 |
| P62495 | 4 | Eukaryotic peptide chain release factor subunit 1 | ETF1 |
| Q07666 | 4 | KH domain-containing, RNA-binding, signal transduction-associated protein 1 | KHDRBS1 |
| P26640 | 4 | Valine-tRNA ligase | VARS1 |
| P09496 | 4 | Clathrin light chain A | CLTA |
| P07814 | 4 | Bifunctional glutamate/proline-tRNA ligase | EPRS1 |
| Q15019 | 4 | Septin-2 | SEPTIN2 |
| P35228 | 4 | Nitric oxide synthase, inducible | NOS2 |
| Q92820 | 4 | Gamma-glutamyl hydrolase | GGH |
| P53999 | 4 | Activated RNA polymerase II transcriptional coactivator p15 | SUB1 |
| Q9Y277 | 4 | Voltage-dependent anion-selective channel protein 3 | VDAC3 |
| Q9BS26 | 4 | Endoplasmic reticulum resident protein 44 | ERP44 |
| Q9Y4L1 | 4 | Hypoxia up-regulated protein 1 | HYOU1 |
| O14980 | 4 | Exportin-1 | XPO1 |
| Q92945 | 4 | Far upstream element-binding protein 2 | KHSRP |
| P50914 | 4 | Large ribosomal subunit protein eL14 | RPL14 |
| O15231 | 4 | Zinc finger protein 185 | ZNF185 |
| P60903 | 4 | Protein S100-A10 | S100A10 |
| P31930 | 4 | Cytochrome b-c1 complex subunit 1, mitochondrial | UQCRC1 |
| O95433 | 4 | Activator of 90 kDa heat shock protein ATPase homolog 1 | AHSA1 |
| P15586 | 4 | N-acetylglucosamine-6-sulfatase | GNS |
| Q16543 | 4 | Hsp90 co-chaperone Cdc37 | CDC37 |
| Q9Y2Z0 | 4 | Protein SGT1 homolog | SUGT1 |
| Q00325 | 4 | Solute carrier family 25 member 3 | SLC25A3 |
| Q99623 | 4 | Prohibitin-2 | PHB2 |
| P20042 | 4 | Eukaryotic translation initiation factor 2 subunit 2 | EIF2S2 |
| P07954 | 4 | Fumarate hydratase, mitochondrial | FH |
| P20674 | 4 | Cytochrome c oxidase subunit 5A, mitochondrial | COX5A |
| Q02790 | 4 | Peptidyl-prolyl cis-trans isomerase FKBP4 | FKBP4 |
| P49748 | 4 | Very long-chain specific acyl-CoA dehydrogenase, mitochondrial | ACADVL |
| O00560 | 4 | Syntenin-1 | SDCBP |
| P05186 | 4 | Alkaline phosphatase, tissue-nonspecific isozyme | ALPL |
| Q99829 | 4 | Copine-1 | CPNE1 |
| P55064 | 4 | Aquaporin-5 | AQP5 |
| Q86UX7 | 4 | Fermitin family homolog 3 | FERMT3 |
| O75955 | 4 | Flotillin-1 | FLOT1 |
| P0CG39 | 4 | POTE ankyrin domain family member J | POTEJ |
| O75369 | 5 | Filamin-B | FLNB |
| P48147 | 5 | Prolyl endopeptidase | PREP |
| P05091 | 5 | Aldehyde dehydrogenase, mitochondrial | ALDH2 |
| Q9P2E9 | 5 | Ribosome-binding protein 1 | RRBP1 |
| P26639 | 5 | Threonine-tRNA ligase 1, cytoplasmic | TARS1 |
| Q01082 | 5 | Spectrin beta chain, non-erythrocytic 1 | SPTBN1 |
| P43034 | 5 | Platelet-activating factor acetylhydrolase IB subunit beta | PAFAH1B1 |
| P35221 | 5 | Catenin alpha-1 | CTNNA1 |
| Q15008 | 5 | 26S proteasome non-ATPase regulatory subunit 6 | PSMD6 |
| P17858 | 5 | ATP-dependent 6-phosphofructokinase, liver type | PFKL |
| O43242 | 5 | 26S proteasome non-ATPase regulatory subunit 3 | PSMD3 |
| P09622 | 5 | Dihydrolipoyl dehydrogenase, mitochondrial | DLD |
| P62195 | 5 | 26S proteasome regulatory subunit 8 | PSMC5 |
| Q13347 | 5 | Eukaryotic translation initiation factor 3 subunit I | EIF3I |
| Q92841 | 5 | Probable ATP-dependent RNA helicase DDX17 | DDX17 |
| Q9Y3C8 | 5 | Ubiquitin-fold modifier-conjugating enzyme 1 | UFC1 |
| P53004 | 5 | Biliverdin reductase A | BLVRA |
| Q13561 | 5 | Dynactin subunit 2 | DCTN2 |
| P05026 | 5 | Sodium/potassium-transporting ATPase subunit beta-1 | ATP1B1 |
| O95777 | 5 | U6 snRNA-associated Sm-like protein LSm8 | LSM8 |
| P46776 | 5 | Large ribosomal subunit protein uL15 | RPL27A |
| P22695 | 5 | Cytochrome b-c1 complex subunit 2, mitochondrial | UQCRC2 |
| P18621 | 5 | Large ribosomal subunit protein uL22 | RPL17 |
| Q99729 | 5 | Heterogeneous nuclear ribonucleoprotein A/B | HNRNPAB |
| P68402 | 5 | Platelet-activating factor acetylhydrolase IB subunit alpha2 | PAFAH1B2 |
| Q9Y6N5 | 5 | Sulfide:quinone oxidoreductase, mitochondrial | SQOR |
| Q99436 | 5 | Proteasome subunit beta type-7 | PSMB7 |
| O00233 | 5 | 26S proteasome non-ATPase regulatory subunit 9 | PSMD9 |
| P35232 | 5 | Prohibitin 1 | PHB1 |
| P48556 | 5 | 26S proteasome non-ATPase regulatory subunit 8 | PSMD8 |
| P68431 | 5 | Histone H3.1 | H3C1 |
| Q16836 | 5 | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | HADH |
| Q15942 | 5 | Zyxin | ZYX |
| P68366 | 6 | Tubulin alpha-4A chain | TUBA4A |
| P48735 | 6 | Isocitrate dehydrogenase [NADP], mitochondrial | IDH2 |
| Q3LXA3 | 6 | Triokinase/FMN cyclase | TKFC |
| Q15149 | 6 | Plectin | PLEC |
| Q9BZQ8 | 6 | Protein Niban 1 | NIBAN1 |
| P10768 | 6 | S-formylglutathione hydrolase | ESD |
| Q9NTK5 | 6 | Obg-like ATPase 1 | OLA1 |
| P35998 | 6 | 26S proteasome regulatory subunit 7 | PSMC2 |
| P35606 | 6 | Coatomer subunit beta’ | COPB2 |
| P05388 | 6 | Large ribosomal subunit protein uL10 | RPLP0 |
| P46459 | 6 | Vesicle-fusing ATPase | NSF |
| P55884 | 6 | Eukaryotic translation initiation factor 3 subunit B | EIF3B |
| P62424 | 6 | Large ribosomal subunit protein eL8 | RPL7A |
| P53618 | 6 | Coatomer subunit beta | COPB1 |
| P30084 | 6 | Enoyl-CoA hydratase, mitochondrial | ECHS1 |
| P21796 | 6 | Non-selective voltage-gated ion channel VDAC1 | VDAC1 |
| P58107 | 6 | Epiplakin | EPPK1 |
| P61758 | 6 | Prefoldin subunit 3 | VBP1 |
| O14773 | 6 | Tripeptidyl-peptidase 1 | TPP1 |
| P04843 | 6 | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit 1 | RPN1 |
| O43852 | 6 | Calumenin | CALU |
| Q99102 | 6 | Mucin-4 | MUC4 |
| O43390 | 7 | Heterogeneous nuclear ribonucleoprotein R | HNRNPR |
| P11216 | 7 | Glycogen phosphorylase, brain form | PYGB |
| P67809 | 7 | Y-box-binding protein 1 | YBX1 |
| P22234 | 7 | Bifunctional phosphoribosylaminoimidazole carboxylase/phosphoribosylaminoimidazole succinocarboxamide synthetase | PAICS |
| P22061 | 7 | Protein-L-isoaspartate(D-aspartate) O-methyltransferase | PCMT1 |
| P62277 | 7 | Small ribosomal subunit protein uS15 | RPS13 |
| Q07960 | 7 | Rho GTPase-activating protein 1 | ARHGAP1 |
| P04066 | 7 | Tissue alpha-L-fucosidase | FUCA1 |
| Q02543 | 7 | Large ribosomal subunit protein eL20 | RPL18A |
| P55058 | 7 | Phospholipid transfer protein | PLTP |
| P22307 | 7 | Sterol carrier protein 2 | SCP2 |
| P49327 | 8 | Fatty acid synthase | FASN |
| P45974 | 8 | Ubiquitin carboxyl-terminal hydrolase 5 | USP5 |
| Q6XQN6 | 8 | Nicotinate phosphoribosyltransferase | NAPRT |
| P20073 | 8 | Annexin A7 | ANXA7 |
| Q15370 | 8 | Elongin-B | ELOB |
| Q15691 | 8 | Microtubule-associated protein RP/EB family member 1 | MAPRE1 |
| P13473 | 8 | Lysosome-associated membrane glycoprotein 2 | LAMP2 |
| Q9UII2 | 8 | ATPase inhibitor, mitochondrial | ATP5IF1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Araújo, C.P.M.; Vieira, C.M.; Ohse, K.C.; Silva, A.S.; Cavalcante, S.A.; Naveca, F.G.; Oliveira, F.N.; Crainey, J.L.; Lacerda, M.V.G.; Melo, G.C.; et al. Nasopharyngeal Proteomic Profiles from Patients Hospitalized Due to COVID-19 in Manaus, Amazonas, Brazil. COVID 2025, 5, 192. https://doi.org/10.3390/covid5110192
Araújo CPM, Vieira CM, Ohse KC, Silva AS, Cavalcante SA, Naveca FG, Oliveira FN, Crainey JL, Lacerda MVG, Melo GC, et al. Nasopharyngeal Proteomic Profiles from Patients Hospitalized Due to COVID-19 in Manaus, Amazonas, Brazil. COVID. 2025; 5(11):192. https://doi.org/10.3390/covid5110192
Chicago/Turabian StyleAraújo, Cláudia P. M., Carolina M. Vieira, Ketlen C. Ohse, Alessandra S. Silva, Sofia A. Cavalcante, Felipe G. Naveca, Fernanda N. Oliveira, James L. Crainey, Marcus V. G. Lacerda, Gisely C. Melo, and et al. 2025. "Nasopharyngeal Proteomic Profiles from Patients Hospitalized Due to COVID-19 in Manaus, Amazonas, Brazil" COVID 5, no. 11: 192. https://doi.org/10.3390/covid5110192
APA StyleAraújo, C. P. M., Vieira, C. M., Ohse, K. C., Silva, A. S., Cavalcante, S. A., Naveca, F. G., Oliveira, F. N., Crainey, J. L., Lacerda, M. V. G., Melo, G. C., Sampaio, V. S., Batista, M., Camillo-Andrade, A. C., Santos, M. D. M., Lima, D. B., Fischer, J. d. S. G., Carvalho, P. C., & Aquino, P. F. (2025). Nasopharyngeal Proteomic Profiles from Patients Hospitalized Due to COVID-19 in Manaus, Amazonas, Brazil. COVID, 5(11), 192. https://doi.org/10.3390/covid5110192








