Machine Learning Models to Discriminate COVID-19 Severity with Biomarkers Available in Brazilian Public Health
Abstract
1. Introduction
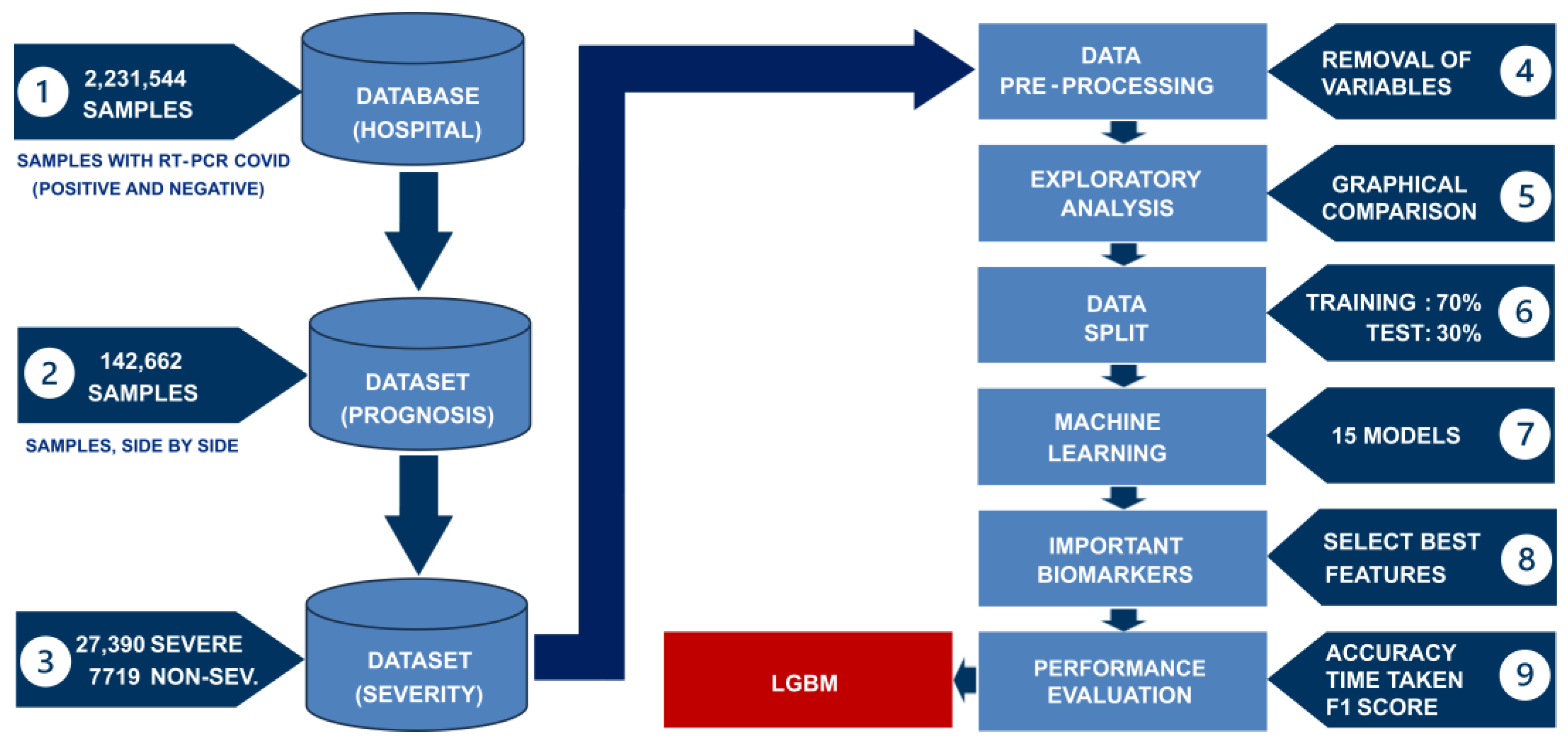
2. Materials and Methods
2.1. Study Location
2.2. Collected Data
2.3. Data Processing
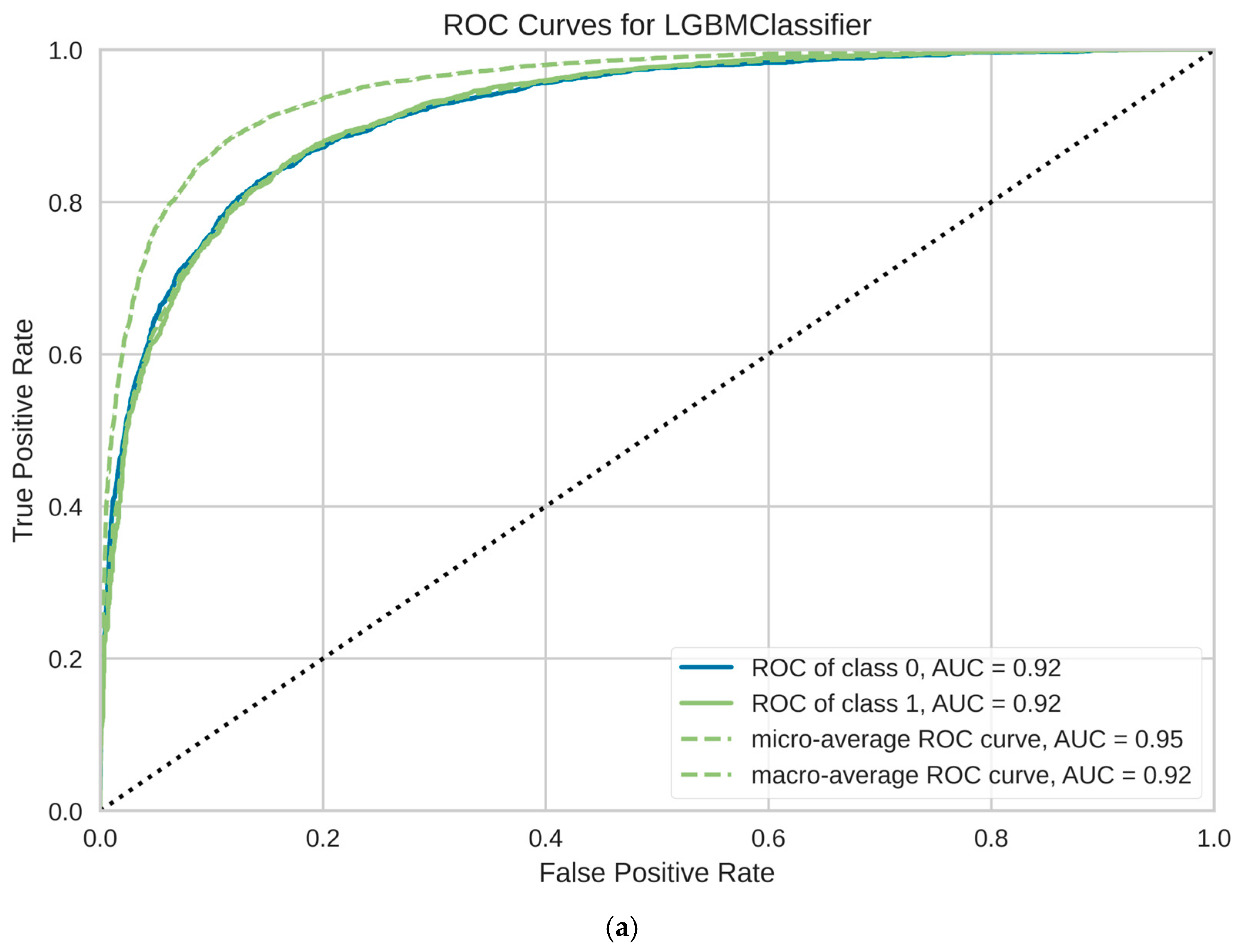
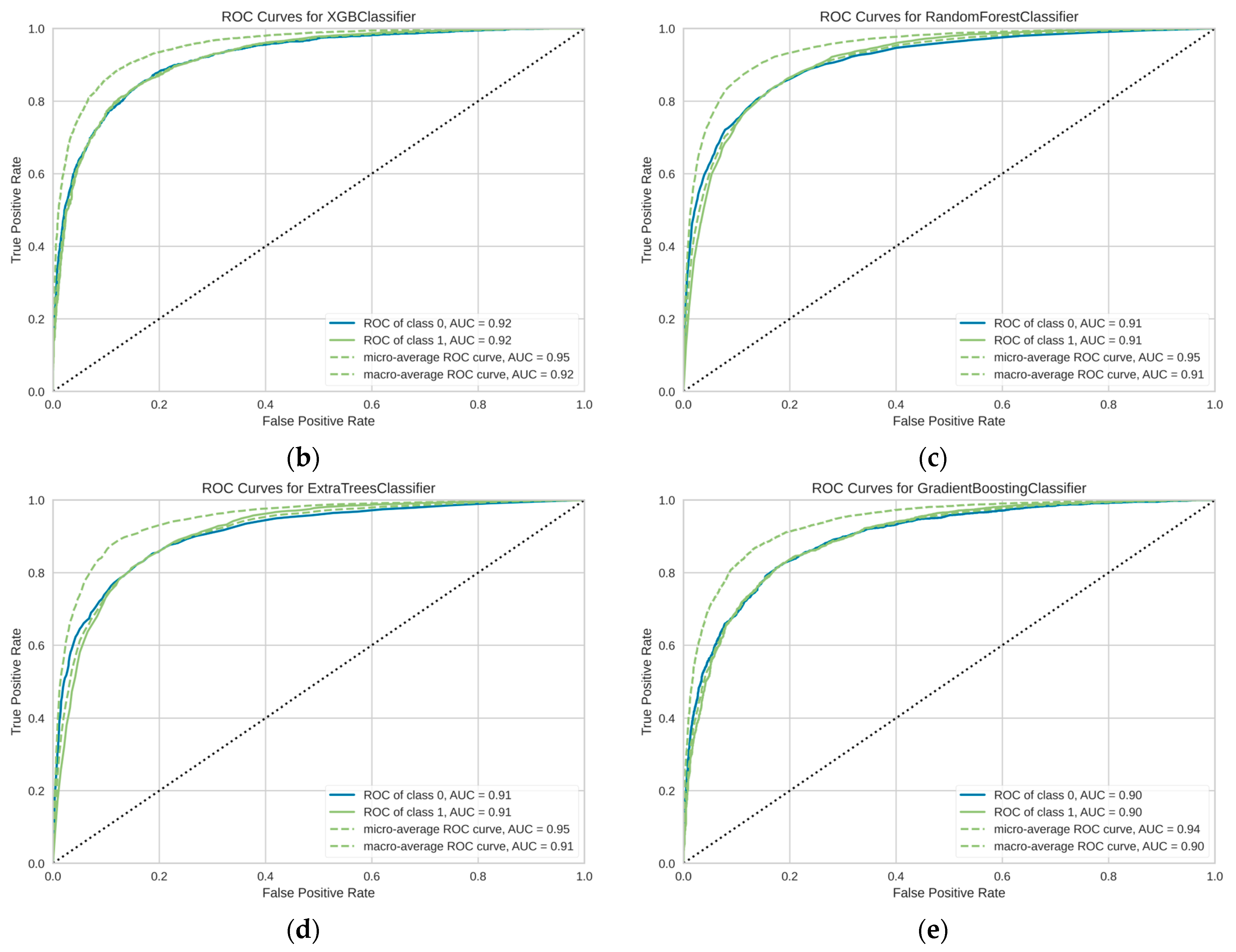
3. Results
4. Discussion
4.1. Biomarkers
4.2. Comparison with Previous Studies
4.3. Limitations
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HC-UFPR | Hospital de Clínicas da Universidade Federal do Paraná |
| IBGE | Instituto Brasileiro de Geografia e Estatística |
| ICU | Intensive Care Unit |
| INPI | Instituto Nacional da Propriedade Industrial |
| LGBM | Light Gradient Boosting Machine |
| ML | Machine Learning |
| SUS | Sistema Único de Saúde |
| WHO | World Health Organization |
Appendix A
| Results | Confusion Matrix | |
|---|---|---|
| Positive | Negative | |
| Positive | a True Positive | b False Positive |
| Negative | c False Negative | d True Negative |
References
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Thevarajan, I.; Nguyen, T.H.O.; Koutsakos, M.; Druce, J.; Caly, L.; van de Sandt, C.E.; Jia, X.; Nicholson, S.; Catton, M.; Cowie, B.; et al. Breadth of concomitant immune responses prior to patient recovery: A case report of mild to moderate COVID-19. Nat. Med. 2020, 26, 453–455. [Google Scholar] [CrossRef]
- Mohamadian, M.; Chiti, H.; Shoghli, A.; Biglari, S.; Parsamanesh, N.; Esmaeilzadeh, A. COVID-19: Virology, biology and novel laboratory diagnosis. J. Gene Med. 2021, 23, e3303. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Post COVID-19 Condition (Long COVID): Facts Sheets. 7 December 2022. Geneva–SZ. 2022. Available online: https://www.who.int/europe/news-room/fact-sheets/item/post-covid-19-condition (accessed on 9 September 2024).
- Davis, H.E.; McCorkell, L.; Vogel, J.M.; Topol, E.J. Long COVID: Major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 2023, 21, 133–146. [Google Scholar] [CrossRef]
- Alizargar, J. Risk of reactivation or reinfection of novel coronavirus (COVID-19). J. Formos. Med. Assoc. 2020, 119, 1123. [Google Scholar] [CrossRef]
- Sahu, P.; Pinkalwar, N.; Dubey, R.D.; Paroha, S.; Chatterjee, S.; Chatterjee, T. Biomarkers: An Emerging Tool for Diagnosis of a Disease and Drug Development. Asian J. Res. Pharm. Sci. 2011, 1, 9–16. [Google Scholar]
- Bodaghi, A.; Fattahi, N.; Ramazani, A. Biomarkers: Promising and valuable tools towards diagnosis, prognosis and treatment of COVID-19 and other diseases. Heliyon 2023, 9, e13323. [Google Scholar] [CrossRef]
- Lippi, G.; Plebani, M. Laboratory abnormalities in patients with COVID-2019 infection. Clin. Chem. Lab. Med. (CCLM) 2020, 58, 1131–1134. [Google Scholar] [CrossRef]
- Xavier, A.R.; Silva, J.S.; Almeida, J.P.C.L.; Conceição, J.F.F.; Lacerda, G.S.; Kanaan, S. COVID-19: Clinical and laboratory manifestations in novel coronavirus infection. J. Bras. Patol. Med. Lab. 2020, 56, e3232020. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. Jama 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Russell, S.J.; Norvig, P. Artificial Intelligence: A Modern Approach, 4th ed.; Pearson Education, Inc.: Hoboken, NJ, USA, 2021; 1168p. [Google Scholar]
- Albuquerque Paiva de Oliveira, R.F.; Bastos Filho, C.J.A.; A. M. V. F. de Medeiros, A.C.; Buarque Lins dos Santos, P.J.; Lopes Freire, D. Machine learning applied in SARS-CoV-2 COVID 19 screening using clinical analysis parameters. IEEE Lat. Am. Trans. 2021, 19, 978–985. [Google Scholar] [CrossRef]
- Zhang, J.; Dong, X.; Cao, Y.; Yuan, Y.; Yang, Y.; Yan, Y.; Akdis, C.A.; Gao, Y. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy 2020, 75, 1730–1741. [Google Scholar] [CrossRef]
- Microsoft. LightGBM’s Documentation. 2025. Available online: https://lightgbm.readthedocs.io/en/latest/index.html (accessed on 25 January 2025).
- Beeching, N.; Fletcher, T.; Fowler, R. Coronavirus Disease 2019 (COVID-19)—Symptoms, diagnosis, and treatment. BMJ Best Practice. 2025. Available online: https://bestpractice.bmj.com/topics/en-gb/3000201 (accessed on 17 October 2024).
- World Health Organization. Clinical Management of COVID-19: Living Guideline (Edition 12). 2023. Available online: https://www.who.int/publications/i/item/WHO-2019-nCoV-clinical-2023.1 (accessed on 17 October 2024).
- Bohn, M.K.; Lippi, G.; Horvath, A.; Sethi, S.; Koch, D.; Ferrari, M.; Wang, C.; Mancini, N.; Steele, S.; Adeli, K. Molecular, serological, and biochemical diagnosis and monitoring of COVID-19: IFCC taskforce evaluation of the latest evidence. Clin. Chem. Lab. Med. (CCLM) 2020, 58, 1037–1052. [Google Scholar] [CrossRef]
- Instituto Brasileiro de Geografia e Estatística. Cidades e Estados: Curitiba. Rio de Janeiro: IBGE. 2021. Available online: https://www.ibge.gov.br/cidades-e-estados/pr/curitiba.html (accessed on 11 November 2024).
- EBSERV, Empresa Brasileira de Serviços Hospitalares. Hospital de Clínicas. Curitiba: EBSERV. 2020. Available online: https://www.gov.br/ebserh/pt-br/hospitais-universitarios/regiao-sul/chc-ufpr/comunicacao/noticias/hospital-de-clinicas-2013-ufpr-ebserh-uma-historia-construida-a-muitas-maos-1 (accessed on 11 November 2024).
- Nelli, F. Python Data Analytics: With Pandas, Numpy, and Matplotlib; Apress Access Books; Apress: Berkeley, CA, USA, 2018; p. 569. [Google Scholar] [CrossRef]
- Ali, M. PYCARET: An Open-Source, Low-Cod Machine Learning Library in Python. 2025. Available online: https://pycaret.org (accessed on 11 November 2024).
- Python Software Foundation. Python Language Site: Documentation. 2025. Available online: https://www.python.org/ (accessed on 11 November 2024).
- Nunes, L.N.; Klück, M.M.; Fachel, J.M.G. Uso da imputação múltipla de dados faltantes: Uma simulação utilizando dados epidemiológicos. Cad. De Saúde Pública 2009, 25, 268–278. [Google Scholar] [CrossRef]
- Prado, A.L.; Cobre, A.F.; Volanski, W.; Signorini, L.; Valdameri, G.; Moure, V.R.; Alves, A.S.C.; Rego, F.G.M.; Picheth, G. COVID-19 Severity: Script in Python for a Model Machine Learning; GitHub, Inc.: San Fracisco, CA, USA, 2025; Available online: https://github.com/COVID-19-Severity (accessed on 18 September 2025).
- Han, J.; Kamber, M.; Pei, J. Data Mining: Concepts and Techniques, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Liu, T.Y. LightGBM: A highly efficient gradient boosting decision tree. In Proceedings of the 31st Conference on Neural Information Processing System, Long Beach, CA, USA, 4–9 December 2017; pp. 1–9. Available online: https://proceedings.neurips.cc/paper_files/paper/2017/file/6449f44a102fde848669bdd9eb6b76fa-Paper.pdf (accessed on 9 September 2024).
- Zhu, W.; Zeng, N.; Wang, N. Sensitivity, Specificity, Accuracy. Associated Confidence Interval and ROC Analysis with Practical SAS® implementations. In Proceedings of the Health Care and Life Sciences. NESUG Proceedings, Baltimore, MD, USA, 14–17 November 2010; pp. 1–9. Available online: https://www.lexjansen.com/nesug/nesug10/hl/hl07.pdf (accessed on 11 November 2024).
- Casalicchio, G.; Molnar, C.; Bischl, B. Visualizing the Feature Importance for Black Box Models; Springer: Cham, Switzerland, 2019; pp. 655–670. [Google Scholar] [CrossRef]
- Buenrostro-Mariscal, R.; Montesinos-López, O.A.; Gonzalez-Gonzalez, C. Predicting Hospitalization in Older Adults Using Machine Learning. Geriatrics 2025, 10, 6. [Google Scholar] [CrossRef]
- Fajgenbaum, D.C.; June, C.H. Cytokine Storm. N. Engl. J. Med. 2020, 383, 2255–2273. [Google Scholar] [CrossRef] [PubMed]
- Siavoshi, F.; Safavi-Naini, S.A.A.; Shirzadeh Barough, S.; Azizmohammad Looha, M.; Hatamabadi, H.; Ommi, D.; Jalili Khoshnoud, R.; Fatemi, A.; Pourhoseingholi, M.A. On-admission and dynamic trend of laboratory profiles as prognostic biomarkers in COVID-19 inpatients. Sci. Rep. 2023, 13, 6993. [Google Scholar] [CrossRef] [PubMed]
- Battaglini, D.; Lopes-Pacheco, M.; Castro-Faria-Neto, H.C.; Pelosi, P.; Rocco, P.R.M. Laboratory Biomarkers for Diagnosis and Prognosis in COVID-19. Front. Immunol. 2022, 13, 857573. [Google Scholar] [CrossRef] [PubMed]
- Pezzullo, A.M.; Axfors, C.; Contopoulos-Ioannidis, D.G.; Apostolatos, A.; Ioannidis, J.P. Age-stratified infection fatality rate of COVID-19 in the non-elderly population. Environ. Res. 2023, 216, 114655. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Underlying Conditions and the Higher Risk for Severe COVID-19. COVID-19. 18 July 2024. Available online: https://www.cdc.gov/covid/hcp/clinical-care/underlying-conditions.html (accessed on 11 November 2024).
- Lippi, G.; Plebani, M.; Henry, B.M. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: A meta-analysis. Clin. Chim. Acta 2020, 506, 145–148. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Luo, R.; Wang, K.; Zhang, M.; Wang, Z.; Dong, L.; Xu, G. Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. 2020, 97, 829–838. [Google Scholar] [CrossRef]
- Taneri, P.E.; Gómez-Ochoa, S.A.; Llanaj, E.; Raguindin, P.F.; Rojas, L.Z.; Roa-Díaz, Z.M.; Franco, O.H. Anemia and iron metabolism in COVID-19: A systematic review and meta-analysis. Eur. J. Epidemiol. 2020, 35, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Sahu, B.R.; Kampa, R.K.; Padhi, A.; Panda, A.K. C-reactive protein: A promising biomarker for poor prognosis in COVID-19 infection. Clin. Chim. Acta 2020, 509, 91–94. [Google Scholar] [CrossRef]
- Huang, I.; Pranata, R. Lymphopenia in severe coronavirus disease-2019 (COVID-19): Systematic review and meta-analysis. J. Intensive Care 2020, 8, 36. [Google Scholar] [CrossRef]
- Violi, F.; Cangemi, R.; Romiti, G.F.; Ceccarelli, G.; Oliva, A.; Alessandri, F.; Pirro, M.; Pignatelli, P.; Lichtner, M.; Carraro, A.; et al. Is albumin predictor of mortality in COVID-19? Antioxid. Redox Signal. 2021, 35, 139–142. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; She, Z.G.; Cheng, X.; Qin, J.J.; Zhang, X.J.; Cai, J.; Li, H. Association of blood glucose control and outcomes in patients with COVID-19 and pre-existing type 2 diabetes. Cell Metab. 2020, 31, 1068–1077.e3. [Google Scholar] [CrossRef]
- Frontera, J.A.; Valdes, E.; Huang, J.M.; Lewis, A.; Lord, A.S.; Zhou, T.; Kahn, D.E.D.; Melmed, K.; Czeisler, B.M.; Yaghi, S.; et al. Prevalence and impact of hyponatremia in patients with coronavirus disease 2019 in New York City. Crit. Care Med. 2020, 48, e1211–e1217. [Google Scholar] [CrossRef]
- Cabitza, F.; Campagner, A.; Ferrari, D.; Di Resta, C.; Ceriotti, D.; Sabetta, E.; Colombini, A.; De Vecchi, E.; Banfi, G.; Locatelli, M.; et al. Development, evaluation, and validation of machine learning models for COVID-19 detection based on routine blood tests. Clin. Chem. Lab. Med. (CCLM) 2021, 59, 421–431. [Google Scholar] [CrossRef]
- Cobre, A.d.F.; Stremel, D.P.; Noleto, G.R.; Fachi, M.M.; Surek, M.; Wiens, A.; Tonin, F.S.; Pontarolo, R. Diagnosis and prediction of COVID-19 severity: Can biochemical tests and machine learning be used as prognostic indicators? Comput. Biol. Med. 2021, 134, 104531. [Google Scholar] [CrossRef]
- Kukar, M.; Gunčar, G.; Vovko, T.; Podnar, S.; Černelč, P.; Brvar, M.; Zalaznik, M.; Notar, M.; Moškon, S.; Notar, M. COVID-19 diagnosis by routine blood tests using machine learning. Sci. Rep. 2021, 11, 10738. [Google Scholar] [CrossRef]
- Huyut, M.T.; Velichko, A.; Belyaev, M. Detection of Risk Predictors of COVID-19 Mortality with Classifier Machine Learning Models Operated with Routine Laboratory Biomarkers. Appl. Sci. 2022, 12, 12180. [Google Scholar] [CrossRef]
- Baik, S.M.; Hong, K.S.; Park, D.J. Application and utility of boosting machine learning model based on laboratory test in the differential diagnosis of non-COVID-19 pneumonia and COVID-19. Clin. Biochem. 2023, 118, 110584. [Google Scholar] [CrossRef]
- Çubukçu, H.C.; Topcu, D.I.; Bayraktar, N.; Gülşen, M.; Sarı, N.; Arslan, A.H. Detection of COVID-19 by Machine Learning Using Routine Laboratory Tests. Am. J. Clin. Pathol. 2021, 157, 758–766. [Google Scholar] [CrossRef]
- Huang, R.; Xie, L.; He, J.; Dong, H.; Liu, T. Association between the peripheral blood eosinophil counts and COVID-19: A meta-analysis. Medicine 2021, 100, e26047. [Google Scholar] [CrossRef]
- Mu, T.; Yi, Z.; Wang, M.; Wang, J.; Zhang, C.; Chen, H.; Bai, M.; Jiang, L.; Zhang, Y. Expression of eosinophil in peripheral blood of patients with COVID-19 and its clinical significance. J. Clin. Lab. Anal. 2021, 35, e23620. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wu, C.; Wang, X.; Yu, J.; Sun, Z. The impact of COVID-19 on blood glucose: A systematic review and meta-analysis. Front. Endocrinol. 2020, 11, 574541. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Guo, H.; Zhou, Y.; Guo, J.; Wang, T.; Zhao, X.; Li, H.; Sun, Y.; Bian, X.; Fang, C. The associations between fasting plasma glucose levels and mortality of COVID-19 in patients without diabetes. Diabetes Res. Clin. Pract. 2020, 169, 108448. [Google Scholar] [CrossRef] [PubMed]
- Prado, A.L.; Volanski, W.; Signorini, L.; Valdameri, G.; Moure, V.R.; Rego, F.G.M.; Picheth, G. COVID-19 Severity. BR512024002371-2. Instituto Nacional da Propriedade Industrial, Diretoria de Patentes, Programas de Computador, 2793 ed., p. 13. 2024. Available online: https://revistas.inpi.gov.br/pdf/Programa_de_computador2793.pdf (accessed on 18 September 2024).

| Profile (Class) | Biomarker |
|---|---|
| Glycemic | Glucose |
| HbA1c | |
| Liver | Alanine transaminase |
| Aspartate transaminase | |
| Albumin | |
| Total Protein | |
| Globulin | |
| Ferritin | |
| C-reactive protein | |
| Lipid | Total Cholesterol |
| HDL-C | |
| LDL-C | |
| Triglycerides | |
| Pancreatic | Amylase |
| Lipase | |
| Renal | Creatinine |
| Urea | |
| Potassium | |
| Sodium | |
| Acid-Base Balance | pCO2 |
| pO2 | |
| sO2 | |
| pH | |
| HCO3-standard | |
| HCO3-actual | |
| BE-CF | |
| BE-B | |
| CTCO2 | |
| Coagulation | Prothrombin Time |
| Prothrombin Time–Relation | |
| Prothrombin Time—International Normalised Ratio | |
| Partial Thromboplastin Time | |
| Partial Thromboplastin Time–Relation | |
| D-Dimer | |
| Complete Blood Count | Erythrocytes |
| Haemoglobin | |
| Leukocytes | |
| Mature Neutrophils | |
| Immature Neutrophils | |
| Neutrophils | |
| Basophils | |
| Eosinophils | |
| Lymphocytes | |
| Atypical Lymphocytes | |
| Monocytes | |
| Platelets | |
| Inflammatory | Procalcitonin |
| Cardiac | Troponin I (cTnI) |
| Model | Complete Name | ACC | AUC | Recall | Prec. | F1 | Kappa | MCC | TT (s) |
|---|---|---|---|---|---|---|---|---|---|
| lightgbm | Light Gradient Boosting Machine | 0.8808 | 0.9177 | 0.8808 | 0.8757 | 0.8757 | 0.6250 | 0.6314 | 6.9560 |
| xgboost | Extreme Gradient Boosting | 0.8796 | 0.9153 | 0.8796 | 0.8747 | 0.8751 | 0.6247 | 0.6297 | 0.7510 |
| rf | Random Forest Classifier | 0.8786 | 0.9060 | 0.8786 | 0.8732 | 0.8725 | 0.6136 | 0.6220 | 5.2900 |
| et | Extra Trees Classifier | 0.8779 | 0.9047 | 0.8779 | 0.8726 | 0.8714 | 0.6098 | 0.6191 | 4.7040 |
| gbc | Gradient Boosting Classifier | 0.8636 | 0.8939 | 0.8636 | 0.8572 | 0.8525 | 0.5460 | 0.5645 | 5.7600 |
| ada | Ada Boost Classifier | 0.8481 | 0.8744 | 0.8481 | 0.8383 | 0.8375 | 0.5024 | 0.5153 | 1.3780 |
| dt | Decision Tree Classifier | 0.8144 | 0.7358 | 0.8144 | 0.8168 | 0.8155 | 0.4656 | 0.4658 | 0.5530 |
| lda | Linear Discriminant Analysis | 0.8087 | 0.7924 | 0.8087 | 0.7868 | 0.7794 | 0.3057 | 0.3401 | 0.2300 |
| lr | Logistic Regression | 0.8031 | 0.7783 | 0.8031 | 0.7785 | 0.7668 | 0.2619 | 0.3045 | 4.4330 |
| ridge | Ridge Classifier | 0.8029 | 0.7927 | 0.8029 | 0.7818 | 0.7587 | 0.2326 | 0.2909 | 0.1790 |
| knn | K Neighbors Classifier | 0.7882 | 0.6947 | 0.7882 | 0.7582 | 0.7616 | 0.2545 | 0.2740 | 1.0320 |
| dummy | Dummy Classifier | 0.7802 | 0.5000 | 0.7802 | 0.6086 | 0.6838 | 0.0000 | 0.0000 | 0.1610 |
| svm | SVM—Linear Kernel | 0.5547 | 0.5315 | 0.5547 | 0.7456 | 0.4531 | 0.0113 | 0.0414 | 1.0880 |
| qda | Quadratic Discriminant Analysis | 0.4920 | 0.7272 | 0.4920 | 0.7780 | 0.4170 | 0.0281 | 0.0930 | 0.3640 |
| nb | Naive Bayes | 0.2816 | 0.7064 | 0.2816 | 0.7608 | 0.2069 | 0.0263 | 0.0890 | 0.1780 |
| Machine Learning Model | Severity Classification Based of the Samples | |
|---|---|---|
| LGBM (n = 10,533) | (0) Mild to Moderate (20%) | (1) Severe (80%) |
| (0) Mild to Moderate | 1462 (14%) | 854 (5%) |
| (1) Severe | 362 (6%) | 7855 (75%) |
| XGBoost (n = 10,533) | (0) Mild to Moderate (18%) | (1) Severe (82%) |
| (0) Mild to Moderate | 1475 (14%) | 841 (8%) |
| (1) Severe | 425 (4%) | 7792 (74%) |
| Random Forest (n = 10,533) | (0) Mild to Moderate (17%) | (1) Severe (83%) |
| (0) Mild to Moderate | 1422 (13%) | 894 (9%) |
| (1) Severe | 387 (4%) | 7830 (74%) |
| Extra Trees (n = 10,533) | (0) Mild to Moderate (16%) | (1) Severe (84%) |
| (0) Mild to Moderate | 1396 (13%) | 920 (9%) |
| (1) Severe | 364 (3%) | 7853 (75%) |
| Gradient Boosting (n = 10,533) | (0) Mild to Moderate (15%) | (1) Severe (85%) |
| (0) Mild to Moderate | 1213 (12%) | 1103 (10%) |
| (1) Severe | 326 (3%) | 7891 (75%) |
| Parameters (%) | LGBM | XGBoost | Random Forest | Extra Trees | Gradient Boosting |
|---|---|---|---|---|---|
| Sensitivity | 80% | 78% | 79% | 79% | 79% |
| Specificity | 90% | 90% | 90% | 89% | 88% |
| Positive Predictive Value | 63% | 64% | 61% | 60% | 52% |
| Negative Predictive Value | 95% | 95% | 95% | 96% | 96% |
| Accuracy | 88% | 88% | 88% | 88% | 86% |
| Matthews Correlation Coefficient | 64% | 63% | 62% | 62% | 57% |
| Age Groups | n | AUC (%) | Sensibility (%) | Specificity (%) | Accuracy (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|
| 0–17 y | 832 | 91 | 82 | 87 | 85 | 72 | 92 |
| 18–29 y | 852 | 91 | 77 | 90 | 87 | 69 | 93 |
| 30–39 y | 1025 | 91 | 75 | 90 | 87 | 63 | 94 |
| 40–49 y | 1295 | 93 | 83 | 91 | 90 | 70 | 96 |
| 50–64 y | 3326 | 91 | 76 | 91 | 89 | 62 | 95 |
| 65–74 y | 1975 | 90 | 75 | 90 | 87 | 62 | 94 |
| 75–84 y | 1028 | 88 | 71 | 90 | 88 | 47 | 96 |
| >85 y | 197 | 87 | 76 | 89 | 87 | 63 | 94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prado, A.L.d.; Cobre, A.d.F.; Volanski, W.; Signorini, L.; Valdameri, G.; Moure, V.R.; Alves, A.d.S.C.; Rego, F.G.d.M.; Picheth, G. Machine Learning Models to Discriminate COVID-19 Severity with Biomarkers Available in Brazilian Public Health. COVID 2025, 5, 167. https://doi.org/10.3390/covid5100167
Prado ALd, Cobre AdF, Volanski W, Signorini L, Valdameri G, Moure VR, Alves AdSC, Rego FGdM, Picheth G. Machine Learning Models to Discriminate COVID-19 Severity with Biomarkers Available in Brazilian Public Health. COVID. 2025; 5(10):167. https://doi.org/10.3390/covid5100167
Chicago/Turabian StylePrado, Ademir Luiz do, Alexandre de Fátima Cobre, Waldemar Volanski, Liana Signorini, Glaucio Valdameri, Vivian Rotuno Moure, Alexessander da Silva Couto Alves, Fabiane Gomes de Moraes Rego, and Geraldo Picheth. 2025. "Machine Learning Models to Discriminate COVID-19 Severity with Biomarkers Available in Brazilian Public Health" COVID 5, no. 10: 167. https://doi.org/10.3390/covid5100167
APA StylePrado, A. L. d., Cobre, A. d. F., Volanski, W., Signorini, L., Valdameri, G., Moure, V. R., Alves, A. d. S. C., Rego, F. G. d. M., & Picheth, G. (2025). Machine Learning Models to Discriminate COVID-19 Severity with Biomarkers Available in Brazilian Public Health. COVID, 5(10), 167. https://doi.org/10.3390/covid5100167






