Incubation Periods of SARS-CoV-2 Wild-Type, Delta, and Omicron Variants–Dominant Periods in Singapore
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Setting and Data Source
2.2. Identification of SARS-CoV-2 Variants
2.3. Determining the Various Incubation Periods of Variants
2.4. Statistical Analysis
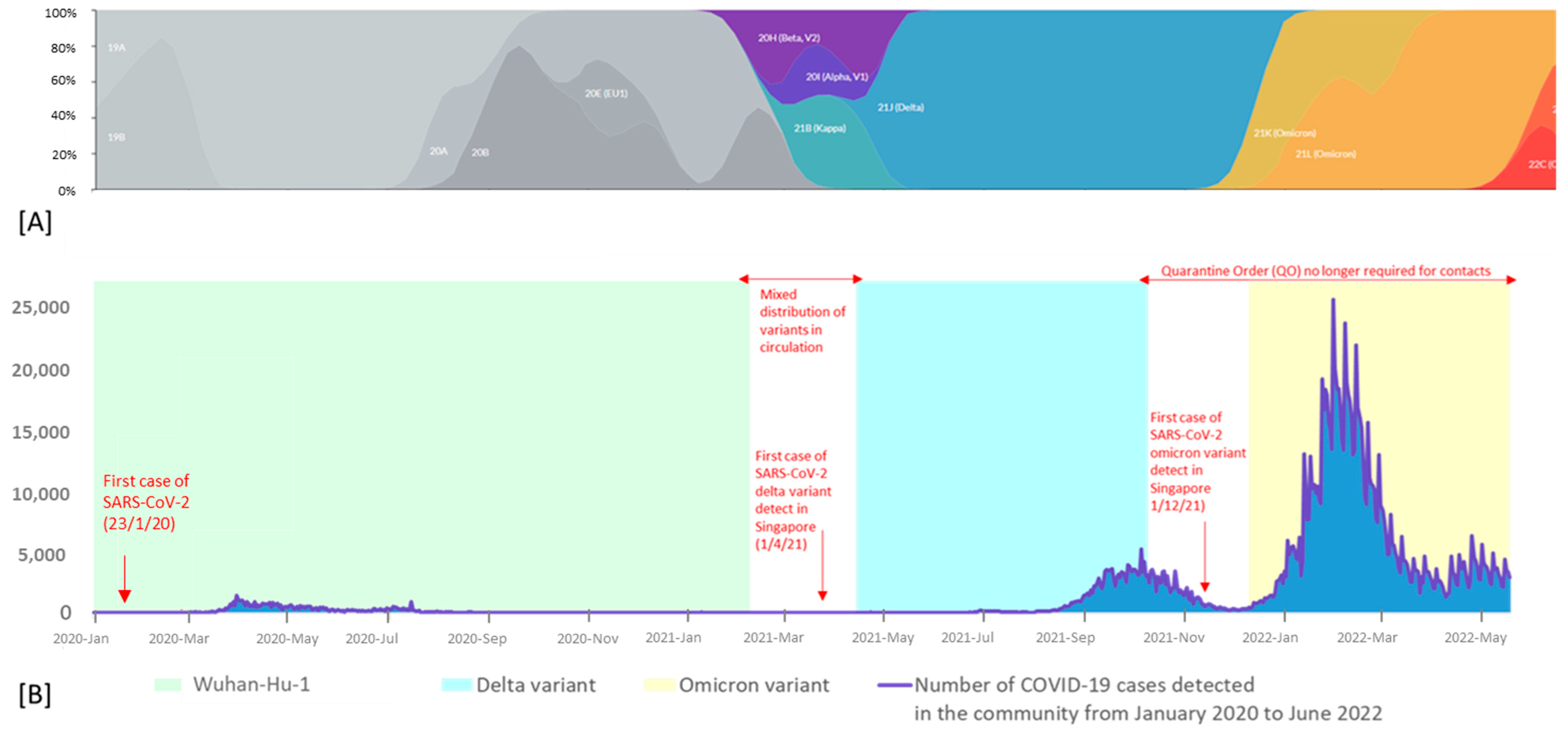
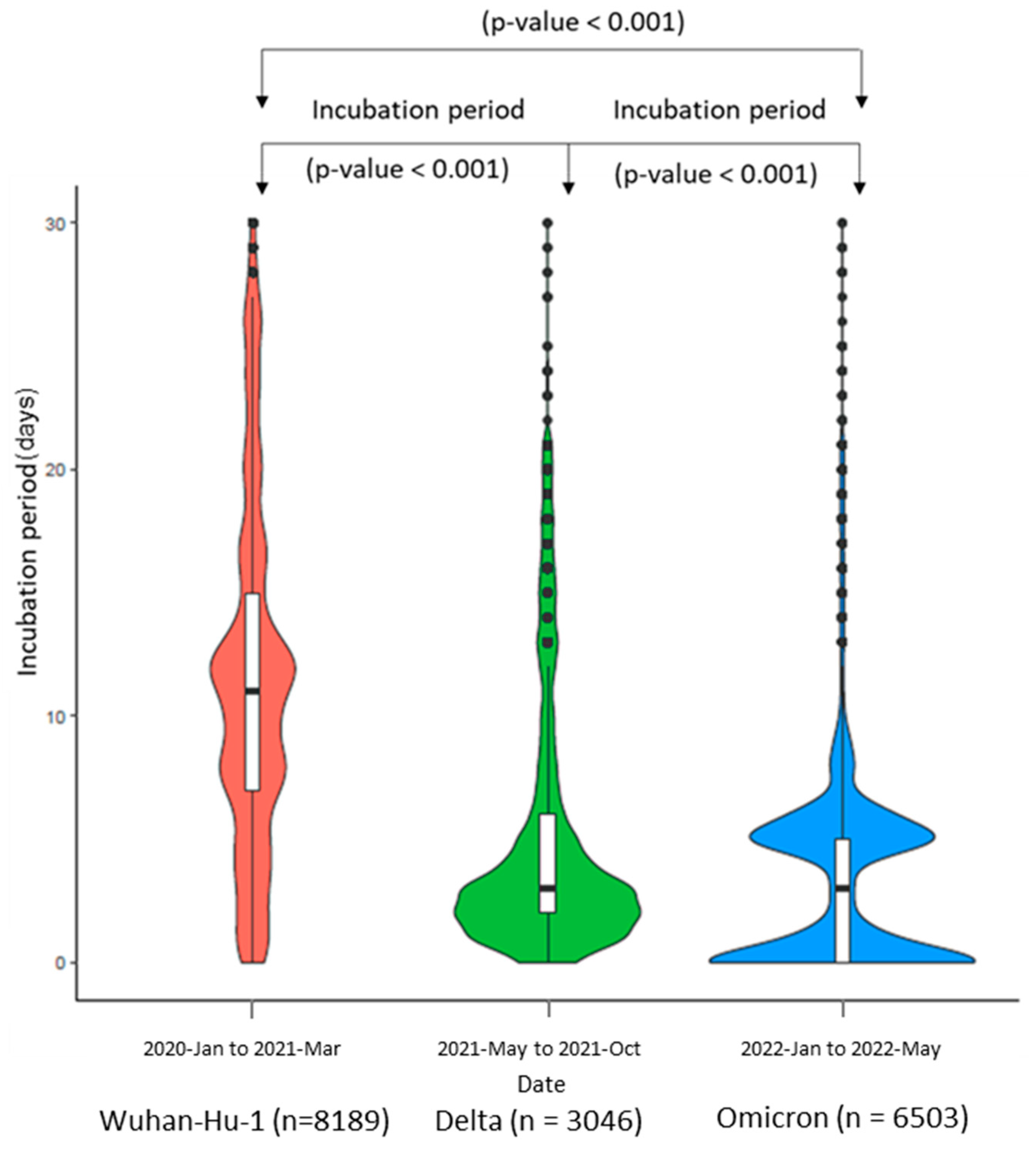
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tan, J.B.; Cook, M.J.; Logan, P.; Rozanova, L.; Wilder-Smith, A. Singapore’s Pandemic Preparedness: An Overview of the First Wave of COVID-19. Int. J. Environ. Res. Public Health 2020, 18, 252. [Google Scholar] [CrossRef] [PubMed]
- Pfefferle, S.; Huang, J.; Nörz, D.; Indenbirken, D.; Lütgehetmann, M.; Oestereich, L.; Günther, T.; Grundhoff, A.; Aepfelbacher, M.; Fischer, N. Complete genome sequence of a SARS-CoV-2 strain isolated in Northern Germany. Microbiol. Resour. Announc. 2020, 9, 10–128. [Google Scholar] [CrossRef] [PubMed]
- Aleem, A.; Akbar Samad, A.B.; Vaqar, S. Emerging Variants of SARS-CoV-2 and Novel Therapeutics Against Coronavirus (COVID-19). In StatPearls; StatPearls: Treasure Island, FL, USA, 2023. [Google Scholar]
- Xin, H.; Wong, J.Y.; Murphy, C.; Yeung, A.; Ali, S.T.; Wu, P.; Cowling, B.J. The Incubation Period Distribution of Coronavirus Disease 2019: A Systematic Review and Meta-analysis. Clin. Infect. Dis. 2021, 73, 2344–2352. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Kang, L.; Guo, Z.; Liu, J.; Liu, M.; Liang, W. Incubation Period of COVID-19 Caused by Unique SARS-CoV-2 Strains: A Systematic Review and Meta-analysis. JAMA Netw. Open 2022, 5, e2228008. [Google Scholar] [CrossRef] [PubMed]
- Lauer, S.A.; Grantz, K.H.; Bi, Q.; Jones, F.K.; Zheng, Q.; Meredith, H.R.; Azman, A.S.; Reich, N.G.; Lessler, J. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application. Ann. Intern Med. 2020, 172, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Ogata, T.; Tanaka, H. SARS-CoV-2 Incubation Period during the Omicron BA.5-Dominant Period in Japan. Emerg. Infect. Dis. 2023, 29, 595–598. [Google Scholar] [CrossRef] [PubMed]
- Khare, S.; Gurry, C.; Freitas, L.; Schultz, M.B.; Bach, G.; Diallo, A.; Akite, N.; Ho, J.; Lee, R.T.; Yeo, W.; et al. GISAID’s Role in Pandemic Response. China CDC Wkly. 2021, 3, 1049–1051. [Google Scholar] [CrossRef] [PubMed]
- MOH|Regulations, Guidelines and Circulars [Internet]. Moh.gov.sg. 2023. Available online: https://www.moh.gov.sg/licensing-and-regulation/regulations-guidelines-and-circulars/details/list-of-covid-19-swab-providers (accessed on 18 August 2024).
- COVID-19 Test Repository (CTR) [Internet]. Synapxe.sg. 2024. Available online: https://www.synapxe.sg/healthtech/covid-19/covid-19-test-repository-ctr (accessed on 18 August 2024).
- MOH|News Highlights [Internet]. Moh.gov.sg. 2022. Available online: https://www.moh.gov.sg/news-highlights/details/supervised-self-administered-antigen-rapid-test-available-at-test-centres_15Feb2022 (accessed on 18 August 2024).
- Lai, D.; Cai, Y.; Chan, T.H.; Gan, D.; Hurson, A.N.; Zhang, Y.D. How to organise travel restrictions in the new future: Lessons from the COVID-19 response in Hong Kong and Singapore. BMJ Glob. Health 2022, 7, e006975. [Google Scholar] [CrossRef] [PubMed]
- Yau, J.W.; Lee, M.Y.; Lim, E.Q.; Tan, J.Y.; Tan, K.B.; Chua, R.S. Genesis, evolution and effectiveness of Singapore’s national sorting logic and home recovery policies in handling the COVID-19 Delta and Omicron waves. Lancet Reg. Health–West. Pac. 2023, 35, 100719. [Google Scholar] [CrossRef] [PubMed]
- Lessler, J.; Reich, N.G.; Brookmeyer, R.; Perl, T.M.; Nelson, K.E.; Cummings, D.A. Incubation periods of acute respiratory viral infections: A systematic review. Lancet Infect. Dis. 2009, 9, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Leo, Y.S.; Tambyah, P.A. (Eds.) COVID-19 Pandemic in Singapore; World Scientific: Singapore, 2022. [Google Scholar]
- Guo, C.-X.; He, L.; Yin, J.-Y.; Meng, X.-G.; Tan, W.; Yang, G.-P.; Bo, T.; Liu, J.-P.; Lin, X.-J.; Chen, X. Epidemiological and clinical features of pediatric COVID-19. BMC Med. 2020, 18, 250. [Google Scholar] [CrossRef] [PubMed]
- Chow, B.W.; Lim, Y.D.; Poh, R.C.; Ko, A.; Hong, G.H.; Zou, S.W.; Cheah, J.; Ho, S.; Lee, V.J.; Ho, M.Z. Use of a digital contact tracing system in Singapore to mitigate COVID-19 spread. BMC Public Health 2023, 23, 2253. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Ng, S.T.; Farwin, A.; Low, A.P.T.; Chang, C.M.; Lim, J. Health equity considerations in COVID-19: Geospatial network analysis of the COVID-19 outbreak in the migrant population in Singapore. J. Travel. Med. 2021, 28, taaa159. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wu, X.; Jiang, X.; Xu, K.; Ying, L.; Ma, C.; Li, S.; Wang, H.; Zhang, S.; Gao, H.; et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: Retrospective case series. BMJ 2020, 368, m606. [Google Scholar] [CrossRef] [PubMed]
- Pung, R.; Cook, A.R.; Chiew, C.J.; Clapham, H.E.; Sun, Y.; Li, Z.; Dickens, B.L.; Ma, S.; Mak, K.; Tan, C.C.; et al. Effectiveness of containment measures against COVID-19 in Singapore: Implications for other national containment efforts. Epidemiology 2021, 32, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Menon, D.; Zhang, L.; Lim, Y.J.; Li, W.; Li, X.; Zhao, Y. Management of the COVID-19 pandemic in Singapore from 2020 to 2021: A revisit. Reports 2022, 5, 35. [Google Scholar] [CrossRef]
- Puhach, O.; Meyer, B.; Eckerle, I. SARS-CoV-2 viral load and shedding kinetics. Nat. Rev. Microbiol. 2023, 21, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Mbunge, E. Integrating emerging technologies into COVID-19 contact tracing: Opportunities, challenges and pitfalls. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 1631–1636. [Google Scholar] [CrossRef] [PubMed]
- Seah, B.Z.; Jailani, R.I.; Law, P.Y.; Teo, R.S.; Chong, X.Y.; Law, O.; Chong, S.J. COVID-19 Close Contact Management: An Evolution of Operations Harnessing the Digital Edge. J. Med. Syst. 2023, 47, 24. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conceicao, E.P.; Xu, Y.; Chan, S.L.; Jin Yee, S.; Yue, Y.; Arora, S.; Ong, M.E.H.; Sim, J.X.Y.; Venkatachalam, I. Incubation Periods of SARS-CoV-2 Wild-Type, Delta, and Omicron Variants–Dominant Periods in Singapore. COVID 2024, 4, 1578-1584. https://doi.org/10.3390/covid4100109
Conceicao EP, Xu Y, Chan SL, Jin Yee S, Yue Y, Arora S, Ong MEH, Sim JXY, Venkatachalam I. Incubation Periods of SARS-CoV-2 Wild-Type, Delta, and Omicron Variants–Dominant Periods in Singapore. COVID. 2024; 4(10):1578-1584. https://doi.org/10.3390/covid4100109
Chicago/Turabian StyleConceicao, Edwin Philip, Yingqi Xu, Sze Ling Chan, Shoon Jin Yee, Yang Yue, Shalvi Arora, Marcus Eng Hock Ong, Jean Xiang Ying Sim, and Indumathi Venkatachalam. 2024. "Incubation Periods of SARS-CoV-2 Wild-Type, Delta, and Omicron Variants–Dominant Periods in Singapore" COVID 4, no. 10: 1578-1584. https://doi.org/10.3390/covid4100109
APA StyleConceicao, E. P., Xu, Y., Chan, S. L., Jin Yee, S., Yue, Y., Arora, S., Ong, M. E. H., Sim, J. X. Y., & Venkatachalam, I. (2024). Incubation Periods of SARS-CoV-2 Wild-Type, Delta, and Omicron Variants–Dominant Periods in Singapore. COVID, 4(10), 1578-1584. https://doi.org/10.3390/covid4100109






