A Global Network Analysis of COVID-19 Vaccine Distribution to Predict Breakthrough Cases among the Vaccinated Population
Abstract
1. Introduction
2. Related Studies
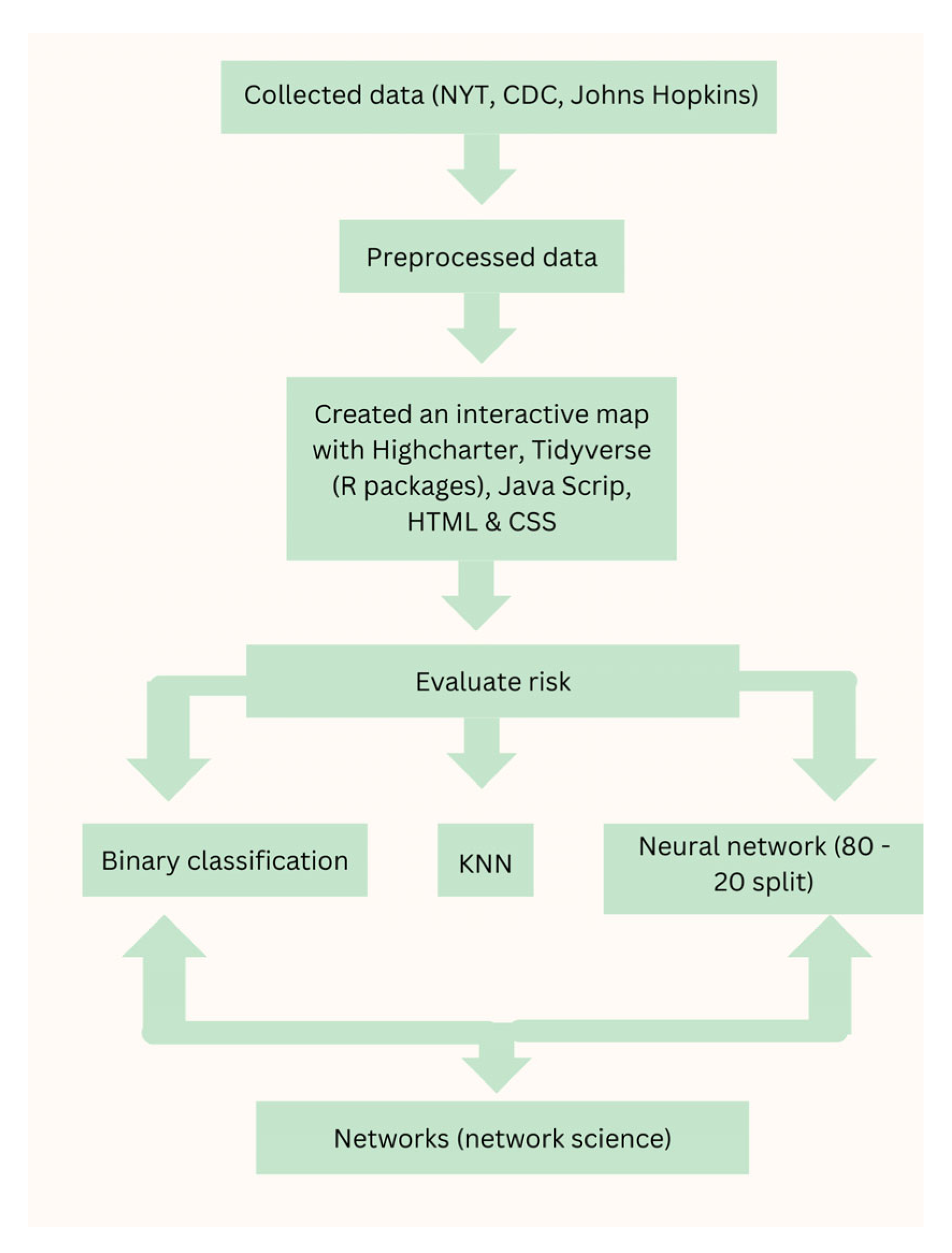
3. Methodology
- Step 1: Data Collection
- (b)
- Due to limited data on some vaccine candidates used in only a few countries, we excluded those candidates from the model. We relied on the CDC, NYT, and JHCRC for global data, assuming they accurately represent worldwide trends. However, some potential bias may exist due to data inconsistencies.
- Step 2: Data Translation
- (a)
- We transferred the collected data from Table 1 into Google spreadsheets to facilitate the development of an interactive map.
- Step 3: Data Preprocessing
- (a)
- We preprocessed the data in Table 2 to compute the percentage of people vaccinated per country per manufacturer. The steps involved and results from preprocessing are as follows:
- Given: Total doses of vaccine (per manufacturer), the population of each country, the number of people vaccinated from each country, and the total number of doses for all vaccines for each country
- We calculated the average number of doses per person by dividing the total number of vaccine doses by the number of vaccinated individuals.
- Example: If country “A” had 1000 doses of vaccine given out and had 500 people vaccinated, then 1000/500 = 2. This means country “A” has an average of 2 doses of vaccine per person
- Using the average number of doses per person and the number of doses per manufacturer we found the number of people vaccinated per manufacturer.
- We then divided the result by the population of the country which gave us the percentage of people vaccinated categorized by vaccine manufacturer per country
- Example: If country “A” had 500 doses of Pfizer and 500 doses of Moderna, then dividing by the average number of doses per person would give us 500/2 = 250. This means 250 people were vaccinated with the Pfizer vaccine and 250 people were vaccinated with Moderna. If country “A” had a population of 2000, then 250/2000 = 0.125. This means that 12.5% of the people were vaccinated with the Pfizer vaccine and another 12.5% with Moderna
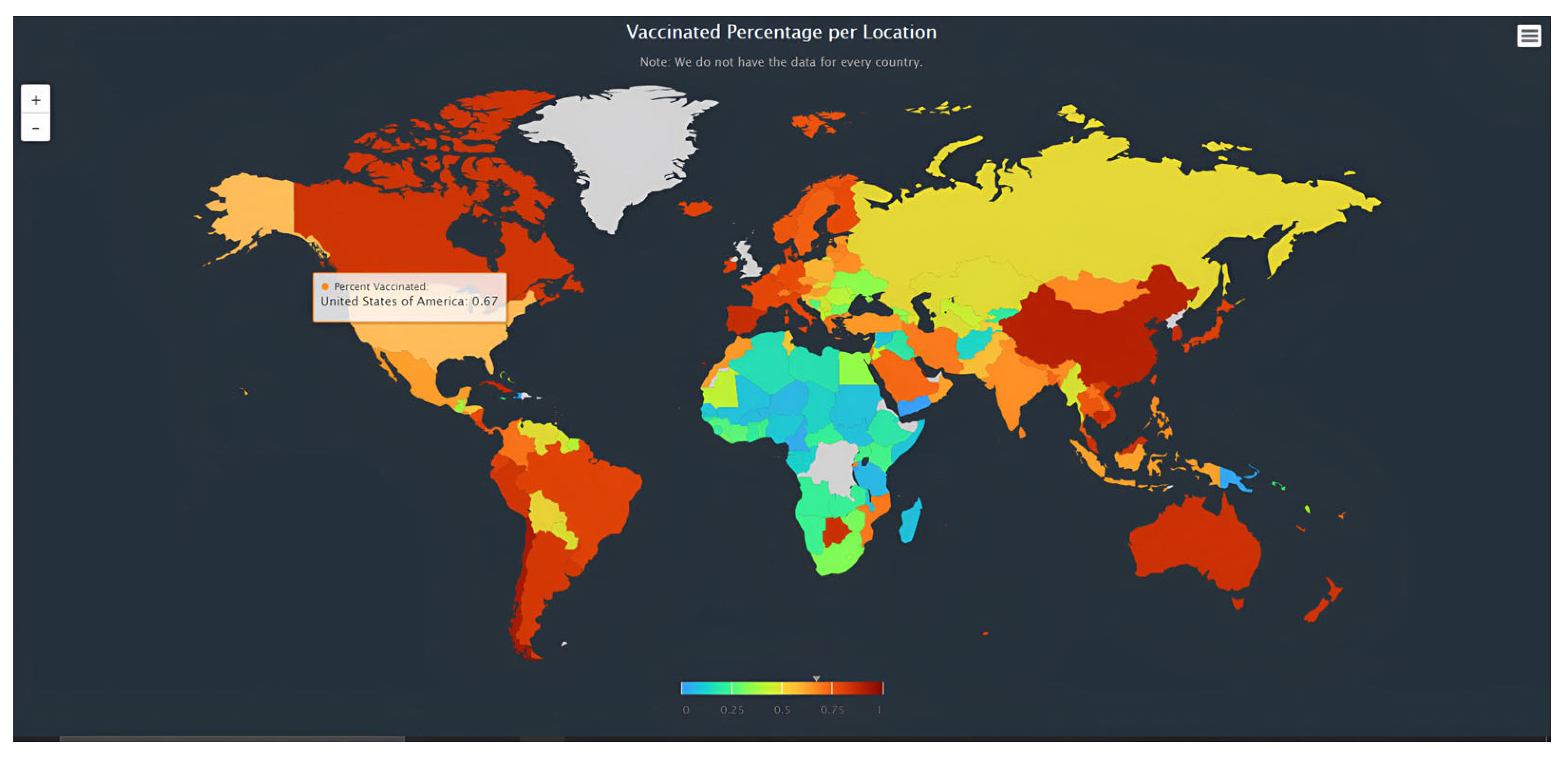
- Step 4: Interactive Map Development
- (a)
- We used two R packages in version 4.3.3, Highcharter and Tidyverse, which allowed us to create an interactive map. We mapped the processed data on the interactive global map with data imported and integrated into the map. Using the Highcharter package, we were able to combine the R code with the JavaScript, HTML, and CSS code, allowing us to make the interactive world map. For the HTML part, we drew in the Highcharter JavaScript code, brought in a few modules, and created a “website” for the map to run on. This “website” was what the map was generated on as we ran the code. For the JavaScript portion, we used the “worldgeojson” data package from Highcharter and integrated a series map. This package uses ISO codes and a map image to illustrate the map. This also added some map navigation with the scroll wheel function, allowing users to actively interact with the map.
- Step 5: Machine Learning Model Implementation
- (a)
- We applied machine learning algorithms to assess the risk of infection for each vaccine candidate:
- The binary classification model uses a supervised machine learning algorithm to categorize the countries as high-risk and low-risk for vulnerability to a COVID-19 pandemic [29] The algorithm used for the binary classification model was logistic regression, a method for fitting a regression curve to the data for predicting a label for a set of data [30].
- The K-nearest neighbors is another supervised machine-learning model that uses the proximity of data points to make classifications. A K-nearest neighbors algorithm is a lazy-learning algorithm, meaning that a model is not created until a query is performed.
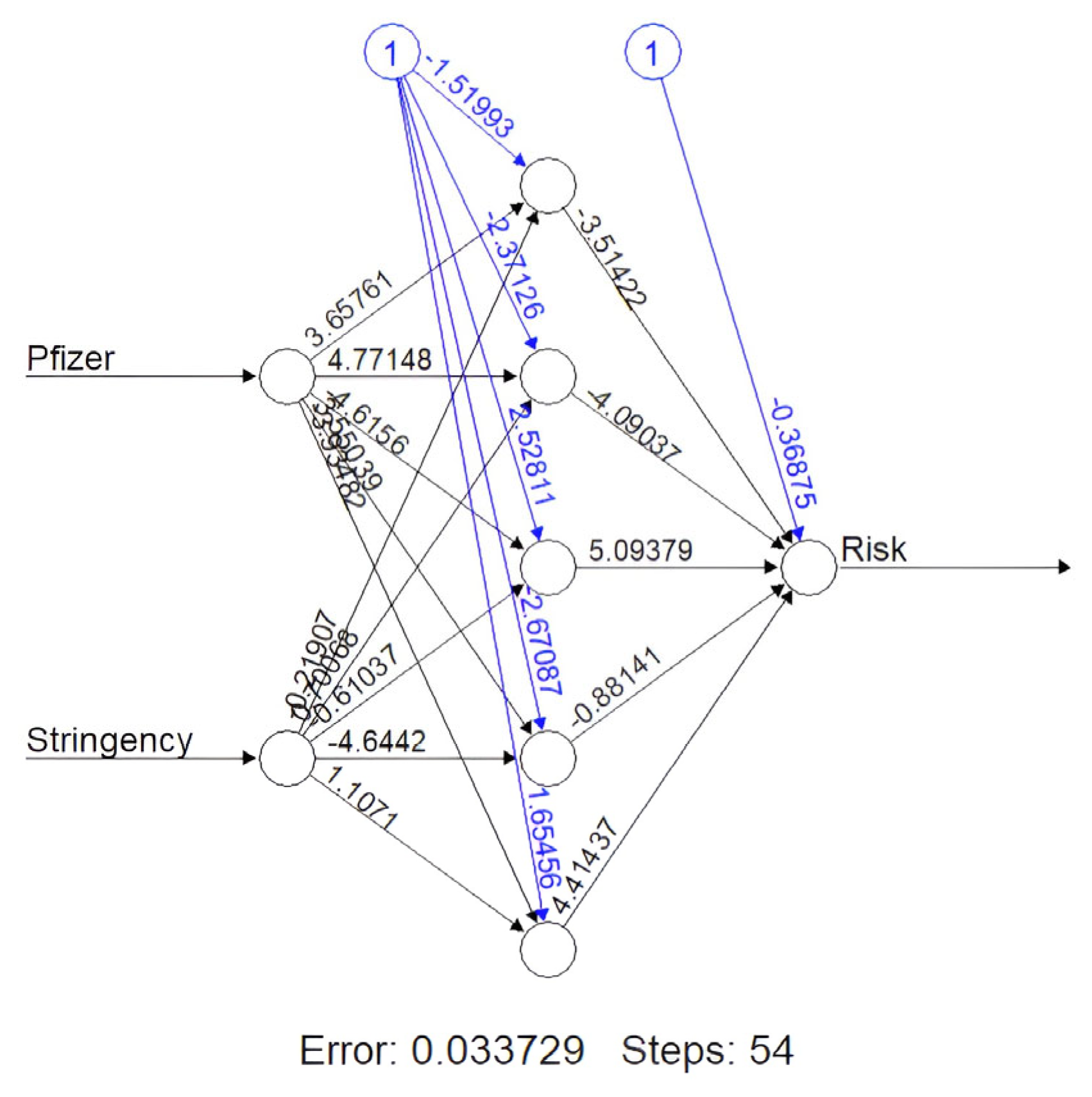
- The final machine learning method used was a neural network algorithm called NeuralNet (NN). NN uses backpropagation to estimate the neural network’s weights [31]. NeuralNet is a package from R that consists of many functions Figure 1 necessary for neural network training and testing, including customizing the number of layers in our model and predicting categories with a test dataset. The model features were vaccination percentages for the specific candidate and stringency rate. These vaccination data were split into a 70–30 train–test split, and the data underwent min–max normalization. The NN consisted of five hidden layers and used resilient backpropagation with weight backtracking. The NN demonstrated the highest accuracy, ranging from 94.3% to 99.02% depending on the vaccine candidate after the data were divided into a 70–30 split. We generated a NeuralNet function (Figure 2) for each vaccine candidate; however, the one shown below is for Pfizer.
- Step 6: Neural network model training
- (a)
- The training data were categorized as 1 (high risk) and 0 (low risk) based on the threshold we set. We set the threshold to be 50% for all vaccine candidates.
- (b)
- Step 7: Network Science for Global Vaccine Connectome
- (a)
- We used network science to visualize global vaccine distribution using Gephi and google sheets. We had a sheet recording the countries, an assigned number, and their vaccine percentages. The nodes represent vaccine percentages for each country, and the edges represent connections between countries that distribute the same vaccine candidate. Regarding the edges, a thicker edge between countries indicates more social connectedness between those two countries. The network was visualized using Gephi, and the connectome was ranked according to vaccine percentage numbers. A set of specific color codes was used to represent the risks of infection. Red edges and nodes meant a low risk of infection, light yellow edges and nodes meant a medium risk of infection, and blue edges and nodes meant a high risk of infection. Counties at a higher risk were assigned if the stringency was low and the vaccine percentage was high for a specific vaccine candidate. The threshold for determining high or low risk was selected by comparing the countries within each vaccine candidate to each other to determine whether the country was considered to be at a high risk. This was necessary, as each vaccine candidate had a different threshold of high or low risks (J&J for example had its highest at 25%, while Pfizer had its highest at 80%). Additionally, we ran the Fruchterman–Reingold model to form the network into a circular shape to make it easier to visualize.
4. Results and Discussion
5. Conclusions and Limitations
6. Limitations
7. Future Works
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chaudhary, J.K.; Yadav, R.; Chaudhary, P.K.; Maurya, A.; Kant, N.; Rugaie, O.A.; Haokip, H.R.; Yadav, D.; Roshan, R.; Prasad, R.; et al. Insights into COVID-19 vaccine development based on immunogenic structural proteins of SARS-COV-2, host immune responses, and herd immunity. Cells 2023, 10, 2949. [Google Scholar] [CrossRef] [PubMed]
- Seladi-Schulman, J. Coronavirus vs. SARS. Healthline, 15 September 2021. [Google Scholar]
- How viral mutations occur in SARS-COV-2. Yale Medicine, 19 February 2021.
- Johnson, S. What you need to know about SARS (severe acute respiratory syndrome). Healthline, 31 March 2017. [Google Scholar]
- Rabaan, A.A.; Al-Ahmed, S.H.; Haque, S.; Sah, R.; Tiwari, R.; Malik, Y.S.; Dhama, K.; Yatoo, M.I.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. SARS-CoV-2, SARS-CoV, and MERS-COV: A comparative overview. Infez. Med. 2020, 28, 174–184. [Google Scholar] [PubMed]
- De Groot, R.J.; Baker, S.C.; Baric, R.S.; Brown, C.S.; Drosten, C.; Enjuanes, L.; Fouchier, R.A.M.; Galiano, M.; Gorbalenya, A.E.; Memish, Z.A.; et al. Middle East Respiratory Syndrome Coronavirus (MERS-CoV): Announcement of the Coronavirus Study Group. J. Virol. 2013, 87, 7790–7792. [Google Scholar] [CrossRef] [PubMed]
- Brant, A.C.; Tian, W.; Majerciak, V.; Yang, W.; Zheng, Z.M. SARS-COV-2: From its discovery to genome structure, transcription, and replication. Cell Biosci. 2023, 11, 136. [Google Scholar]
- Uddin, S.; Haque, I.; Lu, H.; Moni, M.A.; Gide, E. Comparative performance analysis of K-nearest neighbour (KNN) algorithm and its different variants for disease prediction. Sci. Rep. 2022, 12, 6256. [Google Scholar] [CrossRef]
- Barabási, A.-L. Scale-Free Networks: A Decade and Beyond. Science 2009, 325, 412–413. [Google Scholar] [CrossRef]
- Park, M.-B.; Ranabhat, C.L. COVID-19 trends, public restrictions policies and vaccination status by economic ranking of countries: A longitudinal study from 110 countries. Arch. Public Health 2022, 80, 197. [Google Scholar] [CrossRef]
- Kumari, R.; Srivastava, S.K. Machine Learning: A Review on Binary Classification. Int. J. Comput. Appl. 2017, 160, 11–15. [Google Scholar] [CrossRef]
- Mehlig, B. Machine Learning with Neural Networks; University of Gothenburg: Gothenburg, Sweden, 2021. [Google Scholar]
- Sputnik, V. COVID-19 vaccine candidate appears safe and effective. Lancet 2021, 397, 642–643. [Google Scholar]
- Shah, S.; Mulahuwaish, A.; Ghafoor, K.Z.; Maghdid, H.S. Prediction of global spread of COVID-19 pandemic: A review and research challenges. Artif. Intell. Rev. 2021, 55, 1607–1628. [Google Scholar] [CrossRef]
- Hirschprung, R.S.; Hajaj, C. Prediction model for the spread of the COVID-19 outbreak in the global environment. Heliyon 2021, 7, e07416. [Google Scholar] [CrossRef]
- Wieczorek, M.; Siłka, J.; Woźniak, M. Neural network powered COVID-19 spread forecasting model. Chaos Solitons Fractals 2020, 140, 110203. [Google Scholar] [CrossRef] [PubMed]
- Tomar, A.; Gupta, N. Prediction for the spread of COVID-19 in India and effectiveness of preventive measures. Sci. Total. Environ. 2020, 728, 138762. [Google Scholar] [CrossRef] [PubMed]
- Barda, N.; Riesel, D.; Akriv, A.; Levy, J.; Finkel, U.; Yona, G.; Greenfeld, D.; Sheiba, S.; Somer, J.; Bachmat, E.; et al. Developing a COVID-19 mortality risk prediction model when individual-level data are not available. Nat. Commun. 2020, 11, 4439. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, R.; Li, X.; Dadashova, B.; Zhu, L.; Zhang, K.; Li, Y.; Shen, B. Health-Based Geographic Information Systems for Mapping and Risk Modeling of Infectious Diseases and COVID-19 to Support Spatial Decision-Making. In Translational Informatics. Advances in Experimental Medicine and Biology; Shen, B., Ed.; Springer: Singapore, 2022; Volume 1368. [Google Scholar]
- Pal, R.; Sekh, A.A.; Kar, S.; Prasad, D.K. Neural Network Based Country Wise Risk Prediction of COVID-19. Appl. Sci. 2020, 10, 6448. [Google Scholar] [CrossRef]
- Bird, J.J.; Barnes, C.M.; Premebida, C.; Ekárt, A.; Faria, D.R. Country-level pandemic risk and preparedness classification based on COVID-19 data: A machine learning approach. PLoS ONE 2020, 15, e0241332. [Google Scholar] [CrossRef]
- COVID-19 World Map: Cases, Deaths and Global Trends. The New York Times, 11 March 2023.
- Beck, M.W. NeuralNetTools: Visualization and Analysis Tools for Neural Networks. J. Stat. Softw. 2018, 85, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Albert, R.; Barabási, A.-L. Statistical mechanics of complex networks. Rev. Mod. Phys. 2002, 74, 47–97. [Google Scholar] [CrossRef]
- COVID-19 Map—Johns Hopkins Coronavirus Resource Center; Johns Hopkins Coronavirus Resource Center: Baltimore, MD, USA, 2020.
- Serafino, M.; Monteiro, H.S.; Luo, S.; Reis, S.D.S.; Igual, C.; Neto, A.S.L.; Travizano, M.; Andrade, J.S.; Makse, H.A. Digital contact tracing and network theory to stop the spread of COVID-19 using big-data on human mobility geolocalization. PLoS Comput. Biol. 2022, 18, e1009865. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. CDC COVID Data Tracker. Centers for Disease Control and Prevention; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2021. [Google Scholar]
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef]
- Cramer, J.S. The origins of logistic regression. SSRN Electron. J. 2003, 19, 4. [Google Scholar] [CrossRef]
- Günther, F.; Fritsch, S. neuralnet: Training of Neural Networks. R J. 2010, 2, 30–38. [Google Scholar] [CrossRef]
- Stevens, N.T.; Wilson, J.D. The past, present, and future of network monitoring: A panel discussion. Qual. Eng. 2021, 33, 715–718. [Google Scholar] [CrossRef]


| Entity | Code | Day | Pfizer/BioNTech | Moderna | Oxford/AstraZeneca | Johnson&Johnson | Sputnik V | Sinovac | Sinopharm/Beijing | CanSino | Novavax | Covaxin | Medicago | Sanofi/GSK | SKYCovione | Valneva | Total Population |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Argentina | ARG | 2023-02-11 | 19,724,295 | 17,371,749 | 26,774,900 | 0 | 20,751,112 | 0 | 28,941,172 | 985,243 | 0 | 0 | 0 | 0 | 0 | 0 | 45,847,428 |
| Austria | AUT | 2023-02-03 | 16,763,486 | 1,663,838 | 1,593,159 | 368,380 | 0 | 0 | 0 | 0 | 14,893 | 0 | 0 | 140 | 0 | 2162 | 9,065,484 |
| Belgium | BEL | 2023-02-03 | 21,931,050 | 4,389,147 | 2,849,228 | 428,552 | 0 | 0 | 0 | 0 | 406 | 0 | 0 | 0 | 0 | 0 | 11,658,404 |
| Bulgaria | BGR | 2023-02-10 | 3,090,202 | 511,919 | 478,547 | 530,219 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6,866,274 |
| Canada | CAN | 2023-12-04 | 63,482,165 | 28,804,585 | 2,815,498 | 23,591 | 0 | 0 | 0 | 0 | 28,433 | 0 | 863 | 0 | 0 | 0 | 37,742,154 |
| Chile | CHL | 2023-08-30 | 8,067,724 | 0 | 549,673 | 0 | 0 | 25,943,395 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 19,116,201 |
| Croatia | HRV | 2023-10-21 | 4,045,117 | 520,646 | 568,527 | 204,677 | 0 | 0 | 0 | 0 | 1373 | 0 | 0 | 0 | 0 | 0 | 4,067,642 |
| Cyprus | CYP | 2023-02-03 | 1,309,054 | 199,684 | 254,531 | 31,016 | 0 | 0 | 0 | 0 | 898 | 0 | 0 | 0 | 0 | 0 | 1,220,541 |
| Czechia | CZE | 2023-02-21 | 15,662,393 | 1,641,250 | 886,784 | 413,735 | 0 | 348 | 77 | 0 | 11,055 | 5 | 0 | 0 | 0 | 0 | 10,761,297 |
| Denmark | DNK | 2023-02-10 | 12,188,460 | 1,781,705 | 155,952 | 46,005 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5,825,798 |
| Ecuador | ECU | 2023-01-28 | 8,552,679 | 0 | 5,009,163 | 0 | 0 | 15,812,935 | 0 | 536,882 | 0 | 0 | 0 | 0 | 0 | 0 | 18,033,738 |
| Estonia | EST | 2023-02-03 | 1,632,730 | 244,530 | 238,919 | 79,141 | 0 | 0 | 0 | 0 | 2160 | 0 | 0 | 0 | 0 | 0 | 1,324,323 |
| European Union | 2023-02-21 | 664,200,258 | 154,146,954 | 67,192,141 | 18,693,913 | 1,845,376 | 8780 | 2,319,802 | 0 | 303,799 | 126 | 0 | 3905 | 0 | 9234 | 748,835,153 | |
| Finland | FIN | 2023-02-03 | 10,286,849 | 2,068,166 | 553,905 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5,553,102 |
| France | FRA | 2023-02-19 | 121,155,541 | 24,121,747 | 7,862,962 | 1,090,803 | 0 | 0 | 0 | 0 | 38,662 | 0 | 0 | 3645 | 0 | 0 | 65,520,147 |
| Germany | DEU | 2023-02-20 | 138,189,814 | 31,585,369 | 12,803,049 | 3,761,229 | 0 | 0 | 0 | 0 | 159,859 | 0 | 0 | 0 | 0 | 7072 | 83,975,691 |
| Hong Kong | HKG | 2023-02-19 | 11,848,626 | 0 | 0 | 0 | 0 | 8,832,512 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7,585,785 |
| Hungary | HUN | 2023-02-03 | 9,778,198 | 1,079,204 | 1,252,978 | 345,438 | 1,807,392 | 0 | 2,315,130 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9,618,215 |
| Iceland | ISL | 2023-03-28 | 76,558 | 6493 | 1 | 85 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 344,646 |
| India | IND | ||||||||||||||||
| Ireland | IRL | 2023-02-17 | 9,603,020 | 1,668,816 | 1,217,678 | 241,601 | 0 | 0 | 0 | 0 | 850 | 0 | 0 | 0 | 0 | 0 | 5,008,554 |
| Italy | ITA | 2023-02-21 | 95,718,713 | 34,324,511 | 12,169,281 | 1,508,434 | 0 | 0 | 0 | 0 | 42,983 | 0 | 0 | 88 | 0 | 0 | 60,320,493 |
| Japan | JPN | 2023-02-20 | 297815527 | 83,268,972 | 117,831 | 0 | 0 | 0 | 0 | 0 | 300,250 | 0 | 0 | 0 | 0 | 0 | 125,802,521 |
| Latvia | LVA | 2023-07-11 | 1624384 | 715,409 | 262,043 | 293,843 | 0 | 9 | 27 | 0 | 438 | 0 | 0 | 0 | 0 | 0 | 1,855,735 |
| Liechtenstein | LIE | 2023-01-27 | 22479 | 48,599 | 0 | 264 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 38,416 |
| Lithuania | LTU | 2023-07-08 | 3315576 | 328,959 | 536,495 | 295,566 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2,670,680 |
| Luxembourg | LUX | 2023-02-17 | 745,265 | 342,155 | 105,059 | 41,522 | 0 | 0 | 0 | 0 | 496 | 0 | 0 | 0 | 0 | 0 | 640,202 |
| Malta | MLT | 2023-02-03 | 747,037 | 278,881 | 227,870 | 32,421 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 443,646 |
| Nepal | NPL | 2023-07-04 | 1,797,582 | 7,336,243 | 14,088,349 | 3,693,423 | 0 | 0 | 19,972,478 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 29,960,704 |
| Netherlands | NLD | 2023-09-02 | 22,697,220 | 8,448,384 | 2,473,242 | 755,296 | 0 | 0 | 0 | 0 | 2707 | 0 | 0 | 0 | 0 | 0 | 17,195,298 |
| Norway | NOR | 2023-02-03 | 9,766,924 | 2,386,984 | 148,123 | 7394 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5,492,570 |
| Peru | PER | 2023-02-19 | 51,520,283 | 6,301,449 | 8,186,523 | 0 | 0 | 0 | 21,266,405 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3,358,7011 |
| Poland | POL | 2023-02-10 | 41,943,054 | 3,843,870 | 5,292,339 | 2,728,519 | 0 | 0 | 0 | 0 | 15,746 | 0 | 0 | 0 | 0 | 0 | 33,587,011 |
| Portugal | PRT | 2023-02-10 | 17,796,120 | 3,928,446 | 2,276,083 | 1,138,788 | 0 | 8423 | 4568 | 0 | 201 | 121 | 0 | 32 | 0 | 0 | 10,150,252 |
| Romania | ROU | 2023-06-11 | 12,914,439 | 1,008,835 | 849,559 | 2,054,653 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18,900,064 |
| Slovenia | SVN | 2023-02-03 | 2,325,179 | 236,211 | 322,679 | 135,725 | 0 | 0 | 0 | 0 | 218 | 0 | 0 | 0 | 0 | 0 | 2,079,690 |
| South Africa | ZAF | 2023-12-26 | 28,684,331 | 0 | 0 | 9,366,280 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 69,797,779 |
| South Korea | KOR | 2023-02-21 | 52,280,181 | 13,434,676 | 20,084,922 | 1,516,295 | 0 | 0 | 0 | 0 | 258,451 | 0 | 0 | 0 | 536 | 0 | 51,385,361 |
| Uruguay | URY | 2023-02-15 | 2,565,508 | 0 | 91,200 | 0 | 0 | 3,248,201 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3,505,403 |
| Sweden | SWE | 2023-02-03 | 17,262,652 | 4,174,751 | 1,322,063 | 0 | 0 | 0 | 0 | 0 | 7111 | 0 | 0 | 0 | 0 | 0 | 10,265,156 |
| Switzerland | CHE | 2023-02-20 | 6,210,231 | 10,633,270 | 0 | 63,454 | 0 | 0 | 0 | 0 | 3427 | 0 | 0 | 0 | 0 | 0 | 8,821,366 |
| Ukraine | UKR | 2022-02-23 | 14,774,013 | 3,044,899 | 4,041,487 | 20,680 | 0 | 9,802,231 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 43,042,464 |
| United States | USA | 2023-02-15 | 400,059,167 | 251,049,942 | 0 | 18,976,061 | 0 | 0 | 0 | 0 | 78,683 | 0 | 0 | 0 | 0 | 0 | 336,115,637 |
| Uruguay | URY | 2023-02-15 | 2,565,508 | 0 | 91,200 | 0 | 0 | 3,248,201 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3,505,403 |
| Pfizer/BioNTech% | Moderna% | Oxford/AstraZeneca% | Johnson&Johnson% | Covishield | Sputnik V%% | Sinovac% | Sinopharm/Beijing% | CanSino% | Novavax% | Covaxin% | Medicago% | Sanofi/GSK% | SKYCovione% | Valneva% |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.430215955 | 0.378903458 | 0.584000045 | 0 | 0 | 0.452612347 | 0 | 0.631249631 | 0.021489602 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1.849155103 | 0.183535485 | 0.17573899 | 0.040635448 | 0 | 0 | 0 | 0 | 0 | 0.001642825 | 0 | 0 | 1.5 × 10−5 | 0 | 0.000238487 |
| 1.88113656 | 0.376479233 | 0.244392629 | 0.036759062 | 0 | 0 | 0 | 0 | 0 | 3.5 × 10−5 | 0 | 0 | 0 | 0 | 0 |
| 0.450055154 | 0.074555574 | 0.069695296 | 0.077220775 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1.681996343 | 0.763193987 | 0.074598233 | 0.000625057 | 0 | 0 | 0 | 0 | 0 | 0.000753349 | 0 | 2.3 × 10−5 | 0 | 0 | 0 |
| 0.422035947 | 0 | 0.028754301 | 0 | 0 | 0 | 1.357141777 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 0.994462394 | 0.127997007 | 0.1397682 | 0.050318342 | 0 | 0 | 0 | 0 | 0 | 0.000337542 | 0 | 0 | 0 | 0 | 0 |
| 1.072519481 | 0.163602861 | 0.208539492 | 0.025411682 | 0 | 0 | 0 | 0 | 0 | 0.000735739 | 0 | 0 | 0 | 0 | 0 |
| 1.455437295 | 0.152514144 | 0.082404937 | 0.038446574 | 0 | 0 | 3.2 × 10−5 | 7.15527 × 10−6 | 0 | 0.001027293 | 4.6 × 10−7 | 0 | 0 | 0 | 0 |
| 2.092152869 | 0.305830206 | 0.026769208 | 0.007896772 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 0.474259912 | 0 | 0.277766207 | 0 | 0 | 0 | 0.876852874 | 0 | 0.029770977 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1.232878988 | 0.184645287 | 0.180408405 | 0.05975959 | 0 | 0 | 0 | 0 | 0 | 0.001631022 | 0 | 0 | 0 | 0 | 0 |
| 0.886977935 | 0.205848982 | 0.089728882 | 0.02496399 | 0 | 0.002464329 | 1.2 × 10−5 | 0.003097881 | 0 | 0.000405695 | 1.7 × 10−7 | 0 | 5.2 × 10−6 | 0 | 1.2 × 10−5 |
| 1.852450936 | 0.372434362 | 0.099746952 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1.849134145 | 0.368157706 | 0.120008308 | 0.01664836 | 0 | 0 | 0 | 0 | 0 | 0.000590078 | 0 | 0 | 5.6 × 10−5 | 0 | 0 |
| 1.645593056 | 0.376125146 | 0.152461371 | 0.044789497 | 0 | 0 | 0 | 0 | 0 | 0.001903634 | 0 | 0 | 0 | 0 | 8.4 × 10−5 |
| 1.561951202 | 0 | 0 | 0 | 0 | 0 | 1.164350426 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1.016633336 | 0.112204188 | 0.130271365 | 0.03591498 | 0 | 0.187913454 | 0 | 0.240702667 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 0.222135176 | 0.018839621 | 2.9 × 10−6 | 0.00024663 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 0 | 0 | 0 | 0 | 1.2393363 | 0.062483 | 0 | 0 | 0 | 0 | 0.256657215 | 0 | 0 | 0 | |
| 1.917323842 | 0.333193173 | 0.243119671 | 0.048237675 | 0 | 0 | 0 | 0 | 0 | 0.00016971 | 0 | 0 | 0 | 0 | 0 |
| 1.586835721 | 0.569035651 | 0.201743726 | 0.025006991 | 0 | 0 | 0 | 0 | 0 | 0.000712577 | 0 | 0 | 1.5 × 10−6 | 0 | 0 |
| 2.367325588 | 0.661902252 | 0.000936635 | 0 | 0 | 0 | 0 | 0 | 0 | 0.002386677 | 0 | 0 | 0 | 0 | 0 |
| 0.875331877 | 0.385512479 | 0.141207123 | 0.15834319 | 0 | 0 | 4.8 × 10−6 | 1.5 × 10−5 | 0 | 0.000236025 | 0 | 0 | 0 | 0 | 0 |
| 0.585146814 | 1.265071845 | 0 | 0.006872137 | 0 | 0 | 0 | 0 | 0 | 7.8 × 10−5 | 0 | 0 | 0 | 0 | 0 |
| 1.241472584 | 0.123174248 | 0.200883296 | 0.110670691 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1.164109141 | 0.534448502 | 0.164102893 | 0.064857654 | 0 | 0 | 0 | 0 | 0 | 0.000774755 | 0 | 0 | 0 | 0 | 0 |
| 1.683858301 | 0.628611551 | 0.513630237 | 0.073078536 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 0.059997989 | 0.24486217 | 0.470227569 | 0.123275575 | 0 | 0 | 0 | 0.666622453 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1.319966656 | 0.491319429 | 0.143832459 | 0.043924566 | 0 | 0 | 0 | 0 | 0 | 0.000157427 | 0 | 0 | 0 | 0 | 0 |
| 1.778206559 | 0.434584175 | 0.026967886 | 0.001346182 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1.53393474 | 0.187615653 | 0.243740743 | 0 | 0 | 0 | 0 | 0.633173491 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1.248787932 | 0.114445135 | 0.157571003 | 0.081237327 | 0 | 0 | 0 | 0 | 0 | 0.000468812 | 0 | 0 | 0 | 0 | 0 |
| 1.753268786 | 0.387029406 | 0.224239063 | 0.112193077 | 0 | 0 | 0.000829832 | 0.000450038 | 0 | 1.98025 × 10−5 | 1.19209 × 10−5 | 0 | 3.2 × 10−6 | 0 | 0 |
| 0.683301337 | 0.053377332 | 0.044950059 | 0.108711431 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1.11804115 | 0.113579909 | 0.155157259 | 0.06526213 | 0 | 0 | 0 | 0 | 0 | 0.000104823 | 0 | 0 | 0 | 0 | 0 |
| 0.410963378 | 0 | 0 | 0.134191663 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1.017413909 | 0.261449482 | 0.390868559 | 0.029508307 | 0 | 0 | 0 | 0 | 0 | 0.005029662 | 0 | 0 | 0 | 1.0 × 10−5 | 0 |
| 0.731872484 | 0 | 0.02601698 | 0 | 0 | 0 | 0.926626981 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1.681674589 | 0.406691433 | 0.128791321 | 0 | 0 | 0 | 0 | 0 | 0 | 0.000692732 | 0 | 0 | 0 | 0 | 0 |
| 0.703998791 | 1.205399481 | 0 | 0.007193217 | 0 | 0 | 0 | 0 | 0 | 0.000388489 | 0 | 0 | 0 | 0 | 0 |
| 0.343242734 | 0.070741745 | 0.093895345 | 0.000480456 | 0 | 0 | 0.227733965 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1.190242652 | 0.74691539 | 0 | 0.056456942 | 0 | 0 | 0 | 0 | 0 | 0.000234095 | 0 | 0 | 0 | 0 | 0 |
| 0.731872484 | 0 | 0.02601698 | 0 | 0 | 0 | 0.926626981 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Actual | ||
|---|---|---|
| Predicted | 0—Low Risk | 1—High Risk |
| 0—Low Risk | 8 | 0 |
| 1—High Risk | 1 | 25 |
| Vaccine Candidate | Accuracy Rate | Error |
|---|---|---|
| Pfizer | 0.96551724 | 0.03448276 |
| Moderna | 0.988308 | 0.011692 |
| J&J | 0.965699 | 0.034301 |
| All Vaccines | 0.9431 | 0.056785 |
| Vaccine | Countries at Risk |
|---|---|
| Pfizer (countries with this vaccine are at a higher risk because most of their population has the vaccine—around 75%.) | Japan, Austria, Finland, South Africa, Bulgaria, Ireland, Estonia, Norway, Czechia, Denmark |
| Moderna (countries with this vaccine and at “risk” are not at a very high level of risk, because the highest vaccination rate is around 30%; in other words, not very many people are vaccinated with this vaccine. As long as other vaccine candidates are used, these countries should not be at high risk.) | Latvia, Netherlands, Spain, Canada, US, Switzerland, Italy, Luxembourg, Lichtenstein |
| Johnson & Johnson (countries with this vaccine and at “risk” are not at a very high level of risk, because the highest vaccination rate is around 10%; in other words, not very many people are vaccinated with this vaccine.) | Romania, Bulgaria, Latvia, South Africa |
| Sinovac (countries with this vaccine are at a somewhat high risk because almost half of their population is vaccinated—around 45%.) | Chile, Ecuador |
| Sinopharm (countries with this vaccine are at a higher risk because most of their population has the vaccine—around 50%.) | Argentina |
| Sputnik (countries with this vaccine are at a somewhat high risk because almost half of their population is vaccinated—around 45%.) | Argentina |
| Oxford (countries with this vaccine and at “risk” are not at a very high level of risk, because the highest vaccination rate is around 30%; in other words, not many people are vaccinated with that vaccine.) | Argentina, Nepal, South Korea, Malta |
| Covaxin (countries with this vaccine and at “risk” are not at a very high level of risk, because the highest vaccination rate is around 26%; in other words, not many people are vaccinated with this vaccine.) | India |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bodapati, P.; Zhang, E.; Padmanabhan, S.; Das, A.; Bhattacharya, M.; Jahanikia, S. A Global Network Analysis of COVID-19 Vaccine Distribution to Predict Breakthrough Cases among the Vaccinated Population. COVID 2024, 4, 1546-1560. https://doi.org/10.3390/covid4100107
Bodapati P, Zhang E, Padmanabhan S, Das A, Bhattacharya M, Jahanikia S. A Global Network Analysis of COVID-19 Vaccine Distribution to Predict Breakthrough Cases among the Vaccinated Population. COVID. 2024; 4(10):1546-1560. https://doi.org/10.3390/covid4100107
Chicago/Turabian StyleBodapati, Pragyaa, Eddie Zhang, Sathya Padmanabhan, Anisha Das, Medha Bhattacharya, and Sahar Jahanikia. 2024. "A Global Network Analysis of COVID-19 Vaccine Distribution to Predict Breakthrough Cases among the Vaccinated Population" COVID 4, no. 10: 1546-1560. https://doi.org/10.3390/covid4100107
APA StyleBodapati, P., Zhang, E., Padmanabhan, S., Das, A., Bhattacharya, M., & Jahanikia, S. (2024). A Global Network Analysis of COVID-19 Vaccine Distribution to Predict Breakthrough Cases among the Vaccinated Population. COVID, 4(10), 1546-1560. https://doi.org/10.3390/covid4100107






