Abstract
Background: In Senegal, the incidence of SARS-CoV-2 evolved with four successive epidemic waves. The first wave started in March 2020 with low virus variability, whilst the second outbreak, which started in December 2020, was dominated by the Alpha variant. The third wave took place in June 2021, and the fourth at the end of November 2021. Our interest was to investigate the involvement of variants of concern during these four waves and to track the viral diversity of SARS-CoV-2. Methodology: During the four waves of the pandemic, 276,876 nasopharyngeal swabs were analyzed at the Institut de Recherche en Santé, de Surveillance Epidémiologique et de Formation (IRESSEF). Of these, 22,558 samples tested positive for SARS-CoV-2 by RT-PCR. Then, the virus genomes were sequenced in 817 positive samples using the ARTIC Network of Oxford Nanopore Technologies (ONT). In addition, 10% of the negative samples in RT-PCR new variants were also targeted for the detection of new and previously undescribed variants. Results: Our data have overall shown that the Senegalese strains are very similar to each other or closely related to other strains, such as Gambia, France etc. During the first wave, the most common clade found was 19A (67.5%) and a majority of the samples were of the B.1 (50%) lineage. We noted more diversity during the second wave where clade 20A (38.4%) was more frequent, followed by clade 20B (31.52%) and 20I (9.74%). At the level of lineages, we identified variants of concern as B.1.1.7 (9.74%) and B.1.617.2 (0.86%). In the third wave, we observed at the clade level with mainly 21A (32.63%) and 21J (16.84%). During the fourth wave at the end of November 2021, we mainly identified clade 21K Omicron variant 21K (B.1.1.529 and BA.1) (80.47%) and Delta variant (21A, 21J, and 21I) (AY.103, AY.122, AY.122.1, AY.26, AY.34, AY.36, AY.4, AY.48, AY.57, AY.61, and AY.87) (14.06%). Impact: SARS-CoV-2 diversity may affect the virus’s properties, such as how it spreads, disease severity, or the performance of vaccines, tools, or other public health and social measures. Therefore, such tracking of SARS-CoV-2 variants is not only of public interest, but also highlights the role some African institutes such as IRESSEF with surveillance capabilities through the real-time sequencing of SARS-CoV-2 genomes in the local context. Conclusion: In Senegal, the SARS-CoV-2 pandemic has disrupted the organization of the health system. IRESSEF contributed to put in place strategies to respond effectively to the expectations of medical authorities by providing them with data on the strains circulating in Senegal at each moment of the epidemic.
1. Introduction
In December 2019, scientists in China reported the first cases of COVID-19 in the city of Wuhan, China [1,2]. One month later, an international public health emergency was declared by the World Health Organization (WHO). This was the beginning of a pandemic due to the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), which causes a disease called COVID-19 (Coronavirus Disease 2019) [3].
African countries were the last to register cases of COVID-19 at the start of the pandemic [4]. This late entry of COVID-19 in Africa is justified by low international air traffic and a less dense trade network with Western countries and China [5,6]. Egypt was the first African country to report a case of COVID-19 [7]. Since then, the continent has recorded thousands of COVID-19 positive cases without ever being submerged compared to Western countries, even though South Africa has had similar cases to European countries.
In West Africa, Senegal is the third country with the highest number of cases of COVID-19, but it is the second country recording the most deaths after Nigeria. [8]. Senegal, which reported its first case of COVID-19 on 2 March 2020 [9], has been affected by four successive waves of the epidemic, as is the case for most countries in the world. SARS-CoV-2 is a virus whose numerous mutations have led to a resurgence of the pandemic. The high transmissibility of the virus from human to human has accelerated its distribution throughout the world and the emergence of multiple variants, such as B.1.1.7, B.1.351, P.1, B.1.427, and B.1.429 [10]. This has resulted in an increase of positive cases, deaths, and saturation of hospitalization beds [11]. In Senegal, only the Alpha variant was reported during the first two waves of the epidemic [12]. However, the third wave was dominated by the B.1.617.2 variant (Delta), which is reportedly more transmissible [13]. Recently, the Omicron (B.1.1.529) variant was detected for the first time in South Africa on 24 November 2021 [14].
Currently, the evolutionary dynamics of SARS-CoV-2 are poorly investigated in Senegal. In this study, we evaluate the evolution of the different types of SARS-CoV-2 variants of concern that circulated in Senegal during the four waves and genotype diversity during the COVID-19 pandemic.
2. Material and Methods
Epidemiologic study of variant distribution.
2.1. Study Settings and Design
We analyzed genomic data of SARS-CoV-2 samples collected in Senegal between April 2020 and January 2022. Samples were tested at IRESSEF, an ISO 15,189 Plus accredited laboratory, with the approval of the Senegalese Ministry of Health (following number: 000159/MSAS/CNERS/Sec, on 21 August 2020). Samples were collected from two regions (Dakar and Thiès) of the Western part of Senegal. In Thiès region, 16.03% (131/817) of the samples were from the health district of Mbour, 6.49% (53/817) from the district of Thiès, 4.77% (39/817) from Tivaoune, and 1.71% (14/817) from Popenguine. In Dakar, samples were collected at Ngor (29.86%) (244/817), Diamniadio (37.94%) (310/817), and Rufisque (3.18%) (26/817) (Figure 1).
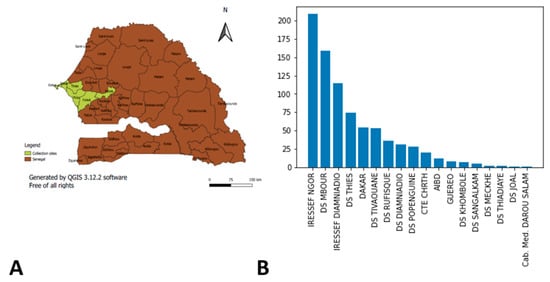
Figure 1.
Samples sites collection. (A) The sites colored in yellow represent the different collection areas. (B) represents the distribution of the number of samples with on the y axis we have the number of samples and the different collection sites on the x axis.
2.2. Study Population and Sampling
The study population consisted of individuals who were suspected of being infected with SARS-CoV-2 and tested at IRESSEF during the study period. In total, 276,876 oropharyngeal and nasopharyngeal swabs were collected in compliance with IRESSEF’s safety and hygiene guidelines. For each sample, a notification form collecting demographic data (name, first name, age, sex, address, type of accommodation), clinical data (presence of symptoms, recent or current illness, and ongoing treatment), and epidemiological data (questionnaire or form) was completed and sent to the laboratory. All participants gave their free and informed consent.
Genome sequencing and data analysis.
2.3. RNA Extraction
To extract the RNA, the oropharyngeal and/or nasopharyngeal samples were first inactivated in a water bath at 90 °C for 30 min. The samples were then aliquoted in 1.5 mL vials, before RNA was extracted using the Kingfisher platform according to the manufacturer’s guidelines and eluted in 50 μL (www.thermofisher.com, accessed on 22 January 2022, SARS-CoV-2 support and solutions KingFisher instruments and MagMAX isolation kit).
2.4. Reverse Transcriptase-Polymerase Chain Reaction
RNA extracted samples were undiluted and plates were stored at 4 °C while the master mix was being prepared. Allplex™ 2019-nCoV assays from Seegene Inc. were used according to the manufacturer’s protocol to perform RT-PCR. Briefly, for one reaction, 5 μL of 2019-nCoV MOM, 5 μL of buffer 5×, 5 μL of RNase-free water, 1 μL of internal control (IC), and 2 μL of enzymes were used. In each well, 18 μL of master mix were distributed and either 8μL of sample added, 8μL of positive control, or 8 μL of RNase-free water for negative control. Plates were then spun down at 2500 rpm for 5 s and analysed on a CFX96 Touch Real-Time PCR from BioRad, Reverse Transcription reaction using the following setting: 1 cycle: 50 °C/20 min–95 °C/15 min, PCR reaction 45 cycles: 94 °C/15 s–60 °C/30 s–72 °C/15 s. Fluorescence was measured at 60 °C and 72 °C using channels FAM (E gene), HEX (IC), Cal Red 610 (RdRP) and Quasar 670 (N gene). Results were compiled and analysed using 2019-nCoV viewer from Seegene Inc. according to the manufacturer’s instructions (Seegene. AllplexTM 2019-nCoV Assay) (Cat no. RP10250X/RP10252W) [15]. Results were defined as positive if the viral genome was detected at threshold cycle (Ct) values ≤35, as indeterminate at Ct values >35 and ≤38, and as negative at Ct values >38 (Figure 2).
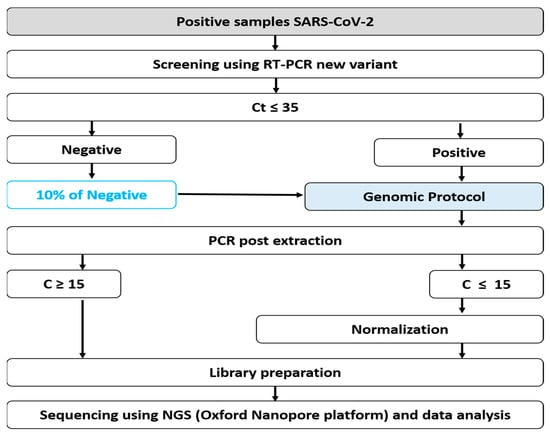
Figure 2.
Genomic sequencing protocol in IRESSEF.
2.5. Whole Genome Sequencing (WGS) of SARS-CoV-2 Using Oxford Nanopore Technology
2.5.1. Library Preparation and Sequencing
Library prep was done following the ARTIC protocol (Version3) (Josh Q). Twenty-four barcoded libraries were sequenced on one flow cell using MinION (Nanopore technology) with expected coverage of more than 65×. We tested the correlation between post-PCR concentration and Ct value and our results showed that the lower, the Ct value the higher the post-PCR concentration (Figure 3).
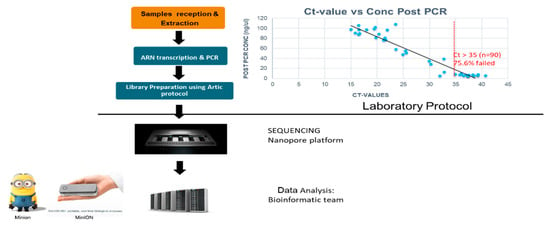
Figure 3.
Protocol for sample selection for Whole Genome Sequencing (WGS).
2.5.2. Data Analysis
We first cleaned and assembled the sequencing data using FASTQ pass on each run. The samples were sequenced on Nanopore technology, which is the reason why we used the artic bioinformatics pipeline workflow for genomic assembly [16]. After assembly, we performed a sequencing quality check and determined the clade of each sequence on the nexclade website [17]. Finally, we determined the lineage of each sequence using the pangolin website [18]. These three procedures above allowed us to clean, assemble, determine the clade, and determine the sub-clade, i.e., the lineage.
The procedure was repeated globally at the end of each batch sequencing. This allowed us to have the different lineages and clades on each wave.
The results allowed us to continuously visualize the evolution of the number of cases and the clades.
For further investigation, we explored the evolution of the lineages over time. We calculated the percentage of each lineage over each month from June 2020 to January 2022, the percentage of which cumulates to 100% to visualize using again the matplotlib module of python version 3.9.0 (Figure 4).
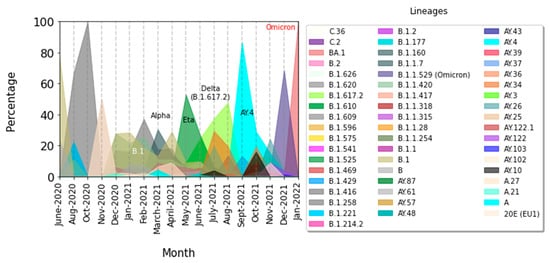
Figure 4.
Evolution over time of the different variants. Finally, we visualized the distribution of SARS-CoV-2 strains by patient’s age using the matplotlib module of python version 3.9.0.
3. Results
In total, 276,876 nasopharyngeal swabs were performed at the Institut de Recherche en Santé, de Surveillance Epidémiologique et de Formation (IRESSEF). Of these, 22,558 samples tested positive for SARS-CoV-2 by RT-PCR. Then, the virus genomes were sequenced for 817 positive samples in RT-PCR Seegene using the ARTIC Network of Oxford Nanopore Technologies (ONT) (percentage of successful sequences is mentioned in supplementary data).
Characteristics of the participants.
3.1. Waves and Variants
Our results show that the majority of our strains have a different distribution during the four epidemic waves of SARS-CoV-2 in Senegal. Figure 5 shows the distribution of clades and lineages on each wave using the matplotlib module of python version 3.9.0. During the first wave, we noted a high prevalence of variant 19A (67.5%) (27/40) and low percentage of 20A (27.5%) (11/40), 20B (2.5%) (1/40), and 19B (2.5%) (1/40). For the lineage, we observed a dominance of B lineages, such as B.1 (50%) (20/40), B.1.416 (22.5%) (9/40), B (17.5%) (7/40), B.1.221 (5%) (2/40), and B.1.1.420 (2.5%) (1/40), and the A lineage, such as A.27 (2.5%) (1/40) (Figure 4). During the second wave, there was a predominance of the 20A (38.4%) (134/349) and 20B (31.52%) (110/349) clade. We also noted a higher proportion of the 20I (Alpha) clade (9.74%) (34/349) compared to the 20E (EU1) (6.02) (21/349) and 19A (7.16) (25/349%) clades combined. The 20G, 21A, 20E, 20C, 20D, and 19B clades were less frequent during this second wave, while B.1 (23.21%) (81/349), B.1.1.420 (18.91%) (66/349), B.1.416 (17.77%) (62/349), and B.1.1.7 (Alpha) (9.74%) (34/349) were the dominant lines (Figure 4 and Figure 6). The Delta variant was detected at the end of the second wave (March) in 0.86% (3/349) of the samples.
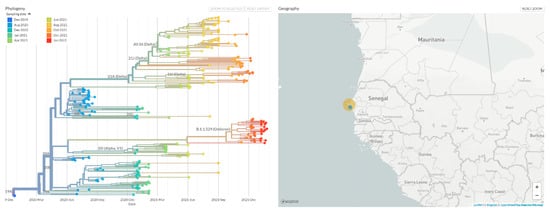
Figure 5.
Phylogenetic three of VOC isolated in IRESSEF. (Phylogeny reconstruction was performed using the nextstrain/ncov tool (https://github.com/nextstrain/ncov, accessed on 22 January 2022, then visualised with Auspice (https://docs.nextstrain.org/projects/auspice/en/stable/, accessed on 22 January 2022. The genome of the original Wuhan-Hu-1 coronavirus isolate (GenBank accession no. NC_045512.2) was added as outgroup. Major (most prevalent) variants are labelled). For this specified analysis, we used sequences with a coverage greater than 65%. Most of them are available on GISAID [19] using the IDs (Supplementary data).
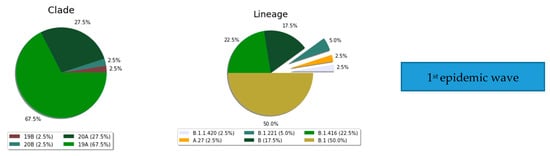
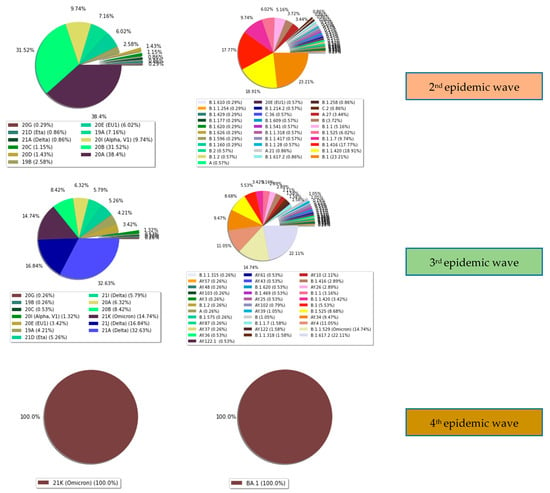
Figure 6.
Distribution of the different epidemic waves observed in Senegal according to the clade and lineage detected.
During the third wave, we identified strains belonging to clade 21A (Delta) (32.63%) (124/380), 21J (Delta) (16.84%) (64/380), 21K (Omicron) (14.74%) (56/380), 20B (8.42%) (32/380), 20A (6.32%) (24/380), 21I (Delta) (5.79%) (22/380), 21D (Eta) (5.26%) (20/380), 19A (4.21%) (16/380), 20E (EU1) (3.42%) (13/380), 20I (1.32%) (5/380), 20C (0.53%) (2/380). 19B (0.26) (1/380), and 20G (0.26) (1/380). The lineages B.1.617.2 (Delta) (22.11%) (84/380), B.1.1.529 (Omicron) (14.74%) (56/380), AY.4 (Delta) (11.05%) (42/380), AY.34 (Delta) (9.47%) (36/380), and B.1.525 (Eta) (8.68%) (33/380) were detected. Incident cases were higher in the third epidemic wave compared to previous waves. Form samples were collected in June. We observed a high prevalence of the variant of interest, Eta (47.69%). The Delta (20%) and Alpha variants (6.17%) were observed as well. In July, the trend was reversed with the increase of the Delta variant in up to 60% of the positive samples (Figure 6). The results suggest the dominance of the delta variant in Senegal in recent months. Lastly, in the fourth wave, the Omicron variant was detected and soon became dominant in the majority of samples since January 2022. We observed an amplitude of the third wave that is about twice as high as that of the second and first waves.
On the other hand, the amplitude of the fourth wave, although higher than that of the first and second waves, is still lower than that of the third wave (Figure 7).
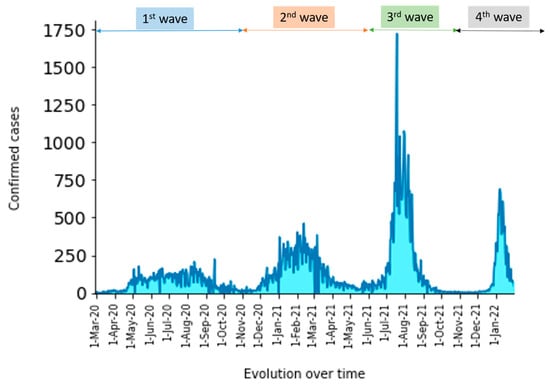
Figure 7.
Evolution over time of confirmed cases per day over time during SARS-CoV-2 pandemic in Senegal.
3.2. Distribution of Variants by Age and Sex
The variants of interest were identified at IRESSEF at different dates, which allowed us to follow the evolution of these variants over time in Senegal (Table 1).

Table 1.
Classification of SARS-CoV-2 lineage.
Of the 817 samples tested, the Alpha variant was more frequent in men (53.85%) than in women (46.15%). The Delta variant was higher in men (50.3%) than in women (49.7%). The Eta variant (60%) was exclusively identified in females. The Omicron variant was higher in men (57.28%) than in women (42.72%)). Finally, other non-VOCs were found in 51.3% of women against 48.7% of men (Table 2).

Table 2.
Distribution of Variant Of Concern (VOC) by sex and age of patients tested at IRESSEF.
Our data showed that patients over the age of 60 were the most infected with the SARS-CoV-2 virus regardless of the variant involved (Figure 8). Young individuals were not affected by the different waves of the COVID-19 epidemic. Similar results were reported in the literature. [20].
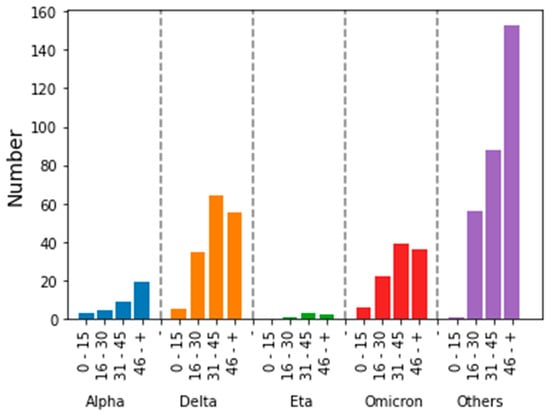
Figure 8.
Distribution of SARS-CoV-2 strains by age of patients tested.
4. Discussion
COVID-19 disease is characterized by a succession of four epidemic waves. These are most often determined by the appearance of new variants of SARS-CoV-2. In Senegal, as in several other countries, there have been four different waves but with different variants emerging each time [21].
Indeed, the predominance of clade 19A during the first wave is obvious. Senegal recorded its first case of COVID-19 in March 2020 [9]. This period corresponds to the expansion of the SARS-CoV-2 virus around the world. Therefore, it is normal that the clade and lineages detected at the beginning can correspond to those found in other countries.
The second wave of the global epidemic was marked by the emergence of the Delta variant. Indeed, the Delta variant represented 97.62% of the global sequences shared during week 32 (3–9 August). This period coincided with the mass detection of the B1.617.2 variant in Senegal. The Alpha variant was found mainly in people over 46 years old (60.8%), as was the Eta variant (45%). The Delta variant affected mainly (60%) individuals between 31 and 45 years old. For the non-VOCs, most cases (53%) were over 46 years old.
This latest variant of interest (Omicron) was first reported to the WHO from South Africa on 19 November 2021 and spread around the world. It was in most cases mainly responsible for the fourth epidemic wave of COVID-19 around the world. The number of cases has increased rapidly in South Africa, reaching proportions of 80% (Ingrid Torjesen). In Senegal, according to the latest data we have from IRESSEF, the contamination rate is over 75%. As part of the response to the COVID-19 pandemic, Senegal has strengthened its surveillance system following the notification by the WHO of the Omicron variant. Our results clearly show the rapid progression of the Omicron variant, and this new variant has become clearly predominant in new infections of SARS-CoV-2 (Figure 4).
This slight delay in the global epidemic can be explained by the fact that Africa, in particular Senegal, was one of the last countries to be invaded by the virus.
Likely, the third wave in Senegal started in July with the emergence of a worrying variant, B.1.525, which made its first appearance in the UK and Nigeria. A large number of cases were observed in Senegal only in early July 2021. Recently, another new variant of concern emerged in South Africa and spread rapidly around the world, causing all state authorities to panic.
5. Conclusions
The COVID-19 pandemic persists with a succession of epidemic waves linked to the emergence of new variants. In Senegal as in the rest of the world, four epidemic waves were observed. The Alpha, Beta, Delta, and Omicron variants most often responsible for these SARS-CoV-2 episodes are still covered by the various vaccines marketed to eradicate the disease. Vaccine coverage in Senegal is less than 5%, emphasizing the risk of other waves. It is still necessary to evaluate the effectiveness of the vaccines used on the variants of interest. Nevertheless, it is important to strengthen the capacities for the surveillance and sequencing of SARS-CoV-2 genomes according to the local context and to detect unusual epidemiological events.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/covid2060052/s1.
Author Contributions
A.P., K.G. and S.M. conceived and designed the study. N.L., S.N. (Seyni Ndiaye), N.D.D. and C.K.D. performed the experiments. A.P., C.K.D., S.N. (Samba Ndiour), K.G. and N.D.D. recruited study partici-pants and collected data. A.P. and K.G. analyzed and interpreted the data. S.M., P.A.D., M.M., D.W., N.L., L.K. and G.L. contributed to reagents/materials/analysis tools. A.P., P.A.D., K.G., C.K.D., A.A., A.S.N., A.J.S.N., M.S., M.B. and Y.A.D. participated to study design. A.P. and S.M. partici-pated in study coordination. A.P., K.G., S.N. (Samba Ndiour) and C.I.L. wrote/drafted the manuscript. A.M., D.W., N.L., G.L., C.I.L., M.M., P.A.D., N.C.T.K., S.M., A.A., G.L., P.A.D. and M.C. critically re-viewed the manuscript for important intellectual content. C.S., B.C., S.M., N.C.T.K. and M.C. approved the final version to be published. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the “European and Developing Countries Clinical Trials Partnership” (EDCTP) (Grant Nr: RIA2020EF-2961), the “West African Task Force for the Control of Emerging and Re-emerging Infectious Diseases” (WATER), and “Innovation in Laboratory Engineered Accelerated Diagnosis” (iLEAD) (Grant Nr. OPP1214434/INV-009631). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The views expressed are those of the authors, and the funders are not responsible for any use that may be made of the information contained herein. This work was supported in part through National Institutes of Health USA grant U01 AI151698 for the United World Antivirus Research Network (UWARN). This article was supported by WANETAM as part of the EDCTP program.
Institutional Review Board Statement
This study was approved by the National Ethics Committee for Health Research of Senegal under the following number: 000159/MSAS/CNERS/SN. Free and informed consent is provided by each adult individual who participated in this study.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We gratefully thank the individuals that participated in this study and the IRESSEF’s personnel involved in the SARS-CoV-2 testing and sequencing. We also thank Didier Raoult from the Institut Méditérannée Infection de Marseille and Abdul Sesay from the Medical Research Council of Gambia for their support.
Conflicts of Interest
The authors have declared that no competing interests exist.
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- WHO. Novel Coronavirus—China. Available online: http://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/ (accessed on 16 January 2021).
- WHO. Director-General’s Remarks at the Media Briefing on 2019-nCoV on 11 February 2020. Available online: https://www.who.int/director-general/speeches/detail/who-director-general-s-remarks-at-the-media-briefing-on-2019-ncov-on-11-february-2020 (accessed on 28 February 2021).
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef]
- Haider, N.; Yavlinsky, A.; Simons, D.; Osman, A.Y.; Ntoumi, F.; Zumla, A.; Kock, R. Passengers’ destinations from China: Low risk of Novel Coronavirus (2019-nCoV) transmission into Africa and South America. Epidemiol. Infect. 2020, 148, e41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, M.; Pullano, G.; Pinotti, F.; Valdano, E.; Poletto, C.; Boëlle, P.-Y.; D’Ortenzio, E.; Yazdanpanah, Y.; Eholie, S.P.; Altmann, M.; et al. Preparedness and vulnerability of African countries against importations of COVID-19: A modelling study. Lancet 2020, 395, 871–877. [Google Scholar] [CrossRef] [Green Version]
- Khattab, N.M.; Vermund, S.H.; Hu, Y. How coronavirus disease 2019 entered Africa and the Middle East: A case study from Egypt. Trans. R. Soc. Trop. Med. Hyg. 2020, 114, 715–717. [Google Scholar] [CrossRef] [PubMed]
- Coronavirus-West-Africa—Sahel and West Africa Club Secretariat. Available online: https://www.oecd.org/swac/coronavirus-west-africa/ (accessed on 16 September 2021).
- WHO|Regional Office for Africa. Senegal Reports First COVID-19 Case. Available online: https://www.afro.who.int/news/senegal-reports-first-covid-19-case (accessed on 16 January 2021).
- Volz, E.; Hill, V.; McCrone, J.T.; Price, A.; Jorgensen, D.; O’Toole, Á.; Southgate, J.; Johnson, R.; Jackson, B.; Nascimento, F.F.; et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell 2021, 184, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Aleem, A.; Akbar Samad, A.B.; Slenker, A.K. Emerging Variants of SARS-CoV-2 and Novel Therapeutics Against Coronavirus (COVID-19); StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Padane, A.; Kanteh, A.; Leye, N.; Mboup, A.; Manneh, J.; Mbow, M.; Diaw, P.; Ndiaye, B.; Lo, G.; Lo, C.; et al. First detection of SARS-CoV-2 variant B.1.1.7 in Senegal. New Microbes New Infect. 2021, 41, 100877. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Deng, A.; Li, K.; Hu, Y.; Li, Z.; Xiong, Q.; Liu, Z.; Guo, Q.; Zou, L.; Zhang, H.; et al. Viral infection and transmission in a large well-traced outbreak caused by the Delta SARS-CoV-2 variant. medRxiv 2021. [CrossRef] [PubMed]
- Sahoo, J.P.; Samal, K.C. World on Alert: WHO Designated South African New COVID Strain (Omicron/B.1.1.529) as a Variant of Concern. Biot. Res. Today 2021, 3, 1086–1088. [Google Scholar]
- Freppel, W.; Merindol, N.; Rallu, F.; Bergevin, M. Efficient SARS-CoV-2 detection in unextracted oro-nasopharyngeal specimens by rRT-PCR with the Seegene Allplex™ 2019-nCoV assay. Virol. J. 2020, 17, 196. [Google Scholar] [CrossRef] [PubMed]
- Artic Network. Available online: https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html (accessed on 22 January 2022).
- Nextclade. Available online: https://clades.nextstrain.org (accessed on 22 January 2022).
- COG-UK. Available online: https://pangolin.cog-uk.io/ (accessed on 22 January 2022).
- GISAID—Initiative. Available online: https://www.gisaid.org/ (accessed on 22 January 2022).
- Staerk, C.; Wistuba, T.; Mayr, A. Estimating effective infection fatality rates during the course of the COVID-19 pandemic in Germany. BMC Public Health 2021, 21, 1073. [Google Scholar] [CrossRef] [PubMed]
- Cascella, M.; Rajnik, M.; Aleem, A.; Dulebohn, S.C.; Di Napoli, R. Features, Evaluation, and Treatment of Coronavirus (COVID-19); StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).