Diagnostic Performance of a Rapid Antigen Test Compared with the Reverse Transcription Polymerase Chain Reaction for SARS-CoV-2 Detection in Asymptomatic Individuals Referring to a Drive-in Testing Facility
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Reagents
2.3. Statistical Analysis
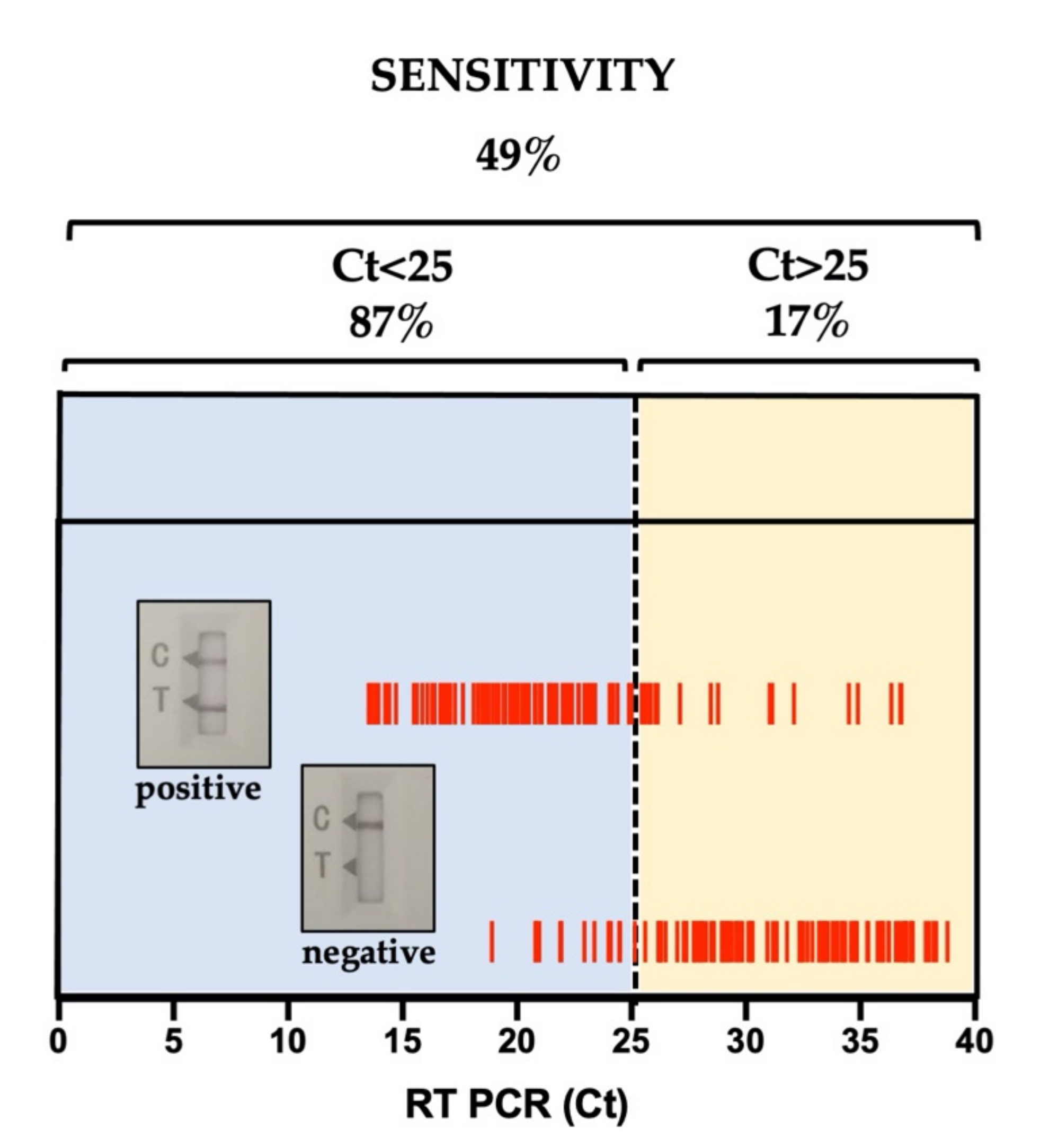
3. Results
Sensitivity and Specificity of Ag-RDT
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Oran, D.P.; Topol, E.J. Prevalence of Asymptomatic SARS-CoV-2 Infection: A Narrative Review. Ann. Intern. Med. 2020, 173, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Lu, S.C.; Bai, C.H.; Wang, P.Y.; Lee, K.Y.; Wang, Y.H. Diagnostic Accuracy of SARS-CoV-2 Antigen Tests for Community Transmission Screening: A Systematic Review and Meta-Analysis. Int, J. Environ. Res. Public Health 2021, 18, 11451. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.; Whiting, P.F.; Brush, J.E. Interpreting a COVID-19 test result. BMJ 2020, 369, m1808. [Google Scholar] [CrossRef] [PubMed]
- Lieberman, J.A.; Pepper, G.; Naccache, S.N.; Huang, M.L.; Jerome, K.R.; Greninger, A.L. Comparison of Commercially Available and Laboratory-Developed Assays for In Vitro Detection of SARS-CoV-2 in Clinical Laboratories. J. Clin. Microbiol. 2020, 58, e00821–e00820. [Google Scholar] [CrossRef]
- Toptan, T.; Eckermann, L.; Pfeiffer, A.E.; Hoehl, S.; Ciesek, S.; Drosten, C.; Corman, V.M. Evaluation of a SARS-CoV-2 rapid antigen test: Potential to help reduce community spread? J. Clin. Virol. 2021, 135, 104713. [Google Scholar] [CrossRef] [PubMed]
- CDC. Interim Guide for Antigen Testing for SARS-CoV-2. 2021. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html (accessed on 12 October 2021).
- Pray, I.W.; Ford, L.; Cole, D.; Lee, C.; Bigouette, J.P.; Abedi, G.R.; Bushman, D.; Delahoy, M.J.; Currie, D.; Cherney, B.; et al. Performance of an Antigen-Based Test for Asymptomatic and Symptomatic SARS-CoV-2 Testing at Two University Campuses—Wisconsin, September-October 2020. MMWR Morb. Mortal. Wkly. Rep. 2021, 69, 1642–1647. [Google Scholar] [CrossRef] [PubMed]
- Peeling, R.W.; Olliaro, P.L.; Boeras, D.I.; Fongwen, N. Scaling up COVID-19 rapid antigen tests: Promises and challenges. Lancet Infect. Dis 2021, 21, e290–e295. [Google Scholar] [CrossRef]
- WHO. Antigen-Detection in the Diagnosis of SARS-CoV-2 Infection using Rapid Immunoassays: Interim Guidance. 2020. Available online: https://apps.who.int/iris/handle/10665/334253 (accessed on 5 September 2021).
- Wagenhauser, I.; Knies, K.; Rauschenberger, V.; Eisenmann, M.; McDonogh, M.; Petri, N.; Andres, O.; Flemming, S.; Gawlik, M.; Papsdorf, M.; et al. Clinical performance evaluation of SARS-CoV-2 rapid antigen testing in point of care usage in comparison to RT-qPCR. EBioMedicine 2021, 69, 103455. [Google Scholar] [CrossRef] [PubMed]
- Kohmer, N.; Toptan, T.; Pallas, C.; Karaca, O.; Pfeiffer, A.; Westhaus, S.; Widera, M.; Berger, A.; Hoehl, S.; Kammel, M.; et al. The Comparative Clinical Performance of Four SARS-CoV-2 Rapid Antigen Tests and Their Correlation to Infectivity In Vitro. J. Clin. Med. 2021, 10, 328. [Google Scholar] [CrossRef] [PubMed]
- Pickering, S.; Batra, R.; Merrick, B.; Snell, L.B.; Nebbia, G.; Douthwaite, S.; Reid, F.; Patel, A.; Ik, M.T.K.; Patel, B.; et al. Comparative performance of SARS-CoV-2 lateral flow antigen tests and association with detection of infectious virus in clinical specimens: A single-centre laboratory evaluation study. Lancet Microbe 2021, 2, e461–e471. [Google Scholar] [CrossRef]
- Stromer, A.; Rose, R.; Schafer, M.; Schon, F.; Vollersen, A.; Lorentz, T.; Fickenscher, H.; Krumbholz, A. Performance of a Point-of-Care Test for the Rapid Detection of SARS-CoV-2 Antigen. Microorganisms 2020, 9, 58. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Haage, V.C.; Bleicker, T.; Schmidt, M.L.; Muhlemann, B.; Zuchowski, M.; Jo, W.K.; Tscheak, P.; Möncke-Buchner, E.; Müller, M.A.; et al. Comparison of seven commercial SARS-CoV-2 rapid point-of-care antigen tests: A single-centre laboratory evaluation study. Lancet Microbe 2021, 2, e311–e319. [Google Scholar] [CrossRef]
- WHO. Emergency Use Listing Procedure for In Vitro Diagnostics. 2021. Available online: https://www.who.int/teams/regulation-prequalification/eul (accessed on 1 September 2021).
- ICGEB. COVID-19/SARS-CoV-2 Resource Page. 2020. Available online: https://www.icgeb.org/covid19-resources/ (accessed on 1 April 2021).
- Rajasekharan, S.; Bonotto, R.M.; Alves, L.N.; Kazungu, Y.; Poggianella, M.; Martinez-Orellana, P.; Skoko, N.; Polez, S.; Marcello, A. Inhibitors of Protein Glycosylation Are Active against the Coronavirus Severe Acute Respiratory Syndrome Coronavirus SARS-CoV-2. Viruses 2021, 13, 808. [Google Scholar] [CrossRef] [PubMed]
- Baba, M.M.; Bitew, M.; Fokam, J.; Lelo, E.A.; Ahidjo, A.; Asmamaw, K.; Beloumou, G.A.; Bulimo, W.D.; Buratti, E.; Chenwi, C.; et al. Diagnostic performance of a colorimetric RT-LAMP for the identification of SARS-CoV-2: A multicenter prospective clinical evaluation in sub-Saharan Africa. EClinicalMedicine 2021, 40, 101101. [Google Scholar] [CrossRef] [PubMed]
- Team CR. A Language and Environment for Statistical Computing. Available online: http://www.r-project.org/ (accessed on 1 April 2021).
- Cohen, J. A coeffcient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Italy. Novel Coronavirus Portal. 2021. Available online: https://www.salute.gov.it/portale/nuovocoronavirus/homeNuovoCoronavirus.jsp?lingua=english (accessed on 5 September 2021).
- FIND. Diagnosis for All. 2021. Available online: https://www.finddx.org/product/novel-corona-virus-sars-cov-2-ag-rapid-test-kit (accessed on 5 September 2021).
- Cevik, M.; Tate, M.; Lloyd, O.; Maraolo, A.E.; Schafers, J.; Ho, A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: A systematic review and meta-analysis. Lancet Microbe 2021, 2, e13–e22. [Google Scholar] [CrossRef]
- Singanayagam, A.; Patel, M.; Charlett, A.; Lopez Bernal, J.; Saliba, V.; Ellis, J.; Ladhani, S.; Zambon, M.; Gopal, R. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill 2020, 25, 32. [Google Scholar] [CrossRef] [PubMed]
- La Scola, B.; Le Bideau, M.; Andreani, J.; Hoang, V.T.; Grimaldier, C.; Colson, P.; Gautret, P.; Raoult, D. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1059–1061. [Google Scholar] [CrossRef] [PubMed]
- Bullard, J.; Dust, K.; Funk, D.; Strong, J.E.; Alexander, D.; Garnett, L.; Boodman, C.; Bello, A.; Hedley, A.; Schiffman, Z.; et al. Predicting Infectious Severe Acute Respiratory Syndrome Coronavirus 2 From Diagnostic Samples. Clin. Infect. Dis 2020, 71, 2663–2666. [Google Scholar] [CrossRef] [PubMed]
| Sensitivity | Specificity | PPV | NPV | N | Precision | Accuracy (95% CI) | p (McNemar’s Test) | Cohen’s Kappa (95% CI) | |
|---|---|---|---|---|---|---|---|---|---|
| Aggregate | 49% | 100% | 100% | 55% | 317 | 100% | 68% (63–74%) | 4.20 × 10−23 | 42% (35–50%) |
| CT < 25 | 87% | NA | NA | NA | 91 | 100% | 87% (78–93%) | 0.0015 | NA |
| CT ≥ 25 | 17% | NA | NA | NA | 106 | 100% | 17% (10–26%) | 1.80 × 10−20 | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lombardo, F.; Triolo, G.; Yang, B.; Liu, Z.; Maiuri, P.; Orsini, E.; Jin, W.; Zacchigna, S.; Marcello, A. Diagnostic Performance of a Rapid Antigen Test Compared with the Reverse Transcription Polymerase Chain Reaction for SARS-CoV-2 Detection in Asymptomatic Individuals Referring to a Drive-in Testing Facility. COVID 2021, 1, 784-789. https://doi.org/10.3390/covid1040063
Lombardo F, Triolo G, Yang B, Liu Z, Maiuri P, Orsini E, Jin W, Zacchigna S, Marcello A. Diagnostic Performance of a Rapid Antigen Test Compared with the Reverse Transcription Polymerase Chain Reaction for SARS-CoV-2 Detection in Asymptomatic Individuals Referring to a Drive-in Testing Facility. COVID. 2021; 1(4):784-789. https://doi.org/10.3390/covid1040063
Chicago/Turabian StyleLombardo, Fabio, Gianluca Triolo, Biao Yang, Zhonghua Liu, Paolo Maiuri, Emanuele Orsini, Wei Jin, Serena Zacchigna, and Alessandro Marcello. 2021. "Diagnostic Performance of a Rapid Antigen Test Compared with the Reverse Transcription Polymerase Chain Reaction for SARS-CoV-2 Detection in Asymptomatic Individuals Referring to a Drive-in Testing Facility" COVID 1, no. 4: 784-789. https://doi.org/10.3390/covid1040063
APA StyleLombardo, F., Triolo, G., Yang, B., Liu, Z., Maiuri, P., Orsini, E., Jin, W., Zacchigna, S., & Marcello, A. (2021). Diagnostic Performance of a Rapid Antigen Test Compared with the Reverse Transcription Polymerase Chain Reaction for SARS-CoV-2 Detection in Asymptomatic Individuals Referring to a Drive-in Testing Facility. COVID, 1(4), 784-789. https://doi.org/10.3390/covid1040063







