A Simple and Fast Method to Sequence the Full-Length Spike Gene for SARS-CoV-2 Variant Identification from Patient Samples
Abstract
:1. Introduction
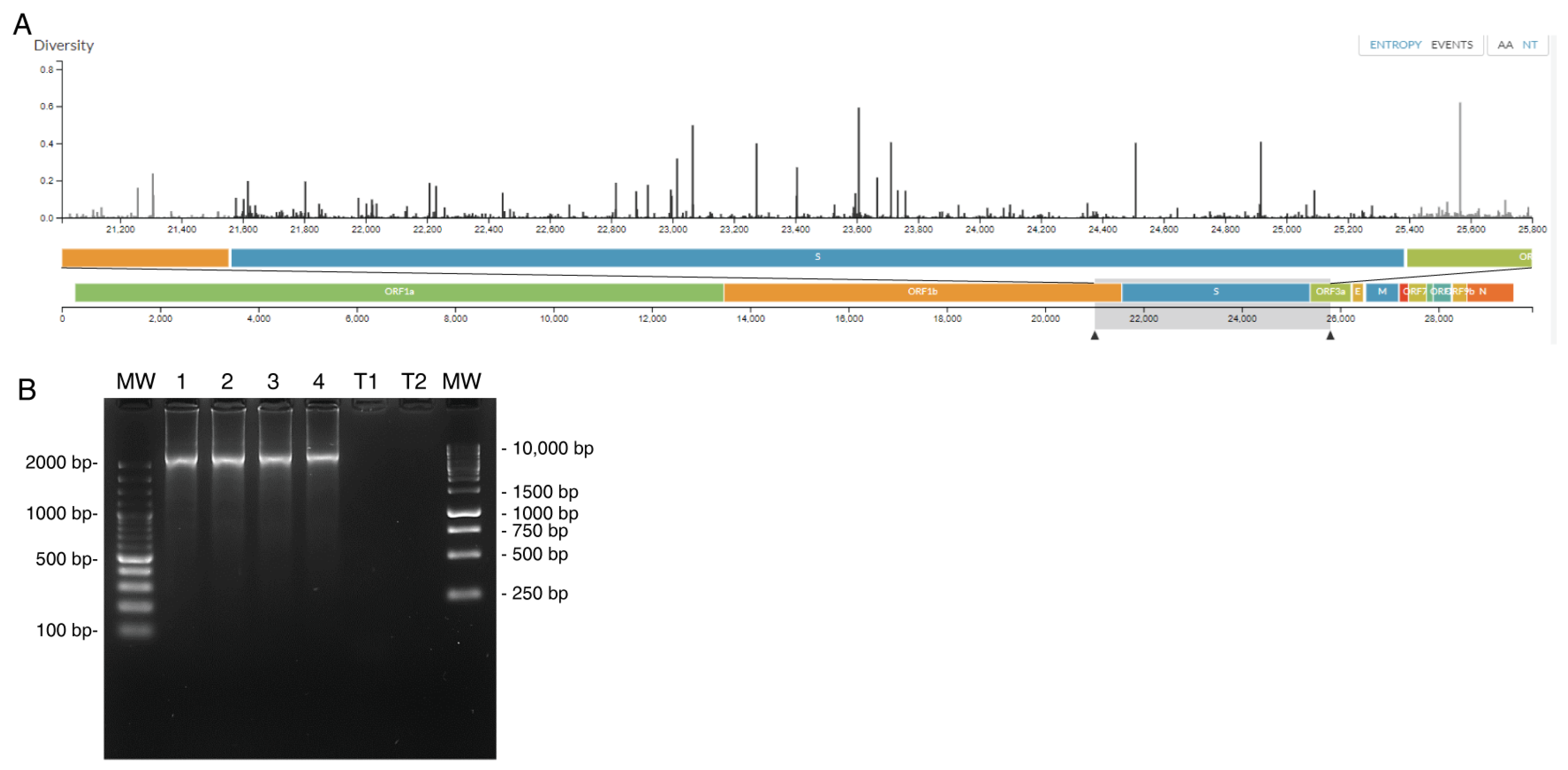
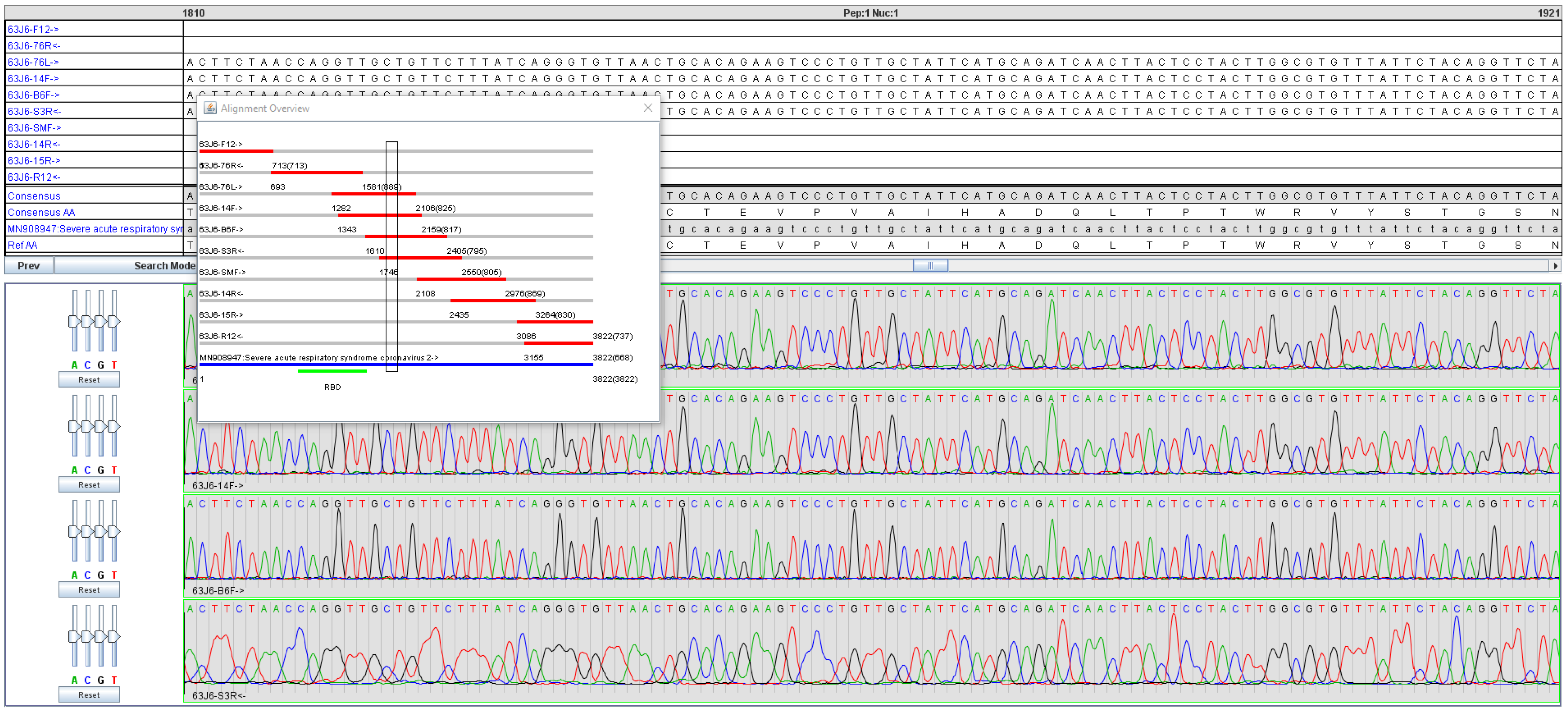
2. Materials and Methods
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html (accessed on 29 March 2021).
- Bosch, B.J.; van der Zee, R.; de Haan, C.A.; Rottier, P.J. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J. Virol. 2003, 77, 8801–8811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piccoli, L.; Park, Y.J.; Tortorici, M.A.; Czudnochowski, N.; Walls, A.C.; Beltramello, M.; Silacci-Fregni, C.; Pinto, D.; Rosen, L.E.; Bowen, J.E.; et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell 2020, 183, 1024–1042.e1021. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e819. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Thao, T.T.N.; Hoffmann, D.; Taddeo, A.; Ebert, N.; Labroussaa, F.; Pohlmann, A.; King, J.; Steiner, S.; Kelly, J.N.; et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 2021, 592, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Weissman, D.; Alameh, M.G.; de Silva, T.; Collini, P.; Hornsby, H.; Brown, R.; LaBranche, C.C.; Edwards, R.J.; Sutherland, L.; Santra, S.; et al. D614G Spike Mutation Increases SARS CoV-2 Susceptibility to Neutralization. Cell Host Microbe 2021, 29, 23–31 e24. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.W.; Tambyah, P.A.; Hui, D.S. Emergence of a new SARS-CoV-2 variant in the UK. J. Infect. 2021, 82, e27–e28. [Google Scholar] [CrossRef] [PubMed]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020. [Google Scholar] [CrossRef]
- Campbell, F.; Archer, B.; Laurenson-Schafer, H.; Jinnai, Y.; Konings, F.; Batra, N.; Pavlin, B.; Vandemaele, K.; Van Kerkhove, M.D.; Jombart, T.; et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill 2021, 26, 2100509. [Google Scholar] [CrossRef]
- Beucher, G.; Blondot, M.-L.; Celle, A.; Pied, N.; Recordon-Pinson, P.; Esteves, P.; Faure, M.; Métifiot, M.; Lacomme, S.; Dacheaux, D.; et al. SARS-CoV-2 transmission via apical syncytia release from primary bronchial epithelia and infectivity restriction in children epithelia. bioRxiv 2021. [Google Scholar] [CrossRef]
- Jary, A.; Leducq, V.; Malet, I.; Marot, S.; Klement-Frutos, E.; Teyssou, E.; Soulie, C.; Abdi, B.; Wirden, M.; Pourcher, V.; et al. Evolution of viral quasispecies during SARS-CoV-2 infection. Clin. Microbiol Infect. 2020, 26, 1560.e1–1560.e4. [Google Scholar] [CrossRef]
| Strain | WIV04/2019 | 20A.EU2 | Alpha | Beta | Gamma | Delta |
|---|---|---|---|---|---|---|
| AA position on spike\# | N/A | 2 | 10 | 12 | 12 | 12 |
| 18 | L | F | F | |||
| 19 | T | R | ||||
| 20 | T | N | ||||
| 26 | P | S | ||||
| 69 | H | - | ||||
| 70 | V | - | ||||
| 80 | D | A | ||||
| 138 | D | Y | ||||
| 142 | G | D | ||||
| 144 | Y | - | ||||
| 154 | E | K | ||||
| 157 | F | - | ||||
| 158 | R | - | ||||
| 190 | R | S | ||||
| 215 | D | G | ||||
| 242 | L | - | ||||
| 243 | A | - | ||||
| 244 | L | - | ||||
| 246 | R | I | ||||
| 417 | K | N | T | |||
| 452 | L | R | ||||
| 477 | S | N | ||||
| 478 | T | K | ||||
| 484 | E | K | K | Q | ||
| 501 | N | Y | Y | Y | ||
| 570 | A | D | ||||
| 614 | D | G | G | G | G | G |
| 655 | H | Y | ||||
| 681 | P | H | R | |||
| 701 | A | V | ||||
| 716 | T | I | ||||
| 950 | D | N | ||||
| 982 | S | A | ||||
| 1027 | T | I | ||||
| 1071 | Q | H | ||||
| 1118 | D | H | ||||
| 1176 | V | F |
| Primer Name | Sequence | Positions (Ref. NC_045512.2) | Reference | |
|---|---|---|---|---|
| RT-PCR | Forward F1-1 | GCATGGTGGACAGCCTTTGT | 21,220 to 21,239 | [7] |
| Reverse F1-1 | TGGGTTTTTGGAACGGCATT | 25,787 to 25,806 | ||
| Nested PCR + sequencing | Forward F1-2 | GGGGTACTGCTGTTATGTCTTT | 21,422 to 21,443 | |
| Reverse F1-2 | CACCCTTGGAGAGTGCTAGT | 25,602 to 25,621 | ||
| Sequencing | nCoV-2019_76_LEFT | AGGGCAAACTGGAAAGATTGCT | 22,798 to 22,819 | ANRS AC43 |
| nCoV-2019_76_RIGHT | ACCTGTGCCTGTTAAACCATTGA | 23,190 to 23,212 | ||
| CV_14_F | CAGGCTGCGTTATAGCTTGGAA | 22,851 to 22,872 | CDC | |
| CV_14_R | AACCAGTGTGTGCCATTTGAA | 24,870 to 24,850 | ||
| CV_15_F | GCAGAAATCAGAGCTTCTGCTAATC | 24,608 to 24,632 | ||
| SARS2_B6F | CCAGCAACTGTTTGTGGACCTA | 23,123 to 23,144 | ||
| SARS SMF | TCAATCCATCATTGCCTACAC | 23,629 to 23,649 | This work | |
| SARS S3R | GAGCAAAGGTGGCAAAACAG | 24,138 to 24,157 |
| Sample Type | Sampling Date | Sex | Ct RTqPCR | Known Spike Mutations | Others Spike Mutations | Variant | ||
|---|---|---|---|---|---|---|---|---|
| N | RdRp | E | ||||||
| Nasal swab | Feb 2021 | M | 28.79 | 28.79 | ND | del69-70, del144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | V6A, Q14Q/P | Alpha |
| Nasal swab | Feb 2021 | F | 31.41 | 31.44 | ND | del69-70, del144, N501Y, A570D, D614G, P681R, T716I, S982A, D1118H | Alpha | |
| Nasal swab | Feb 2021 | F | 17.68 | 19.39 | ND | del69-70, del144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | V47V/I, A831V, T998T/P, T1006T/P | Alpha |
| Nasal swab | Feb 2021 | M | 20.26 | 21.62 | ND | del69-70, del144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | V6A | Alpha |
| Nasal swab | Feb 2021 | F | 24.91 | 25.50 | ND | del69-70, del144, N501Y, A570D, D614G, P681R, T716I, S982A, D1118H | Alpha | |
| Nasal swab | Feb 2021 | F | 21.37 | 20.90 | ND | L18F, D80A, D215G, del242-244, K417N, E484K, N501Y, D614G, A701V | Beta | |
| Nasal swab | Feb 2021 | F | 35.16 | >40 | ND | del69-70, del144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | Alpha | |
| Nasal swab | Feb 2021 | M | 26.90 | 28.65 | ND | L18F, L452R, N501Y, A653V, H655Y, D796Y, G1219V | R346I, Q667H, K1191N | new |
| Nasal swab | Feb 2021 | F | 23.72 | 31.50 | ND | L18F, L452R, N501Y, A653V, H655Y, D796Y, G1219V | R346I, Q667H, K1191N | new |
| Nasal swab | Feb 2021 | M | 35.76 | 31.50 | ND | del69-70, del144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | I197I/V, A831V, D839D/G, T1238T/I | Alpha |
| Nasal swab | Feb 2021 | M | 15.14 | 16.58 | 13.53 | D614G | A222V | WT |
| Nasal swab | Feb 2021 | M | 27.19 | 29.57 | 25.64 | S477N, D614G, G1219C | 20A.EU2 | |
| BAL | April 2021 | M | 17.90 | 19.43 | 16.89 | del69-70, del144, N501Y, A570D, D614G, P681R, T716I, S982A, D1118H | Alpha | |
| BAL | Feb 2021 | F | 15.26 | 16.6 | 13.56 | S477N, D614G | F2L, S459F, K1045E | 20A.EU2 |
| BAL | Feb 2021 | M | 21.95 | 22.12 | 20.29 | D614G | T29I, Q675H | WT |
| Cell culture | May 2020 | - | ND | ND | ND | no mutation | no mutation | WT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Recordon-Pinson, P.; Blondot, M.-L.; Bellecave, P.; Lafon, M.-E.; Tumiotto, C.; Métifiot, M.; Andreola, M.-L. A Simple and Fast Method to Sequence the Full-Length Spike Gene for SARS-CoV-2 Variant Identification from Patient Samples. COVID 2021, 1, 337-344. https://doi.org/10.3390/covid1010028
Recordon-Pinson P, Blondot M-L, Bellecave P, Lafon M-E, Tumiotto C, Métifiot M, Andreola M-L. A Simple and Fast Method to Sequence the Full-Length Spike Gene for SARS-CoV-2 Variant Identification from Patient Samples. COVID. 2021; 1(1):337-344. https://doi.org/10.3390/covid1010028
Chicago/Turabian StyleRecordon-Pinson, Patricia, Marie-Lise Blondot, Pantxika Bellecave, Marie-Edith Lafon, Camille Tumiotto, Mathieu Métifiot, and Marie-Line Andreola. 2021. "A Simple and Fast Method to Sequence the Full-Length Spike Gene for SARS-CoV-2 Variant Identification from Patient Samples" COVID 1, no. 1: 337-344. https://doi.org/10.3390/covid1010028
APA StyleRecordon-Pinson, P., Blondot, M.-L., Bellecave, P., Lafon, M.-E., Tumiotto, C., Métifiot, M., & Andreola, M.-L. (2021). A Simple and Fast Method to Sequence the Full-Length Spike Gene for SARS-CoV-2 Variant Identification from Patient Samples. COVID, 1(1), 337-344. https://doi.org/10.3390/covid1010028






