Observation of the Transition Phenomenon of High-Density Cell Distribution in a Two-Dimensional Microspace of the Unicellular Green Alga Chlamydomonas reinhardtii
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
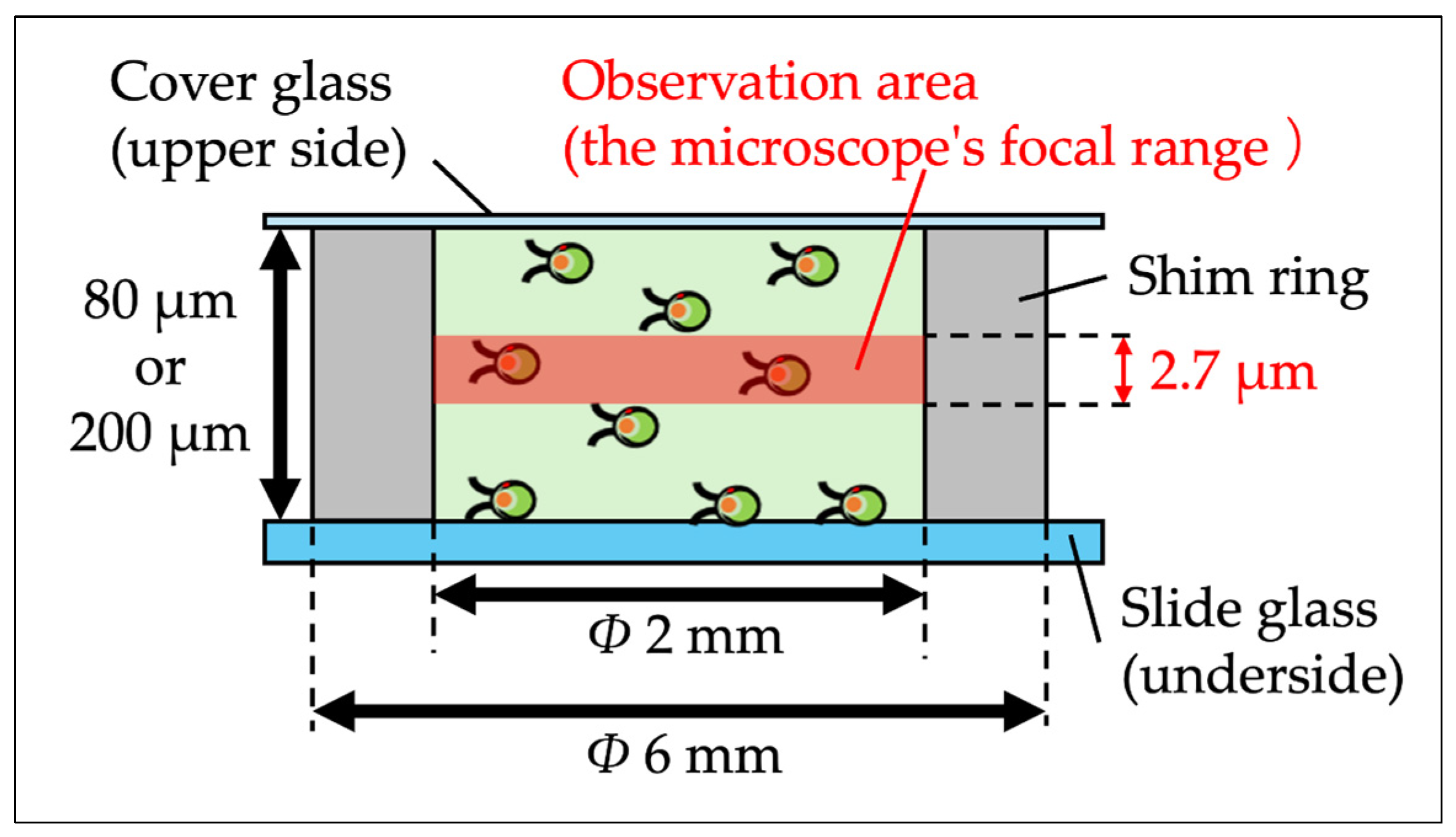
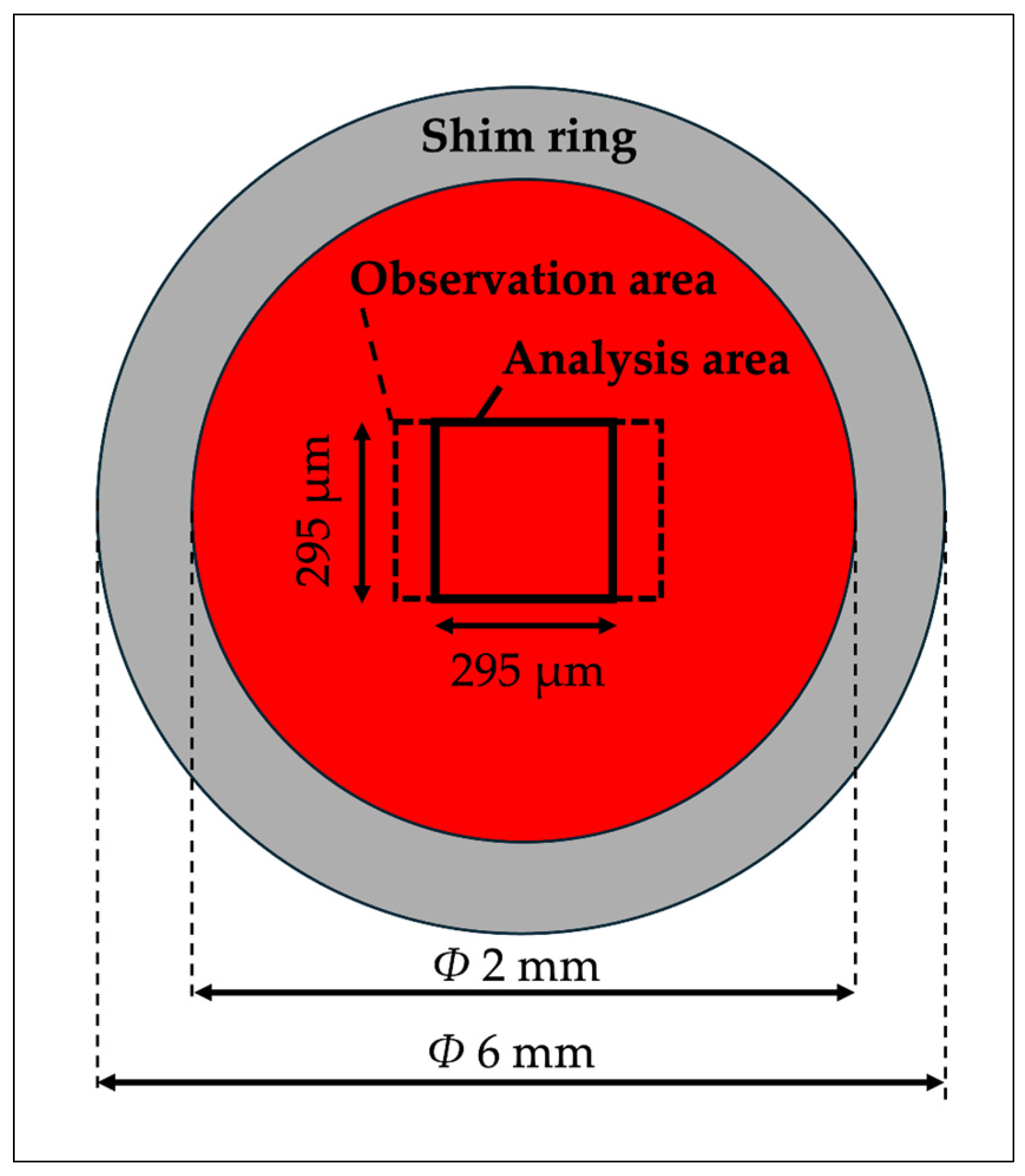
2.2. Quasi-2D Spatial Device
2.3. Experimental Apparatus
2.4. Image Analysis
2.4.1. Frame Interval
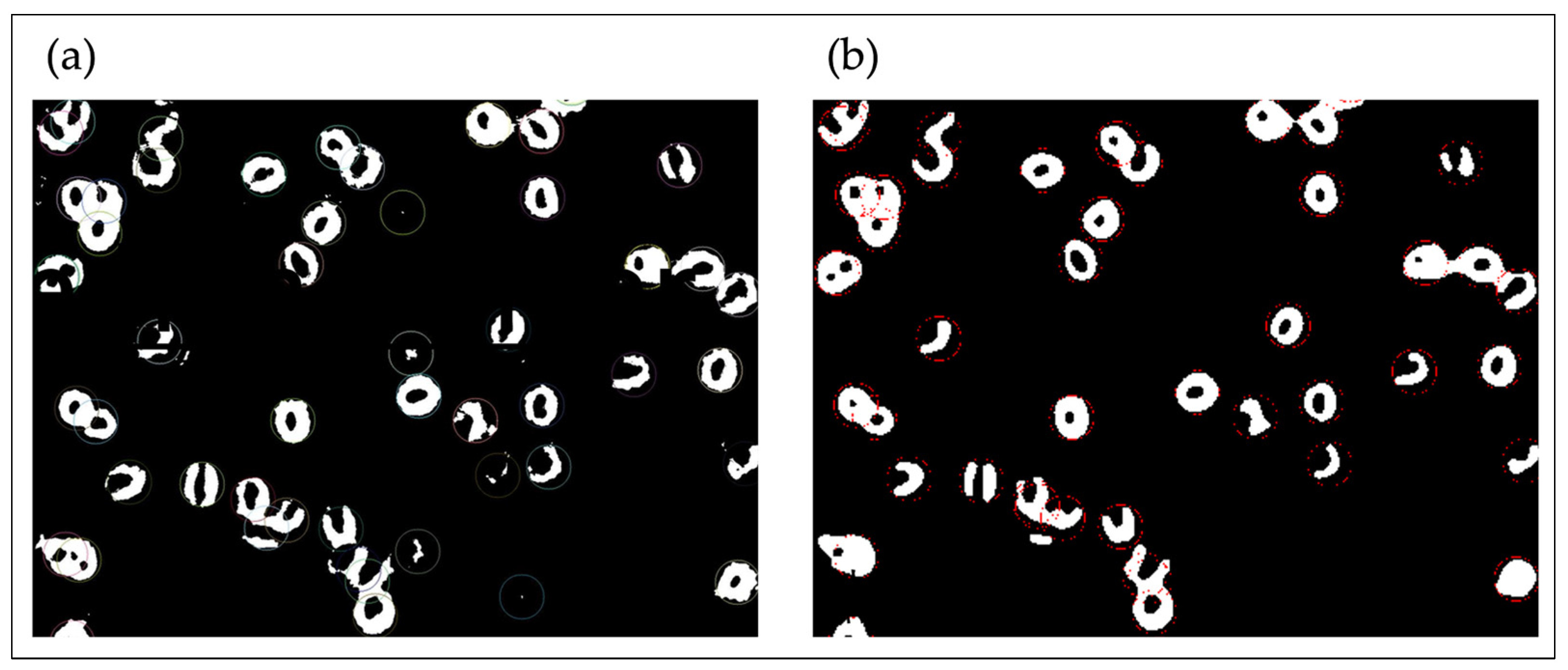
2.4.2. Binarization and Particle Tracking
2.4.3. Cell Counting
2.4.4. Simulation
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A


References
- Fox, M.; Anthony, M. Quantum Optics: An Introduction; Oxford University Press: New York, NY, USA, 2006. [Google Scholar]
- Koyama, K.; Hokunan, H.; Hasegawa, M.; Kawamura, S.; Koseki, S. Do Bacterial Cell Numbers Follow a Theoretical Poisson Distribution? Comparison of Experimentally Obtained Numbers of Single Cells with Random Number Generation via Computer Simulation. Food Microbiol. 2016, 60, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.T. Higher-Order Criteria for Nonclassical Effects in Photon Statistics. Phys. Rev. A 1990, 41, 1721–1723. [Google Scholar] [CrossRef] [PubMed]
- Bechinger, C.; Di Leonardo, R.; Löwen, H.; Reichhardt, C.; Volpe, G.; Volpe, G. Active Particles in Complex and Crowded Environments. Available online: https://arxiv.org/abs/1602.00081v2 (accessed on 10 February 2023).
- Reynolds, C.W. Flocks, Herds and Schools: A Distributed Behavioral Model. ACM SIGGRAPH Comput. Graph. 1987, 21, 25–34. [Google Scholar] [CrossRef]
- Couzin, I.D.; Krause, J.; James, R.; Ruxton, G.D.; Franks, N.R. Collective Memory and Spatial Sorting in Animal Groups. J. Theor. Biol. 2002, 218, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hemelrijk, C.K.; Hildenbrandt, H. Schools of Fish and Flocks of Birds: Their Shape and Internal Structure by Self-Organization. Interface Focus 2012, 2, 726–737. [Google Scholar] [CrossRef] [PubMed]
- Reid, D.A.P.; Hildenbrandt, H.; Padding, J.T.; Hemelrijk, C.K. Fluid Dynamics of Moving Fish in a Two-Dimensional Multiparticle Collision Dynamics Model. Phys. Rev. E 2012, 85, 021901. [Google Scholar] [CrossRef] [PubMed]
- Cresswell, W. Flocking Is an Effective Anti-Predation Strategy in Redshanks, Tringa Totanus. Anim. Behav. 1994, 47, 433–442. [Google Scholar] [CrossRef]
- Krause, J.; Ruxton, G.D. Living in Groups; Oxford University Press: New York, NY, USA, 2002. [Google Scholar]
- Cresswell, W.; Quinn, J.L. Predicting the Optimal Prey Group Size from Predator Hunting Behaviour. J. Anim. Ecol. 2011, 80, 310–319. [Google Scholar] [CrossRef] [PubMed]
- Moeur, M. Characterizing Spatial Patterns of Trees Using Stem-Mapped Data. For. Sci. 1993, 39, 756–775. [Google Scholar] [CrossRef]
- Darnton, N.C.; Turner, L.; Rojevsky, S.; Berg, H.C. Dynamics of Bacterial Swarming. Biophys. J. 2010, 98, 2082–2090. [Google Scholar] [CrossRef]
- Be’er, A.; Ariel, G. A Statistical Physics View of Swarming Bacteria. Mov. Ecol. 2019, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Zuo, W.; Wu, Y. Dynamic Motility Selection Drives Population Segregation in a Bacterial Swarm. Proc. Natl. Acad. Sci. USA 2020, 117, 4693–4700. [Google Scholar] [CrossRef] [PubMed]
- Bees, M.A. Advances in Bioconvection. Annu. Rev. Fluid Mech. 2020, 52, 449–476. [Google Scholar] [CrossRef]
- Nonaka, Y.; Kikuchi, K.; Numayama-Tsuruta, K.; Kage, A.; Ueno, H.; Ishikawa, T. Inhomogeneous Distribution of Chlamydomonas in a Cylindrical Container with a Bubble Plume. Biol. Open 2016, 5, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Jánosi, I.M.; Czirók, A.; Silhavy, D.; Holczinger, A. Is Bioconvection Enhancing Bacterial Growth in Quiescent Environments? Environ. Microbiol. 2002, 4, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Pooley, C.M.; Alexander, G.P.; Yeomans, J.M. Hydrodynamic Interaction between Two Swimmers at Low Reynolds Number. Phys. Rev. Lett. 2007, 99, 228103. [Google Scholar] [CrossRef] [PubMed]
- Knežević, M.; Welker, T.; Stark, H. Collective Motion of Active Particles Exhibiting Non-Reciprocal Orientational Interactions. Sci. Rep. 2022, 12, 19437. [Google Scholar] [CrossRef] [PubMed]
- Aono, T.; Yamashita, K.; Hashimoto, M.; Ishikawa, Y.; Aizawa, K.; Tokunaga, E. Spatial Distribution of Flagellated Microalgae Chlamydomonas reinhardtii in a Quasi-Two-Dimensional Space. Micromachines 2023, 14, 813. [Google Scholar] [CrossRef]
- Xiang, C.; Liu, J.; Ma, L.; Yang, M. Overexpressing Codon-Adapted Fusion Proteins of 4-Coumaroyl-CoA Ligase (4CL) and Stilbene Synthase (STS) for Resveratrol Production in Chlamydomonas reinhardtii. J. Appl. Phycol. 2020, 32, 1669–1676. [Google Scholar] [CrossRef]
- Saifuddin, N.; Ong, M.Y.; Priatharsini, P. Optimization of Photosynthetic Hydrogen Gas Production by Green Alga in Sulfur Deprived Condition. Indian J. Sci. Technol. 2016, 9, 1–13. [Google Scholar] [CrossRef]
- Fragkopoulos, A.A.; Vachier, J.; Frey, J.; Le Menn, F.-M.; Mazza, M.G.; Wilczek, M.; Zwicker, D.; Bäumchen, O. Self-Generated Oxygen Gradients Control Collective Aggregation of Photosynthetic Microbes. J. R. Soc. Interface 2021, 18, 20210553. [Google Scholar] [CrossRef]
- Bentley, S.A.; Laeverenz-Schlogelhofer, H.; Anagnostidis, V.; Cammann, J.; Mazza, M.G.; Gielen, F.; Wan, K.Y. Phenotyping Single-Cell Motility in Microfluidic Confinement. eLife 2022, 11, e76519. [Google Scholar] [CrossRef] [PubMed]
- Ostapenko, T.; Schwarzendahl, F.J.; Böddeker, T.J.; Kreis, C.T.; Cammann, J.; Mazza, M.G.; Bäumchen, O. Curvature-Guided Motility of Microalgae in Geometric Confinement. Phys. Rev. Lett. 2018, 120, 068002. [Google Scholar] [CrossRef]
- Cammann, J.; Schwarzendahl, F.J.; Ostapenko, T.; Lavrentovich, D.; Bäumchen, O.; Mazza, M.G. Emergent Probability Fluxes in Confined Microbial Navigation. Proc. Natl. Acad. Sci. USA 2021, 118, e2024752118. [Google Scholar] [CrossRef] [PubMed]
- Markov, D.; Grigorov, E.; Kirov, B.; Denev, J.A.; Galabov, V.; Marinov, M.B. Low-Cost Three-Dimensionally Printed Inverted Plug and Play Optical Instrument for Microfluidic Imaging. Micro 2023, 3, 537–548. [Google Scholar] [CrossRef]
- Morisita, M. Measuring of Interspecific Association and Similarity between Communities. Mem. Fac. Sci. Kyushu Univ. Ser. E Biol. 1959, 3, 65–80. [Google Scholar]
- Eberhardt, L.L. Some Developments in “Distance Sampling”. Biometrics 1967, 23, 207–216. [Google Scholar] [CrossRef]
- Hines, W.G.S.; Hines, R.J.O. The Eberhardt Statistic and the Detection of Nonrandomness of Spatial Point Distributions. Biometrika 1979, 66, 73–79. [Google Scholar] [CrossRef]
- Kreis, C.T.; Le Blay, M.; Linne, C.; Makowski, M.M.; Bäumchen, O. Adhesion of Chlamydomonas Microalgae to Surfaces Is Switchable by Light. Nat. Phys. 2018, 14, 45–49. [Google Scholar] [CrossRef]
- Benefer, C.M.; D’Ahmed, K.S.; Blackshaw, R.P.; Sint, H.M.; Murray, P.J. The Distribution of Soil Insects across Three Spatial Scales in Agricultural Grassland. Front. Ecol. Evol. 2016, 4, 41. [Google Scholar] [CrossRef]
- Chen, G.; Zhu, N.; Hu, Z.; Liu, L.; Wang, G.-Q.; Wang, G. Motility Changes Rather than EPS Production Shape Aggregation of Chlamydomonas microsphaera in Aquatic Environment. Environ. Technol. 2021, 42, 2916–2924. [Google Scholar] [CrossRef]
- Zhao, R.; Chen, G.; Liu, L.; Zhang, W.; Sun, Y.; Li, B.; Wang, G. Bacterial Foraging Facilitates Aggregation of Chlamydomonas microsphaera in an Organic Carbon Source-Limited Aquatic Environment. Environ. Pollut. 2020, 259, 113924. [Google Scholar] [CrossRef] [PubMed]
- Pinto, E.; Van Nieuwerburgh, L.; Paes de Barros, M.; Pedersén, M.; Colepicolo, P.; Snoeijs, P. Density-Dependent Patterns of Thiamine and Pigment Production in the Diatom Nitzschia microcephala. Phytochemistry 2003, 63, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Palanisamy, K.M.; Bhat, O.A.; Govindan, N.; Rahim, M.H.A.; Maniam, G.P. Impact of Biomass Density on Growth Rates of Spirulina platensis under Different Light Spectra. Maejo Int. J. Energy Environ. Commun. 2023, 5, 1–5. [Google Scholar] [CrossRef]
- Knight, W. A Method of Sequential Estimation Applicable to the Hypergeometric, Binomial, Poisson, and Exponential Distributions. Ann. Math. Stat. 1965, 36, 1494–1503. [Google Scholar] [CrossRef]
- Gnutt, D.; Gao, M.; Brylski, O.; Heyden, M.; Ebbinghaus, S. Excluded-Volume Effects in Living Cells. Angew. Chem. Int. Ed. 2015, 54, 2548–2551. [Google Scholar] [CrossRef] [PubMed]


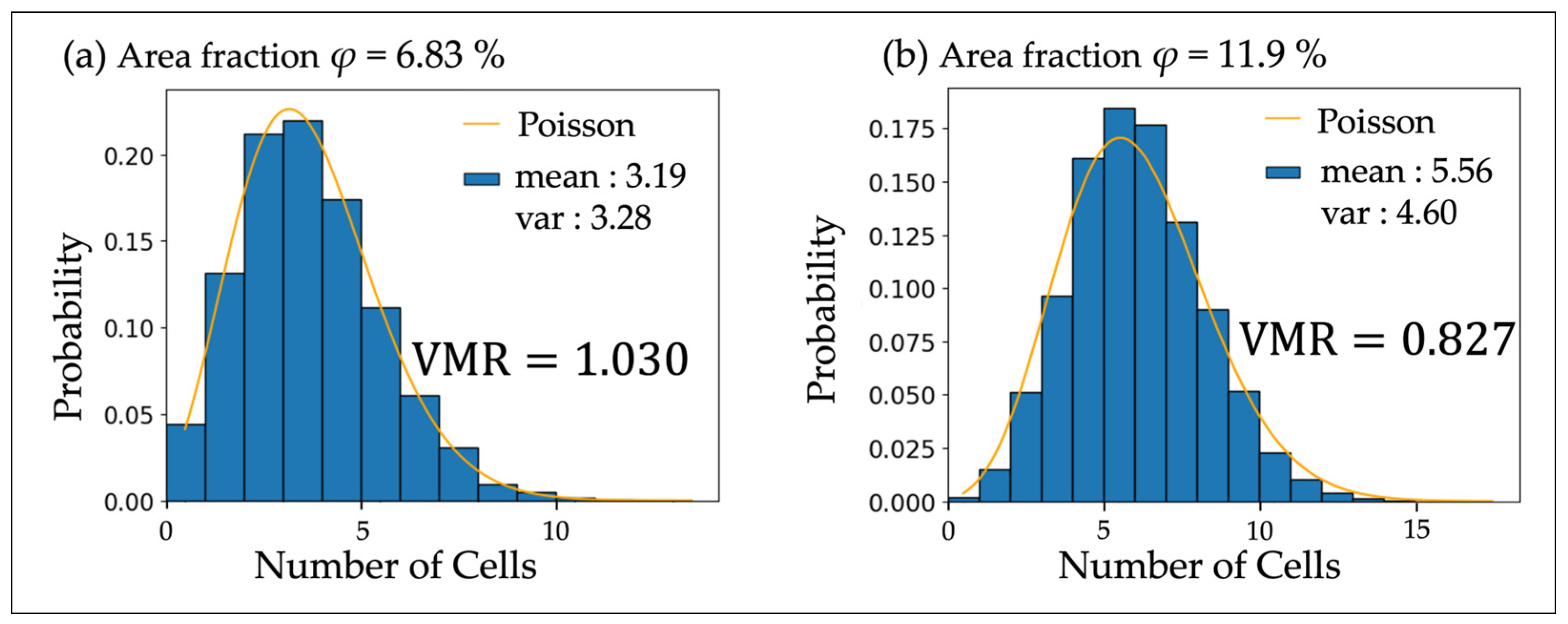

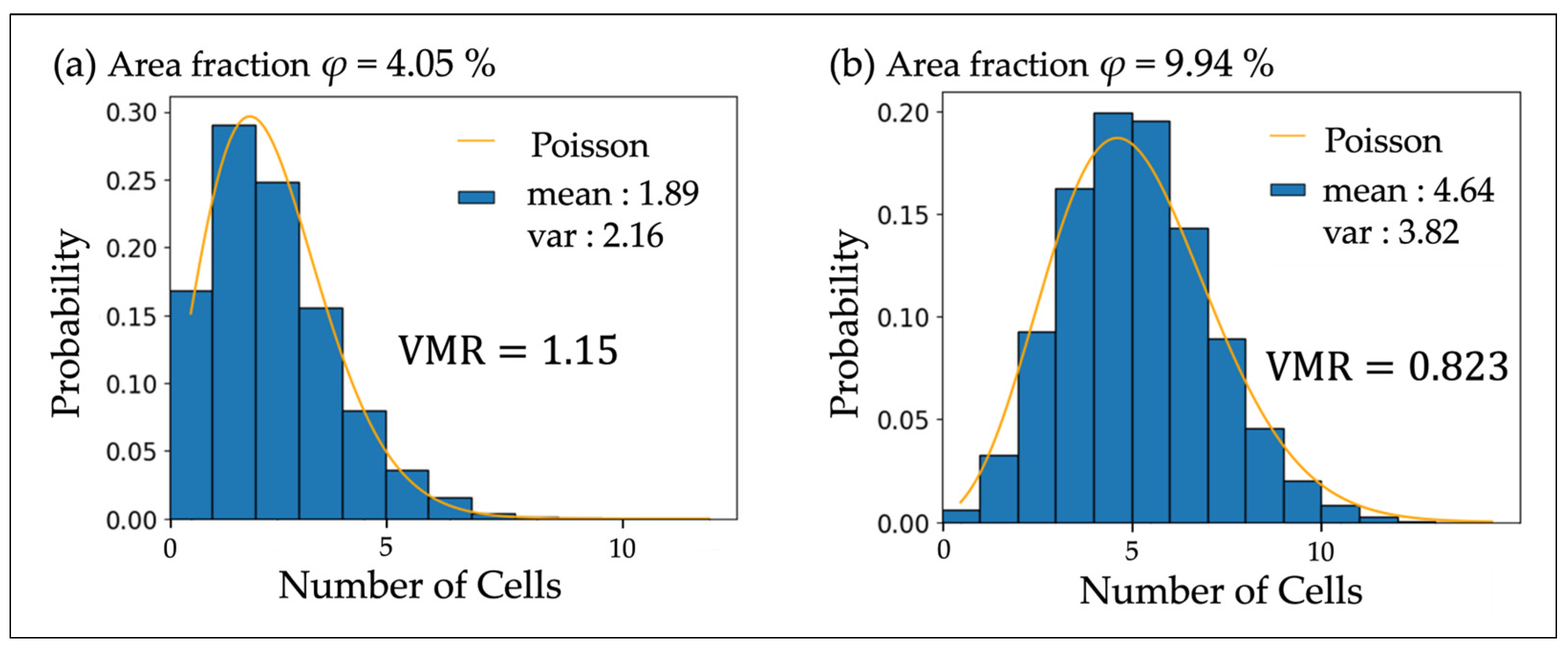
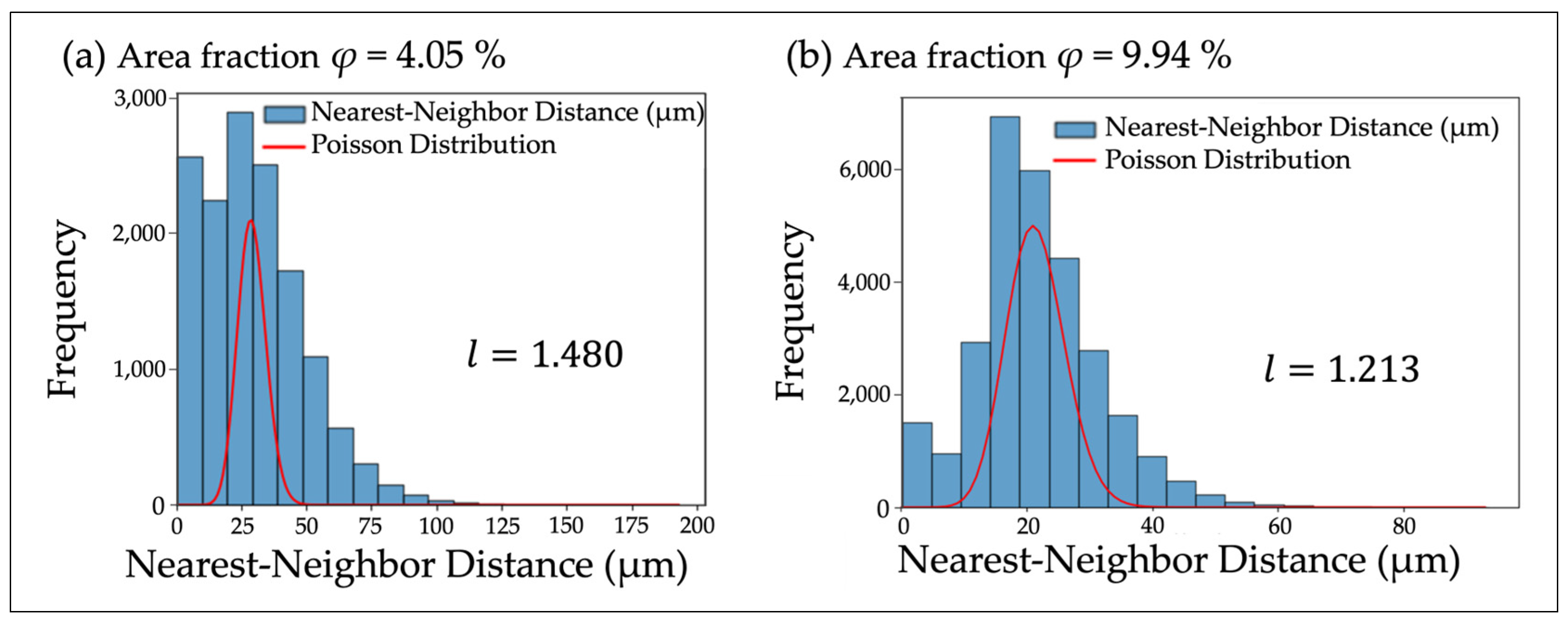
| Area Fraction φ (%) | Measured VMR | Simulated VMR | Eberhardt Analysis l | Cellular Interaction |
|---|---|---|---|---|
| 5.39 | 1.034 | 0.920 ± 0.012 | 1.378 | Swarming |
| 6.24 | 1.008 | 0.908 ± 0.011 | 1.344 | Swarming |
| 6.83 | 1.030 | 0.903 ± 0.004 | 1.412 | Swarming |
| 9.43 | 0.855 | 0.881 ± 0.005 | 1.254 | Dispersal |
| 10.7 | 0.834 | 0.865 ± 0.010 | 1.255 | Dispersal |
| 11.9 | 0.827 | 0.865 ± 0.005 | 1.253 | Dispersal |
| Area Fraction φ (%) | Measured VMR | Simulated VMR | Eberhardt Analysis l | Cellular Interaction |
|---|---|---|---|---|
| 2.86 | 1.131 | 0.940 ± 0.007 | 1.500 | Swarming |
| 3.25 | 1.078 | 0.940 ± 0.007 | 1.427 | Swarming |
| 4.05 | 1.146 | 0.928 ± 0.007 | 1.480 | Swarming |
| 7.80 | 0.886 | 0.889 ±0.009 | 1.249 | Swarming or Dispersal |
| 9.94 | 0.823 | 0.873 ± 0.008 | 1.213 | Dispersal |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goda, Y.; Yamashita, K.; Aono, T.; Aizawa, K.; Hashimoto, M.; Tokunaga, E. Observation of the Transition Phenomenon of High-Density Cell Distribution in a Two-Dimensional Microspace of the Unicellular Green Alga Chlamydomonas reinhardtii. Micro 2024, 4, 412-425. https://doi.org/10.3390/micro4030026
Goda Y, Yamashita K, Aono T, Aizawa K, Hashimoto M, Tokunaga E. Observation of the Transition Phenomenon of High-Density Cell Distribution in a Two-Dimensional Microspace of the Unicellular Green Alga Chlamydomonas reinhardtii. Micro. 2024; 4(3):412-425. https://doi.org/10.3390/micro4030026
Chicago/Turabian StyleGoda, Yuka, Kyohei Yamashita, Tetsuo Aono, Kentaro Aizawa, Masafumi Hashimoto, and Eiji Tokunaga. 2024. "Observation of the Transition Phenomenon of High-Density Cell Distribution in a Two-Dimensional Microspace of the Unicellular Green Alga Chlamydomonas reinhardtii" Micro 4, no. 3: 412-425. https://doi.org/10.3390/micro4030026
APA StyleGoda, Y., Yamashita, K., Aono, T., Aizawa, K., Hashimoto, M., & Tokunaga, E. (2024). Observation of the Transition Phenomenon of High-Density Cell Distribution in a Two-Dimensional Microspace of the Unicellular Green Alga Chlamydomonas reinhardtii. Micro, 4(3), 412-425. https://doi.org/10.3390/micro4030026






