Immunocompetent High-Throughput Gut-on-Chip Model for Intestinal Microbes—Host Interaction Studies
Abstract
1. Introduction
- The possibility of combining several cell types in one assay (e.g., epithelial and immune cells).
- The 3D spatial organization of the tissues.
- The use of human-derived host cells.
2. Materials and Methods
2.1. PBMC
2.2. Intestinal Epithelial Cells
2.3. OrganoPlate Seeding
2.4. Bacteria Co-Culture in OrganoPlate
2.5. Cytokine Secretion
2.6. TEER Measurements
2.7. Immunohistochemistry and Microscopy
2.8. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Buckley, A.; Turner, J.R. Cell Biology of Tight Junction Barrier Regulation and Mucosal Disease. Cold Spring Harb. Perspect. Biol. 2018, 10, a029314. [Google Scholar] [CrossRef]
- Moonwiriyakit, A.; Pathomthongtaweechai, N.; Steinhagen, P.R.; Chantawichitwong, P.; Satianrapapong, W.; Pongkorpsakol, P. Tight junctions: From molecules to gastrointestinal diseases. Tissue Barriers 2023, 11, 2077620. [Google Scholar] [CrossRef]
- Tommaso, D.; Gasbarrini, A.; Ponziani, F.R. Intestinal Barrier in Human Health and Disease. Int. J. Environ. Res. Public Health 2021, 18, 12836. [Google Scholar] [CrossRef]
- Stolfi, C.; Maresca, C.; Monteleone, G.; Laudisi, F. Implication of Intestinal Barrier Dysfunction in Gut Dysbiosis and Diseases. Biomedicines 2022, 10, 289. [Google Scholar] [CrossRef]
- Kinashi, Y.; Hase, K. Partners in Leaky Gut Syndrome: Intestinal Dysbiosis and Autoimmunity. Front. Immunol. 2021, 12, 673708. [Google Scholar] [CrossRef]
- Régnier, M.; Van Hul, M.; Knauf, C.; Cani, P.D. Gut microbiome, endocrine control of gut barrier function and metabolic diseases. J. Endocrinol. 2021, 248, R67–R82. [Google Scholar] [CrossRef] [PubMed]
- Mou, Y.; Du, Y.; Zhou, L.; Yue, J.; Hu, X.; Liu, Y.; Chen, S.; Lin, X.; Zhang, G.; Xiao, H.; et al. Gut Microbiota Interact With the Brain Through Systemic Chronic Inflammation: Implications on Neuroinflammation, Neurodegeneration, and Aging. Front. Immunol. 2022, 13, 796288. [Google Scholar] [CrossRef] [PubMed]
- Kayama, H.; Okumura, R.; Takeda, K. Interaction Between the Microbiota, Epithelia, and Immune Cells in the Intestine. Annu. Rev. Immunol. 2020, 38, 23–48. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Whitley, C.S.; Haribabu, B.; Jala, V.R. Regulation of Intestinal Barrier Function by Microbial Metabolites. Cell. Mol. Gastroenterol. Hepatol. 2021, 11, 1463–1482. [Google Scholar] [CrossRef]
- Neurath, M.F. Cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 2014, 14, 329–342. [Google Scholar] [CrossRef]
- Barbara, G.; Barbaro, M.R.; Fuschi, D.; Palombo, M.; Falangone, F.; Cremon, C.; Marasco, G.; Stanghellini, V. Inflammatory and Microbiota-Related Regulation of the Intestinal Epithelial Barrier. Front. Nutr. 2021, 8, 718356. [Google Scholar] [CrossRef]
- Chelakkot, C.; Ghim, J.; Ryu, S.H. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp. Mol. Med. 2018, 50, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Capaldo, C.T.; Nusrat, A. Cytokine regulation of tight junctions. Biochim. Biophys. Acta 2009, 1788, 864–871. [Google Scholar] [CrossRef]
- Martyniak, A.; Medyńska-Przęczek, A.; Wędrychowicz, A.; Skoczeń, S.; Tomasik, P.J. Prebiotics, Probiotics, Synbiotics, Paraprobiotics and Postbiotic Compounds in IBD. Biomolecules 2021, 11, 1903. [Google Scholar] [CrossRef]
- Tsai, Y.L.; Lin, T.L.; Chang, C.J.; Wu, T.R.; Lai, W.F.; Lu, C.C.; Lai, H.C. Probiotics, prebiotics and amelioration of diseases. J. Biomed. Sci. 2019, 26, 3. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.H.; Siah, K.W.; Lo, A.W. Estimation of clinical trial success rates and related parameters. Biostatistics 2019, 20, 273–286. [Google Scholar] [CrossRef]
- Ciorba, M.A.; Konnikova, L.; Hirota, S.A.; Lucchetta, E.M.; Turner, J.R.; Slavin, A.; Johnson, K.; Condray, C.D.; Hong, S.; Cressall, B.K.; et al. Challenges in IBD Research 2024: Preclinical Human IBD Mechanisms. Inflamm. Bowel Dis. 2024, 30 (Suppl. S2), S5–S18. [Google Scholar] [CrossRef]
- Pocock, K.; Delon, L.; Bala, V.; Rao, S.; Priest, C.; Prestidge, C.; Thierry, B. Intestine-on-a-Chip Microfluidic Model for Efficient in Vitro Screening of Oral Chemotherapeutic Uptake. ACS Biomater. Sci. Eng. 2017, 3, 951–959. [Google Scholar] [CrossRef]
- Chi, M.; Yi, B.; Oh, S.; Park, D.J.; Sung, J.H.; Park, S. A microfluidic cell culture device (μFCCD) to culture epithelial cells with physiological and morphological properties that mimic those of the human intestine. Biomed. Microdevices 2015, 17, 9966. [Google Scholar] [CrossRef] [PubMed]
- Belotserkovsky, I.; Vernochet, C.; Roelens, M.; Beitz, B.; Ben Abdallah, B.; Poissonnier, S.; Bellais, S.; Hesketh, A.; Meza Torres, J.; Mouharib, M.; et al. Bifidobacterium Longum subsp. infantis and Lacticaseibacillus Rhamnosus GG Protect Intestinal Epithelium Against Inflammation-Mediated Damage in an Immunocompetent In-Vitro Model. Appl. Microbiol. 2025, 5, 110. [Google Scholar] [CrossRef]
- Beaurivage, C.; Naumovska, E.; Chang, Y.X.; Elstak, E.D.; Nicolas, A.; Wouters, H.; van Moolenbroek, G.; Lanz, H.L.; Trietsch, S.J.; Joore, J.; et al. Development of a Gut-On-A-Chip Model for High Throughput Disease Modeling and Drug Discovery. Int. J. Mol. Sci. 2019, 20, 5661. [Google Scholar] [CrossRef]
- Gijzen, L.; Marescotti, D.; Raineri, E.; Nicolas, A.; Lanz, H.L.; Guerrera, D.; van Vught, R.; Joore, J.; Vulto, P.; Peitsch, M.C.; et al. An Intestine-on-a-Chip Model of Plug-and-Play Modularity to Study Inflammatory Processes. SLAS Technol. 2020, 25, 585–597. [Google Scholar] [CrossRef]
- Bounab, Y.; Eyer, K.; Dixneuf, S.; Rybczynska, M.; Chauvel, C.; Mistretta, M.; Tran, T.; Aymerich, N.; Chenon, G.; Llitjos, J.F.; et al. Dynamic single-cell phenotyping of immune cells using the microfluidic platform DropMap. Nat. Protoc. 2020, 15, 2920–2955. [Google Scholar] [CrossRef] [PubMed]
- Meslier, V.; Plaza Oñate, F.; Ania, M.; Nehlich, M.; Belotserkovsky, I.; Bellais, S.; Thomas, V. Draft Genome Sequence of Isolate POC01, a Novel Anaerobic Member of the Oscillospiraceae Family, Isolated from Human Feces. Microbiol. Resour. Announc. 2022, 11, e0113421. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Singhal, R.; Shah, Y.M. Oxygen battle in the gut: Hypoxia and hypoxia-inducible factors in metabolic and inflammatory responses in the intestine. J. Biol. Chem. 2020, 295, 10493–10505. [Google Scholar] [CrossRef] [PubMed]
- van Deventer, S.J. Review article: Chemokine production by intestinal epithelial cells: A therapeutic target in inflammatory bowel disease? Aliment. Pharmacol. Ther. 1997, 11 (Suppl. S3), 116–120; discussion 120–121. [Google Scholar] [CrossRef]
- Vebr, M.; Pomahačová, R.; Sýkora, J.; Schwarz, J. A Narrative Review of Cytokine Networks: Pathophysiological and Therapeutic Implications for Inflammatory Bowel Disease Pathogenesis. Biomedicines 2023, 11, 3229. [Google Scholar] [CrossRef]
- Delday, M.; Mulder, I.; Logan, E.T.; Grant, G. Bacteroides thetaiotaomicron Ameliorates Colon Inflammation in Preclinical Models of Crohn’s Disease. Inflamm. Bowel Dis. 2019, 25, 85–96. [Google Scholar] [CrossRef]
- Luo, Y.; Lan, C.; Ren, W.; Wu, A.; Yu, B.; He, J.; Chen, D. Bacteroides thetaiotaomicron: A symbiotic ally against diarrhea along with modulation of gut microbial ecological networks via tryptophan metabolism and AHR-Nrf2 signaling. J. Adv. Res. 2025, in press. [Google Scholar] [CrossRef]
- Pan, M.; Barua, N.; Ip, M. Mucin-degrading gut commensals isolated from healthy faecal donor suppress intestinal epithelial inflammation and regulate tight junction barrier function. Front. Immunol. 2022, 13, 1021094. [Google Scholar] [CrossRef]
- Li, K.; Hao, Z.; Du, J.; Gao, Y.; Yang, S.; Zhou, Y. Bacteroides thetaiotaomicron relieves colon inflammation by activating aryl hydrocarbon receptor and modulating CD4(+)T cell homeostasis. Int. Immunopharmacol. 2021, 90, 107183. [Google Scholar] [CrossRef] [PubMed]
- Buffie, C.G.; Bucci, V.; Stein, R.R.; McKenney, P.T.; Ling, L.; Gobourne, A.; No, D.; Liu, H.; Kinnebrew, M.; Viale, A.; et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015, 517, 205–208. [Google Scholar] [CrossRef]
- Abt, M.C.; McKenney, P.T.; Pamer, E.G. Clostridium difficile colitis: Pathogenesis and host defence. Nat. Rev. Microbiol. 2016, 14, 609–620. [Google Scholar] [CrossRef]
- Daniel, S.L.; Ridlon, J.M. Clostridium scindens: History and current outlook for a keystone species in the mammalian gut involved in bile acid and steroid metabolism. FEMS Microbiol. Rev. 2025, 49, fuaf016. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, Y.-J.; Trapecar, M.; Wright, C.; Schneider, K.; Kemmitt, J.; Hernandez-Gordillo, V.; Yoon, J.H.; Poyet, M.; Alm, E.J.; et al. An immune-competent human gut microphysiological system enables inflammation-modulation by Faecalibacterium prausnitzii. NPJ Biofilms Microbiomes 2024, 10, 31. [Google Scholar] [CrossRef]
- Jalili-Firoozinezhad, S.; Gazzaniga, F.S.; Calamari, E.L.; Camacho, D.M.; Fadel, C.W.; Bein, A.; Swenor, B.; Nestor, B.; Cronce, M.J.; Tovaglieri, A.; et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 2019, 3, 520–531. [Google Scholar] [CrossRef]
- Marzorati, M.; Vanhoecke, B.; De Ryck, T.; Sadabad, M.S.; Pinheiro, I.; Possemiers, S.; Abbeele, P.V.D.; Derycke, L.; Bracke, M.; Pieters, J.; et al. The HMI module: A new tool to study the Host-Microbiota Interaction in the human gastrointestinal tract in vitro. BMC Microbiol. 2014, 14, 133. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Fritz, J.V.; Glaab, E.; Desai, M.S.; Greenhalgh, K.; Frachet, A.; Niegowska, M.; Estes, M.; Jäger, C.; Seguin-Devaux, C.; et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat. Commun. 2016, 7, 11535. [Google Scholar] [CrossRef]
- Shin, W.; Wu, A.; Massidda, M.W.; Foster, C.; Thomas, N.; Lee, D.-W.; Koh, H.; Ju, Y.; Kim, J.; Kim, H.J. A Robust Longitudinal Co-culture of Obligate Anaerobic Gut Microbiome With Human Intestinal Epithelium in an Anoxic-Oxic Interface-on-a-Chip. Front. Bioeng. Biotechnol. 2019, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Fofanova, T.Y.; Karandikar, U.C.; Auchtung, J.M.; Wilson, R.L.; Valentin, A.J.; Britton, R.A.; Grande-Allen, K.J.; Estes, M.K.; Hoffman, K.; Ramani, S.; et al. A novel system to culture human intestinal organoids under physiological oxygen content to study microbial-host interaction. PLoS One 2024, 19, e0300666. [Google Scholar] [CrossRef]
- Singh, U.P.; Singh, N.P.; Murphy, E.A.; Price, R.L.; Fayad, R.; Nagarkatti, M.; Nagarkatti, P.S. Chemokine and cytokine levels in inflammatory bowel disease patients. Cytokine 2016, 77, 44–49. [Google Scholar] [CrossRef]
- Reinecker, H.-C.; Loh, E.Y.; Ringler, D.J.; Mehta, A.; Rombeau, J.L.; MacDermott, R.P. Monocyte-chemoattractant protein 1 gene expression in intestinal epithelial cells and inflammatory bowel disease mucosa. Gastroenterology 1995, 108, 40–50. [Google Scholar] [CrossRef]
- MacDermott, R.P.; Sanderson, I.R.; Reinecker, H.C. The central role of chemokines (chemotactic cytokines) in the immunopathogenesis of ulcerative colitis and Crohn’s disease. Inflamm. Bowel Dis. 1998, 4, 54–67. [Google Scholar] [CrossRef] [PubMed]
- Banks, C.; Bateman, A.; Payne, R.; Johnson, P.; Sheron, N. Chemokine expression in IBD. Mucosal chemokine expression is unselectively increased in both ulcerative colitis and Crohn’s disease. J. Pathol. 2003, 199, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Meitei, H.T.; Jadhav, N.; Lal, G. CCR6-CCL20 axis as a therapeutic target for autoimmune diseases. Autoimmun. Rev. 2021, 20, 102846. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.Y.S.; Eri, R.; Lyons, A.B.; Grimm, M.C.; Korner, H. CC Chemokine Ligand 20 and Its Cognate Receptor CCR6 in Mucosal T Cell Immunology and Inflammatory Bowel Disease: Odd Couple or Axis of Evil? Front. Immunol. 2013, 4, 194. [Google Scholar] [CrossRef]
- Ivanov, A.I.; Parkos, C.A.; Nusrat, A. Cytoskeletal Regulation of Epithelial Barrier Function During Inflammation. Am. J. Pathol. 2010, 177, 512–524. [Google Scholar] [CrossRef]
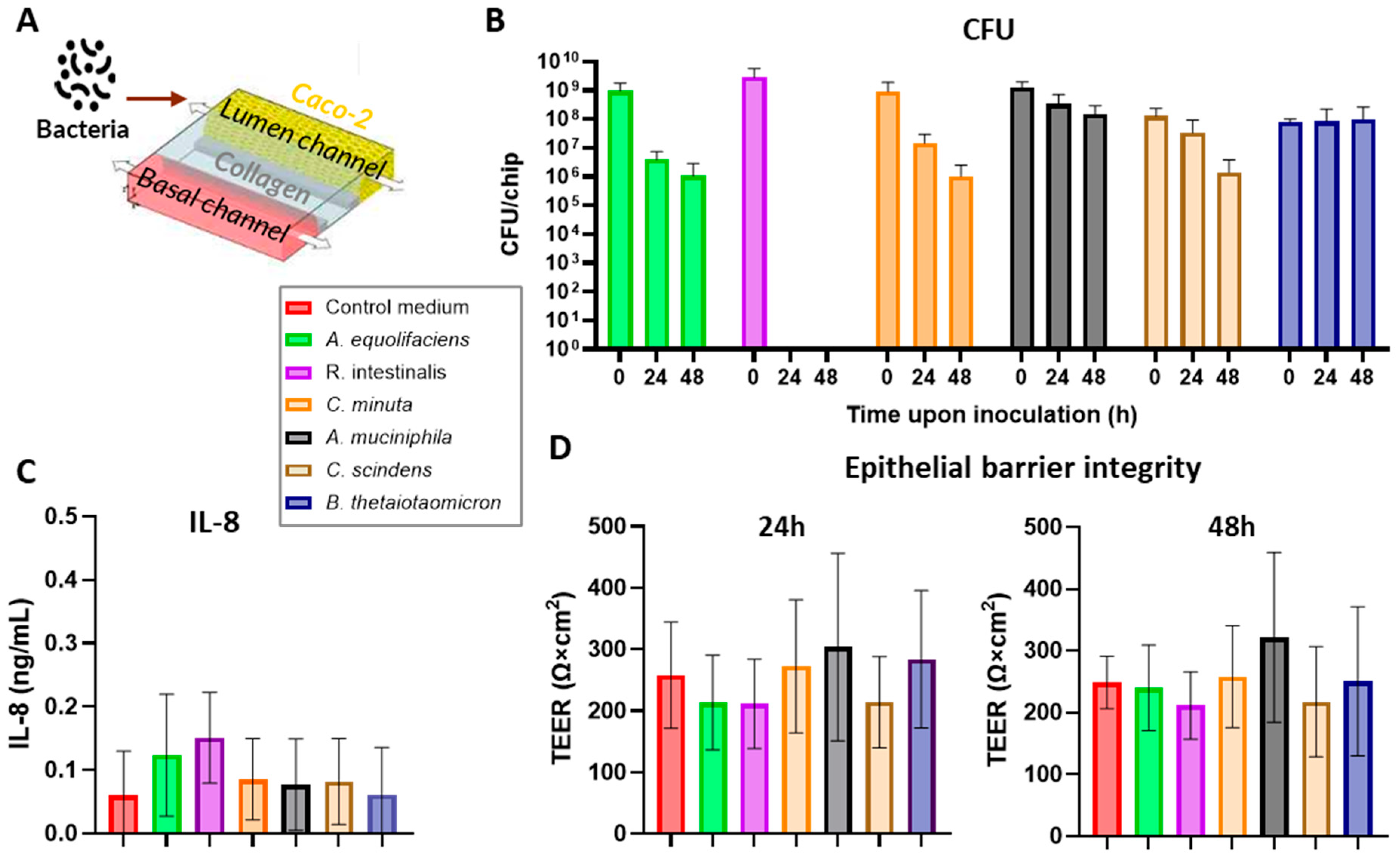
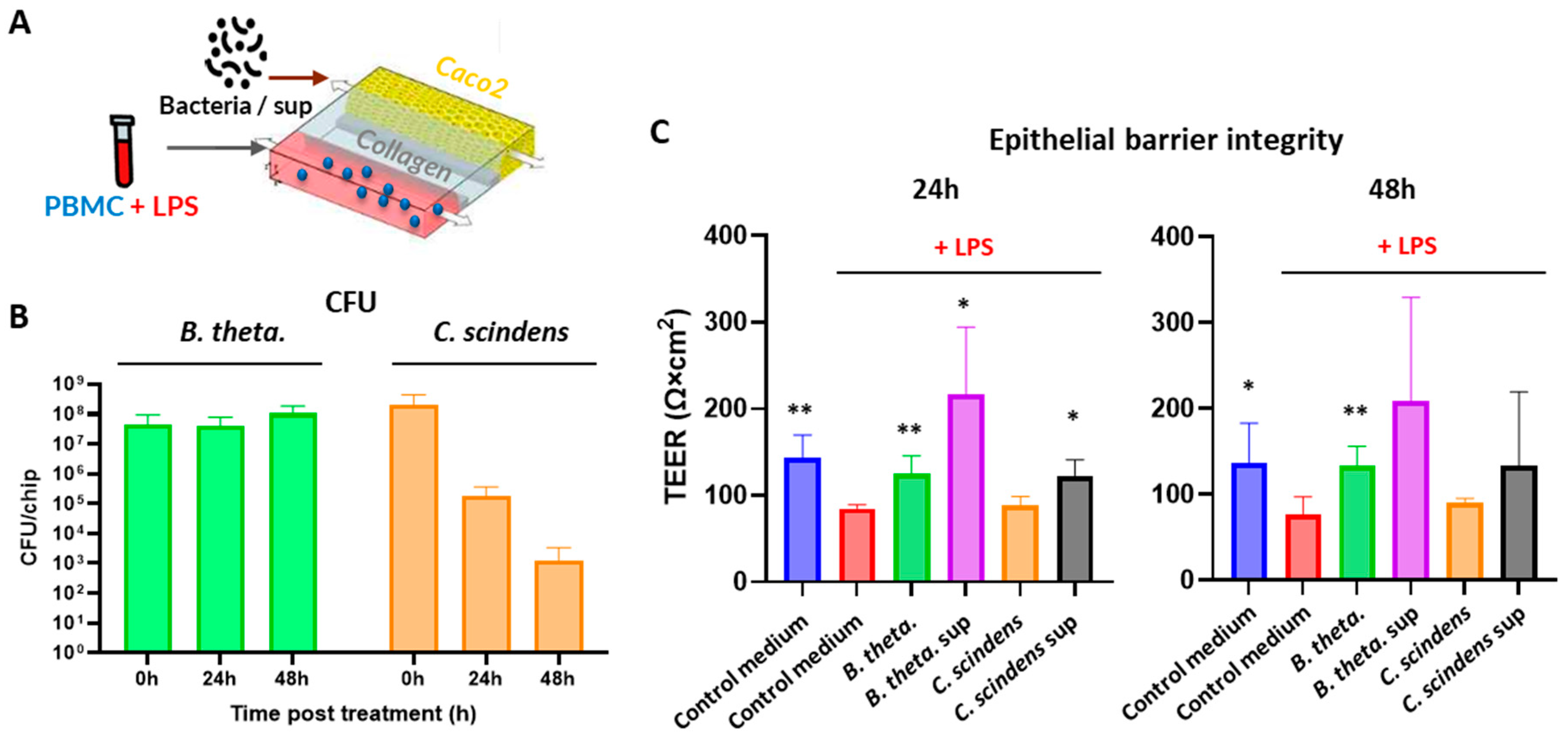
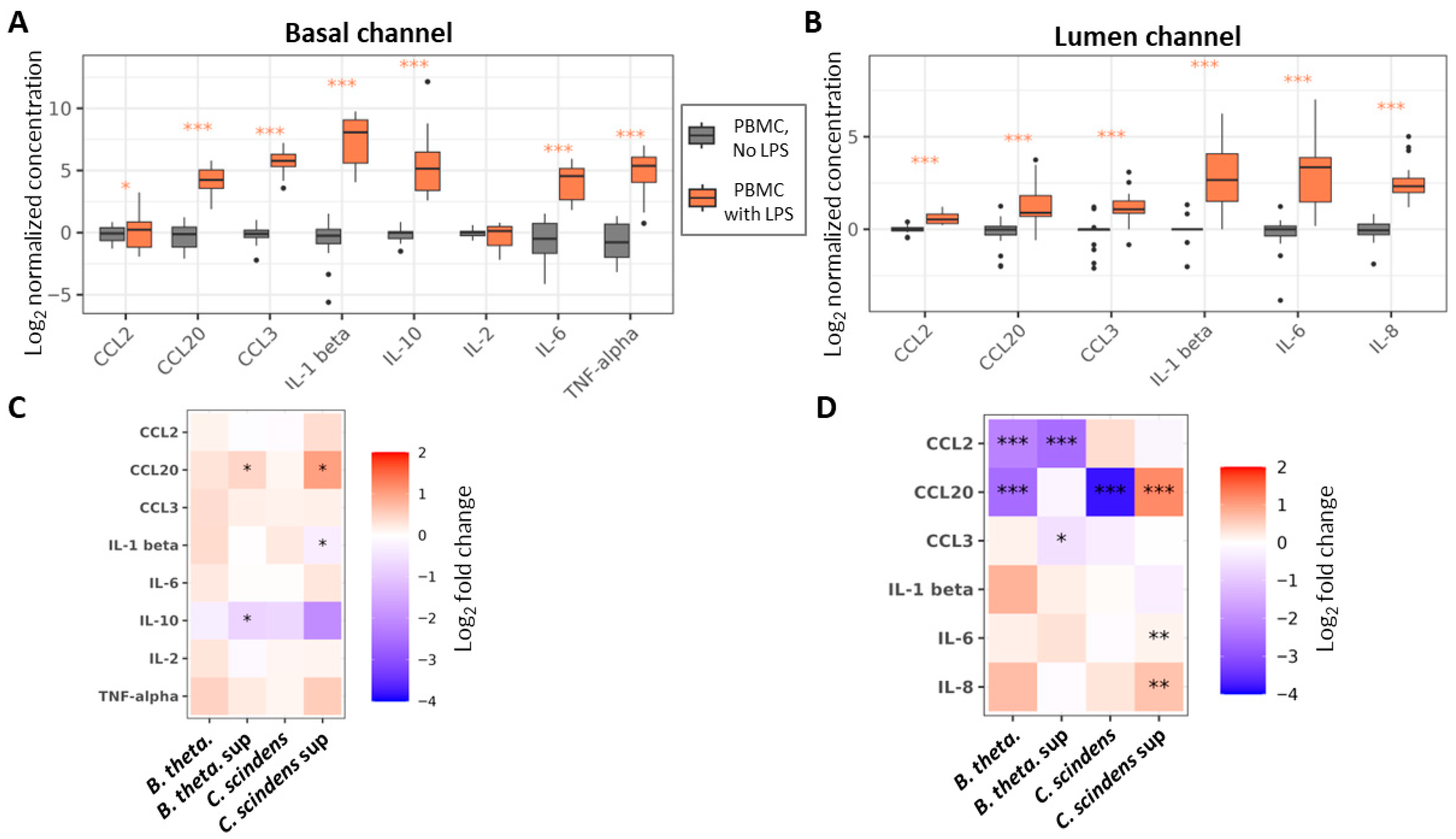
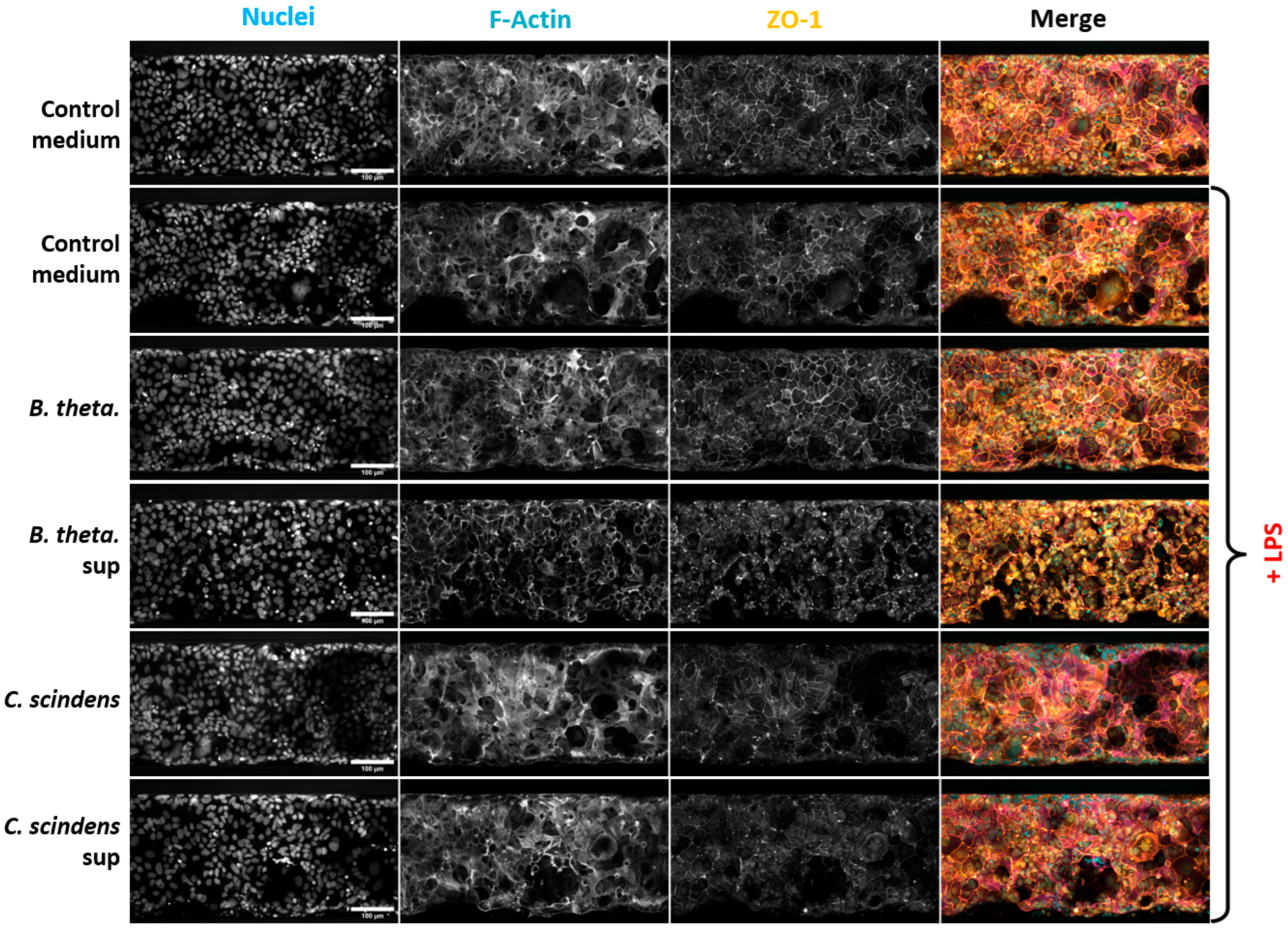
| Species | Strain |
|---|---|
| Bacteroides thetaiotaomicron | DSM 72079 |
| Akkermansia muciniphila | DSM 22959 |
| Christensenella minuta | DSM 22607 |
| Roseburia intestinalis | DSM 14610 |
| Clostridium scindens | DSM 5676 |
| Adlercreutzia equolifaciens | DSM 19450 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Canourgues, N.; Adicéam, E.; Beitz, B.; Atwell, S.; Roelens, M.; Rekiki, A.; Vedrine, C.; Belotserkovsky, I. Immunocompetent High-Throughput Gut-on-Chip Model for Intestinal Microbes—Host Interaction Studies. Appl. Microbiol. 2025, 5, 117. https://doi.org/10.3390/applmicrobiol5040117
Canourgues N, Adicéam E, Beitz B, Atwell S, Roelens M, Rekiki A, Vedrine C, Belotserkovsky I. Immunocompetent High-Throughput Gut-on-Chip Model for Intestinal Microbes—Host Interaction Studies. Applied Microbiology. 2025; 5(4):117. https://doi.org/10.3390/applmicrobiol5040117
Chicago/Turabian StyleCanourgues, Naomi, Emilie Adicéam, Benoît Beitz, Scott Atwell, Maroussia Roelens, Abdessalem Rekiki, Christophe Vedrine, and Ilia Belotserkovsky. 2025. "Immunocompetent High-Throughput Gut-on-Chip Model for Intestinal Microbes—Host Interaction Studies" Applied Microbiology 5, no. 4: 117. https://doi.org/10.3390/applmicrobiol5040117
APA StyleCanourgues, N., Adicéam, E., Beitz, B., Atwell, S., Roelens, M., Rekiki, A., Vedrine, C., & Belotserkovsky, I. (2025). Immunocompetent High-Throughput Gut-on-Chip Model for Intestinal Microbes—Host Interaction Studies. Applied Microbiology, 5(4), 117. https://doi.org/10.3390/applmicrobiol5040117






