Antimicrobial Compounds from Anaerobic Microorganisms: A Review of an Untapped Reservoir
Abstract
1. Introduction
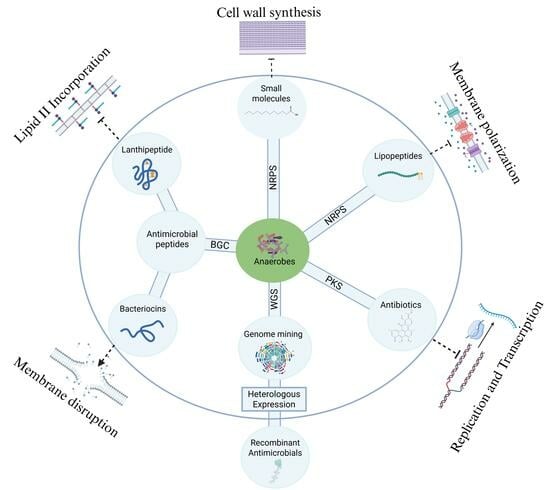
2. Tools and Technologies for Unlocking Antimicrobial Potential in Anaerobes
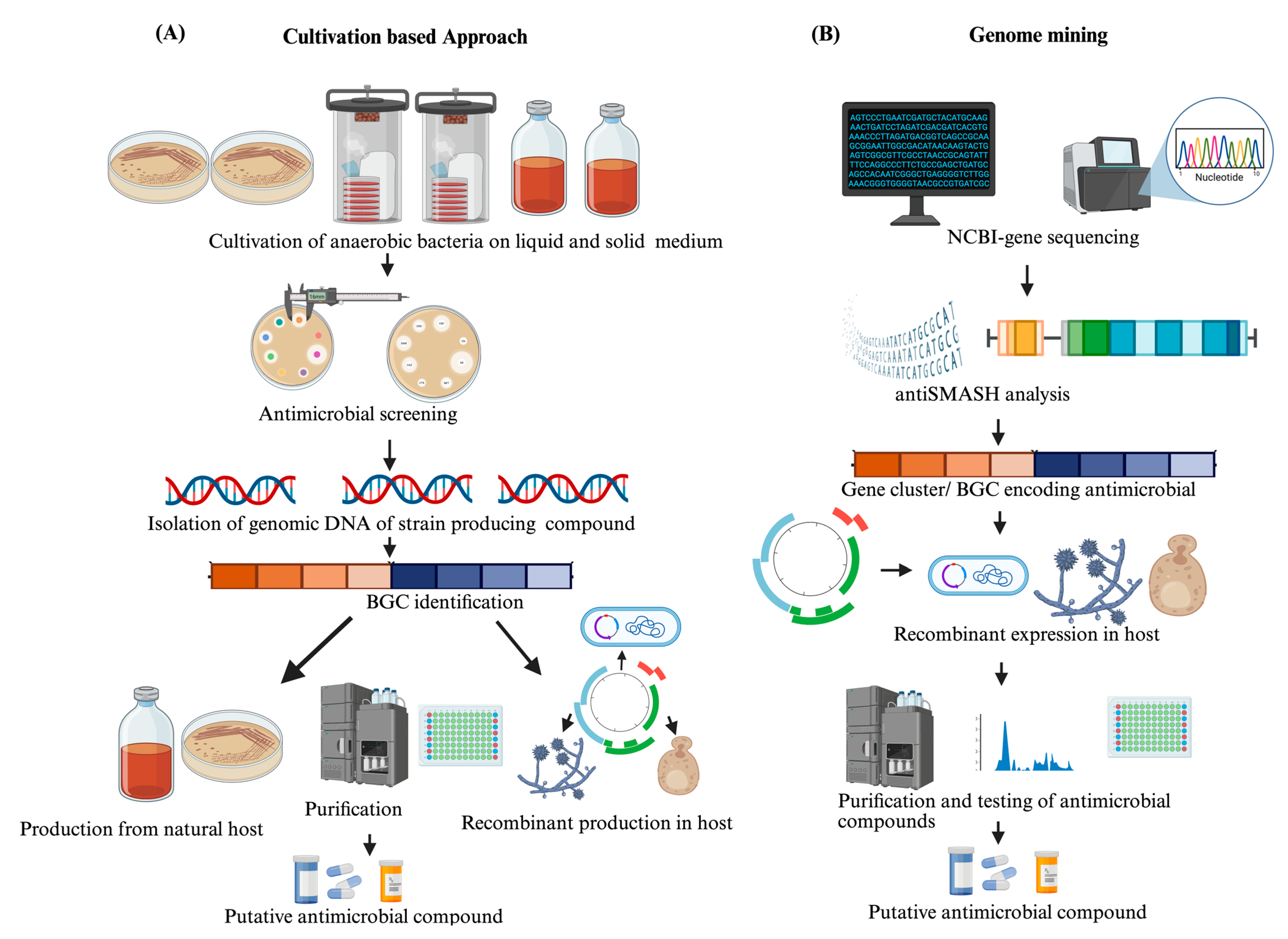
2.1. Cultivation Strategies for Anaerobic Bacteria
2.2. Genome Mining and Bioinformatic Tools
2.3. Heterologous Expression of Anaerobic BGCs
3. Antibiotics from Anaerobes
| S.No. | Compound | Producer Strain | BGC Features (Genes/Size/Type) | Genomic Organization Summary | Classification | MIBiG Accession | Reference |
|---|---|---|---|---|---|---|---|
| 1 | Closthioamide | R. cellulolyticum DSM 5812 | Multiple genes including ctaA–ctaK, noncanonical thiotemplated assembly line (~20 kb) | Modular genes for starter unit biosynthesis, sulfur incorporation, dimerization, and thioamidation; Rc-Sfp PPTase for PCP activation | Nonribosomal peptide-like thiotemplated metabolite | BGC0001891 | [28] |
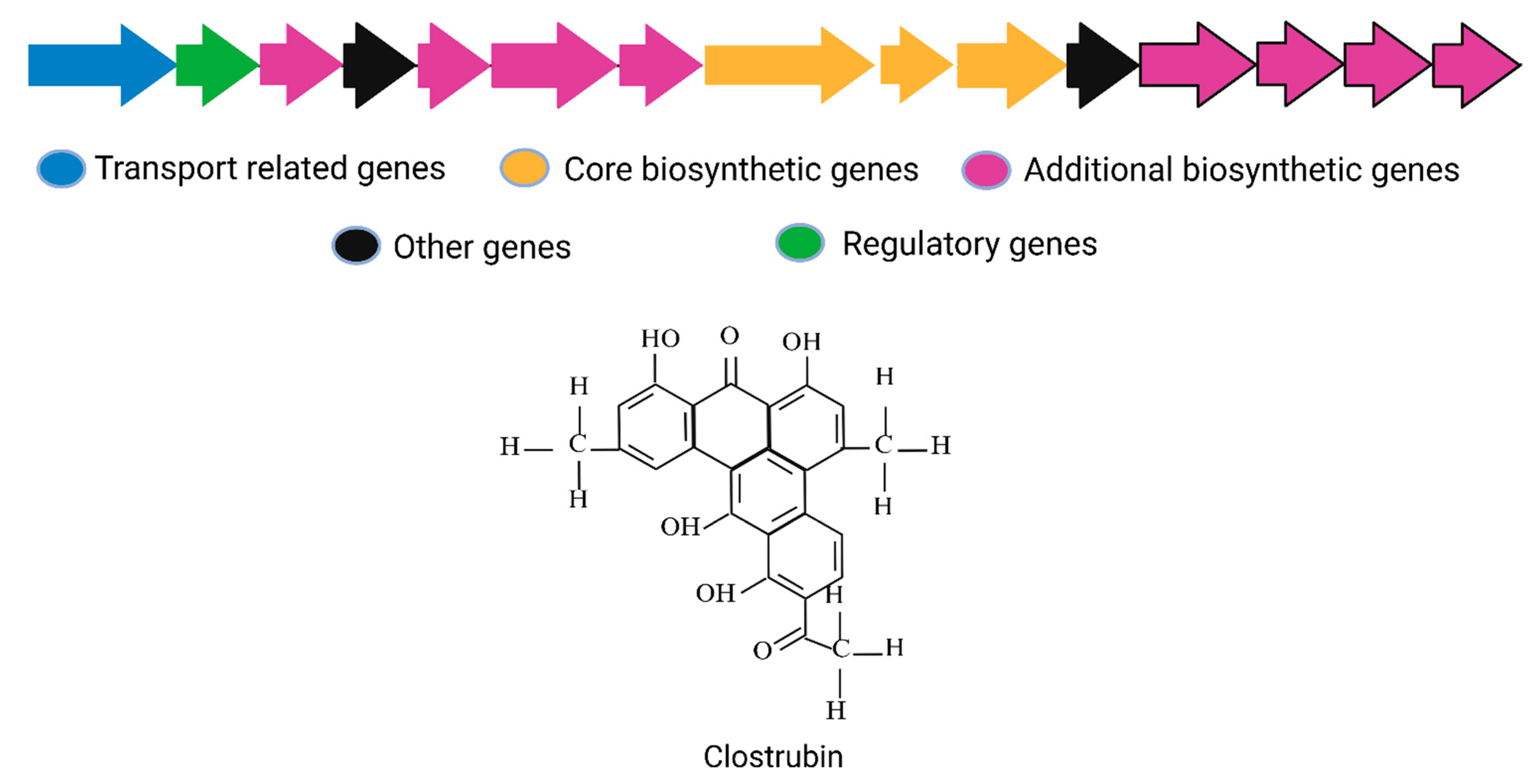
| 2 | Clostrubin | C. beijerinckii HKI0724 | Type II PKS; ~40 kb aromatic polyketide cluster | Multi-module type-II PKS genes arranged with cyclases/oxidases for pentacyclic core formation | Polyketide | NA | [29] |
| 3 | Diffocins | C. difficile CD4, CD16 | Phage tail-like BGC (~20 kb); ORFs CD1359–1376 including structural and receptor-binding proteins | Encodes R-type bacteriocin machinery; regulated by SOS response; ORF1374 determines target specificity | Phage tail-like bacteriocin (R-type) | NA | [30] |
| 4 | Circularin A | C. beijerinckii ATCC 25752 | cirABCDE core genes; includes membrane (cirB, cirC), ATPase (cirD), and immunity (cirE) | Compact gene cluster with overlapping ORFs; circular bacteriocin via head-to-tail peptide bond | Head-to-tail cyclic peptide (Class V bacteriocin) | BGC0000488 | [31] |
| 5 | Barnesin A | S. barnesii SES-3 | Hybrid NRPS–PKS cluster (~20–30 kb); includes tailoring and regulatory enzymes | Modular gene layout with NRPS and PKS modules in tandem; expression validated by transcriptomics | Hybrid NRPS–PKS lipopeptide | BGC0001524 | [11] |
| 6 | Ruminococcin C | R. gnavus E1 | RiPP/sactipeptide-type BGC (~12.8 kb); includes 13 genes organized in 3 operons | Includes three structural genes for RumA, modification enzyme (RumM), and transporter (RumT); expression regulated by trypsin-dependent signaling | Type AII sactipeptide | BGC0002043 | [32] |
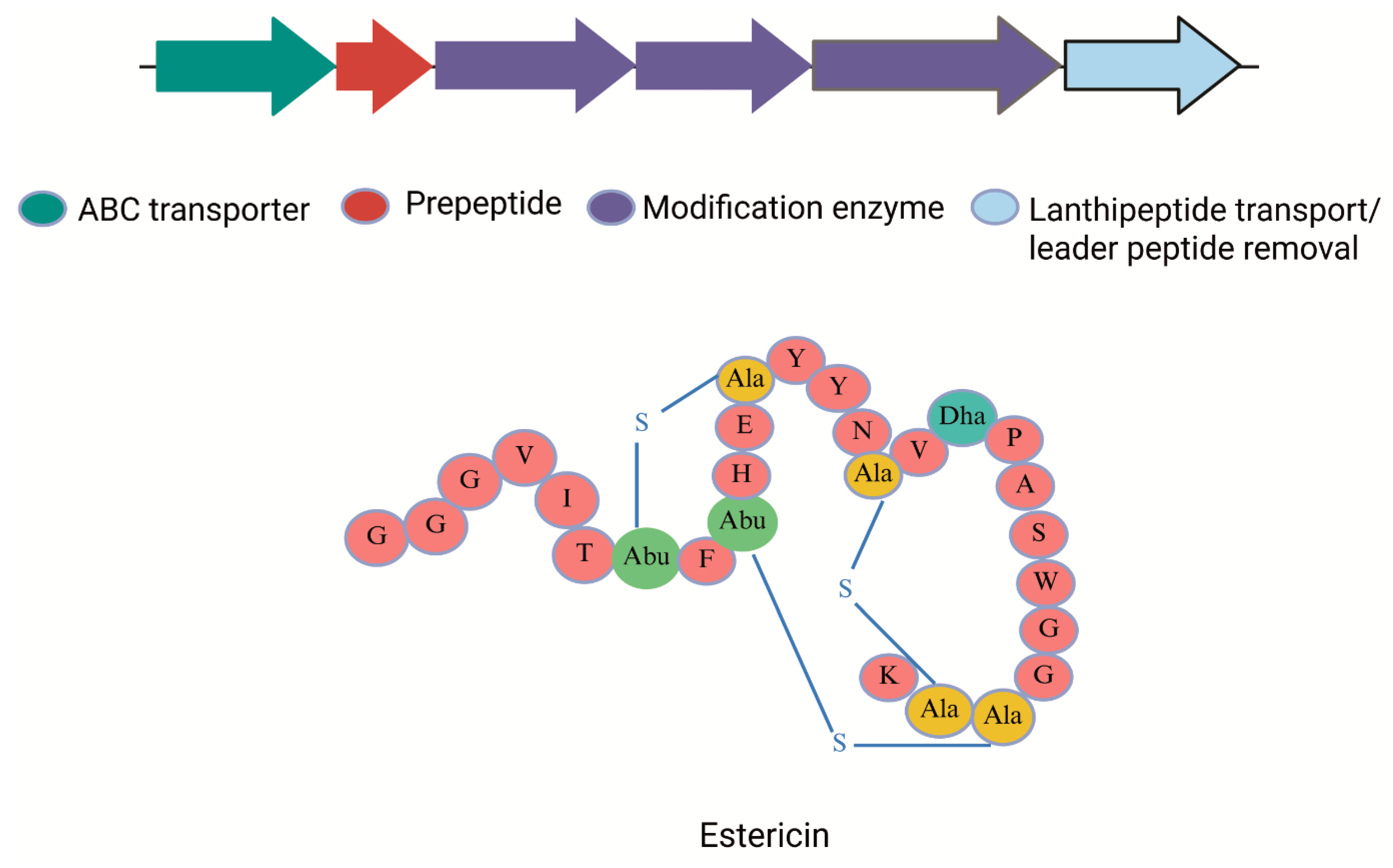
| 7 | Estericin A | C. estertheticum CF016 | RiPP cluster (~25 genes); includes radical SAM enzyme, protease, transporter | Lantibiotic-like BBGC7 cluster on ~180 kb plasmid; high stability against heat and enzymes | Class II lanthipeptide | BGC0002667 | [33] |
3.1. Closthioamide
3.2. Clostrubin
3.3. Clostrindolin
3.4. Naphthalecin
4. Bacteriocins or AMPs from Anaerobes
4.1. Bacteriocins or AMPs from Clostridium Species
4.1.1. Bacteriocin from Clostridium acetobutylicum
4.1.2. Bacteriocin N5
4.1.3. Boticin B and P
4.1.4. CBP22
4.1.5. Closticin 574 and Circularin A
4.1.6. Clostocin A, B, C, D and E
4.1.7. Clostrocyloin
4.1.8. Diffocins
4.1.9. Intestinalin (P30)
4.1.10. Perfrin
4.1.11. Clostocin O
4.2. Bacteriocins or AMPs from Other Anaerobic Genera
4.2.1. Bifidin I
4.2.2. Bifidococcin_664
4.2.3. Nigrescin
4.2.4. Propionicin T1
5. Thiopeptides and Related RiPPs from Anaerobes
5.1. Clostrisin and Cellulosin
5.2. Estericin A
5.3. Pseudocin 196
5.4. Ruminococcin
6. Glycopeptides from Anaerobes
6.1. Clostridocins
6.2. Bacteriocin 28
7. Lipopeptides from Anaerobes
7.1. Barnesin A
7.2. Neutral Lipopeptide
8. Translational Challenges in Clinical Development of Anaerobic-Derived Antimicrobials
9. Conclusions and Future Research Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mast, Y.; Stegmann, E. Actinomycetes: The Antibiotics Producers. Antibiotics 2019, 8, 105. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Zhang, L.; Cao, Y.; Dai, Z.; Ge, X.; Cai, R.; Wang, R.; Hu, Z. Antibiotic Resistance Characterization, Virulence Factors and Molecular Characteristics of Bacillus Species Isolated from Probiotic Preparations in China. J. Glob. Antimicrob. Resist. 2025, 43, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Soltani, S.; Hammami, R.; Cotter, P.D.; Rebuffat, S.; Said, L.B.; Gaudreau, H.; Bédard, F.; Biron, E.; Drider, D.; Fliss, I. Bacteriocins as a New Generation of Antimicrobials: Toxicity Aspects and Regulations. FEMS Microbiol. Rev. 2021, 45, fuaa039. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.S.; Sharma, D.; Baindara, P.; Choksket, S.; Harshvardhan; Mandal, S.M.; Grover, V.; Korpole, S. Characterization and Antimicrobial Studies of Iturin-like and Bogorol-like Lipopeptides from Brevibacillus spp. Strains GI9 and SKDU10. Front. Microbiol. 2021, 12, 729026. [Google Scholar] [CrossRef]
- Singh, S.S.; Sharma, D.; Singh, C.; Kumar, S.; Singh, P.; Sharma, A.; Das, D.K.; Pinnaka, A.K.; Thakur, K.G.; Ringe, R.P.; et al. Brevicillin, a Novel Lanthipeptide from the Genus Brevibacillus with Antimicrobial, Antifungal, and Antiviral Activity. J. Appl. Microbiol. 2023, 134, lxad054. [Google Scholar] [CrossRef]
- Ibrahim, S.A.; Ayivi, R.D.; Zimmerman, T.; Siddiqui, S.A.; Altemimi, A.B.; Fidan, H.; Esatbeyoglu, T.; Bakhshayesh, R.V. Lactic Acid Bacteria as Antimicrobial Agents: Food Safety and Microbial Food Spoilage Prevention. Foods 2021, 10, 3131. [Google Scholar] [CrossRef]
- Singh, S.S.; Akhtar, M.N.; Sharma, D.; Mandal, S.M.; Korpole, S. Characterization of Iturin V, a Novel Antimicrobial Lipopeptide from a Potential Probiotic Strain Lactobacillus sp. M31. Probiotics Antimicrob. Proteins 2021, 13, 1766–1779. [Google Scholar] [CrossRef]
- Butkovich, L.V.; Vining, O.B.; O’Malley, M.A. New Approaches to Secondary Metabolite Discovery from Anaerobic Gut Microbes. Appl. Microbiol. Biotechnol. 2025, 109, 12. [Google Scholar] [CrossRef]
- Bhattacharya, D.; Nanda, P.K.; Pateiro, M.; Lorenzo, J.M.; Dhar, P.; Das, A.K. Lactic Acid Bacteria and Bacteriocins: Novel Biotechnological Approach for Biopreservation of Meat and Meat Products. Microorganisms 2022, 10, 2058. [Google Scholar] [CrossRef]
- Seedorf, H.; Fricke, W.F.; Veith, B.; Brüggemann, H.; Liesegang, H.; Strittmatter, A.; Miethke, M.; Buckel, W.; Hinderberger, J.; Li, F.; et al. The Genome of Clostridium kluyveri, a Strict Anaerobe with Unique Metabolic Features. Proc. Natl. Acad. Sci. USA 2008, 105, 2128–2133. [Google Scholar] [CrossRef]
- Rischer, M.; Raguž, L.; Guo, H.; Beemelmanns, C. Biosynthesis, Synthesis, and Activities of Barnesin A, a NRPS–PKS Hybrid Produced by an Anaerobic Epsilonproteobacterium. ACS Chem. Biol. 2018, 13, 1990–1995. [Google Scholar] [CrossRef] [PubMed]
- Hanišáková, N.; Vítězová, M.; Rittmann, S.K.R. The Historical Development of Cultivation Techniques for Methanogens and Other Strict Anaerobes and Their Application in Modern Microbiology. Microorganisms 2022, 10, 412. [Google Scholar] [CrossRef] [PubMed]
- Lawson, P.A.; Tanner, R.S. Cultivation of Anaerobic Bacteria: Foundations and Principles. Anaerobe 2025, 93, 102951. [Google Scholar] [CrossRef] [PubMed]
- Litti, Y.; Zhuravleva, E.; Kovalev, A. Anaerobic Fermentation and High-Value Bioproducts: A Brief Overview of Recent Progress and Current Challenges. Fermentation 2024, 10, 537. [Google Scholar] [CrossRef]
- Li, J.S.; Barber, C.C.; Zhang, W. Natural products from anaerobes. J. Ind. Microbiol. Biotechnol. 2019, 46, 375–383. [Google Scholar] [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid Identification, Annotation and Analysis of Secondary Metabolite Biosynthesis Gene Clusters in Bacterial and Fungal Genome Sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef]
- Hart, T.; Moffat, J. BAGEL: A Computational Framework for Identifying Essential Genes from Pooled Library Screens. BMC Bioinform. 2016, 17, 164. [Google Scholar] [CrossRef]
- Zdouc, M.M.; Blin, K.; Louwen, N.L.L.; Navarro, J.; Loureiro, C.; Bader, C.D.; Bailey, C.B.; Barra, L.; Booth, T.J.; Bozhüyük, K.A.J.; et al. MIBiG 4.0: Advancing Biosynthetic Gene Cluster Curation Through Global Collaboration. Nucleic Acids Res. 2025, 53, D678–D690. [Google Scholar] [CrossRef]
- Skinnider, M.A.; Dejong, C.A.; Rees, P.N.; Johnston, C.W.; Li, H.; Webster, A.L.; Wyatt, M.A.; Magarvey, N.A. Genomes to Natural Products Prediction Informatics for Secondary Metabolomes (PRISM). Nucleic Acids Res. 2015, 43, 9645–9662. [Google Scholar] [CrossRef]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMPR3: A Database on Sequences, Structures and Signatures of Antimicrobial Peptides. Nucleic Acids Res. 2016, 44, D1094–D1097. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef]
- Singh, S.; Chaudhary, K.; Dhanda, S.K.; Bhalla, S.; Usmani, S.S.; Gautam, A.; Tuknait, A.; Agrawal, P.; Mathur, D.; Raghava, G.P.S. SATPdb: A Database of Structurally Annotated Therapeutic Peptides. Nucleic Acids Res. 2016, 44, D1119–D1126. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yang, Y.; Wang, S.; Yang, X.; Zhou, K.; Xu, C.; Zhang, X.; Fan, J.; Hou, D.; Li, X.; et al. NPASS Database Update 2023: Quantitative Natural Product Activity and Species Source Database for Biomedical Research. Nucleic Acids Res. 2023, 51, D621–D628. [Google Scholar] [CrossRef] [PubMed]
- Poynton, E.F.; van Santen, J.A.; Pin, M.; Contreras, M.M.; McMann, E.; Parra, J.; Showalter, B.; Zaroubi, L.; Duncan, K.R.; Linington, R.G. The Natural Products Atlas 3.0: Extending the Database of Microbially Derived Natural Products. Nucleic Acids Res. 2025, 53, D691–D699. [Google Scholar] [CrossRef] [PubMed]
- Hao, T.; Xie, Z.; Wang, M.; Liu, L.; Zhang, Y.; Wang, W.; Zhang, Z.; Zhao, X.; Li, P.; Guo, Z.; et al. An Anaerobic Bacterium Host System for Heterologous Expression of Natural Product Biosynthetic Gene Clusters. Nat. Commun. 2019, 10, 3665. [Google Scholar] [CrossRef]
- Olano, C.; Méndez, C.; Salas, J.A. Molecular Insights on the Biosynthesis of Antitumour Compounds by Actinomycetes. Microb. Biotechnol. 2011, 4, 144–164. [Google Scholar] [CrossRef]
- Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B.; et al. Towards the Sustainable Discovery and Development of New Antibiotics. Nat. Rev. Chem. 2021, 5, 726–749. [Google Scholar] [CrossRef]
- Dunbar, K.L.; Büttner, H.; Molloy, E.M.; Dell, M.; Kumpfmüller, J.; Hertweck, C. Genome Editing Reveals Novel Thiotemplated Assembly of Polythioamide Antibiotics in Anaerobic Bacteria. Angew. Chem. Int. Ed. 2018, 57, 14080–14084. [Google Scholar] [CrossRef]
- Pidot, S.; Ishida, K.; Cyrulies, M.; Hertweck, C. Discovery of Clostrubin, an Exceptional Polyphenolic Polyketide Antibiotic from a Strictly Anaerobic Bacterium. Angew. Chem. Int. Ed. 2014, 53, 7856. [Google Scholar] [CrossRef]
- Gebhart, D.; Williams, S.R.; Bishop-Lilly, K.A.; Govoni, G.R.; Willner, K.M.; Butani, A.; Sozhamannan, S.; Martin, D.; Fortier, L.C.; Scholl, D. Novel High-Molecular-Weight, R-Type Bacteriocins of Clostridium difficile. J. Bacteriol. 2012, 194, 6240–6247. [Google Scholar] [CrossRef]
- Kemperman, R.; Kuipers, A.; Karsens, H.; Nauta, A.; Kuipers, O.P.; Kok, J. Identification and Characterization of Two Novel Clostridial Bacteriocins, Circularin A and Closticin 574. Appl. Environ. Microbiol. 2003, 69, 1589–1597. [Google Scholar] [CrossRef]
- Gomez, A.; Ladiré, M.; Marcille, F.; Fons, M. Trypsin Mediates Growth Phase-Dependent Transcriptional Regulation of Genes Involved in Biosynthesis of Ruminococcin A, a Lantibiotic Produced by a Ruminococcus gnavus Strain from a Human Intestinal Microbiota. J. Bacteriol. 2002, 184, 18–28. [Google Scholar] [CrossRef]
- Wambui, J.; Stevens, M.J.A.; Sieber, S.; Cernela, N.; Perreten, V.; Stephan, R. Targeted Genome Mining Reveals the Psychrophilic Clostridium estertheticum Complex as a Potential Source for Novel Bacteriocins, Including Cesin A and Estercticin A. Front. Microbiol. 2022, 12, 801467. [Google Scholar] [CrossRef] [PubMed]
- Lincke, T.; Behnken, S.; Ishida, K.; Roth, M.; Hertweck, C. Closthioamide: An Unprecedented Polythioamide Antibiotic from the Strictly Anaerobic Bacterium Clostridium cellulolyticum. Angew. Chem. Int. Ed. 2010, 49, 2011–2013. [Google Scholar] [CrossRef] [PubMed]
- Dunbar, K.L.; Dell, M.; Molloy, E.M.; Büttner, H.; Kumpfmüller, J.; Hertweck, C. An Unexpected Split-Merge Pathway in the Assembly of the Symmetric Nonribosomal Peptide Antibiotic Closthioamide. Angew. Chem. Int. Ed. 2021, 60, 4104–4109. [Google Scholar] [CrossRef] [PubMed]
- Miari, V.F.; Solanki, P.; Hleba, Y.; Stabler, R.A.; Heap, J.T. In Vitro Susceptibility to Closthioamide among Clinical and Reference Strains of Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 2017, 61, e00929-17. [Google Scholar] [CrossRef]
- Yang, M.; Li, J.; Li, A. Total Synthesis of Clostrubin. Nat. Commun. 2015, 6, 6445. [Google Scholar] [CrossRef]
- Schieferdecker, S.; Shabuer, G.; Knuepfer, U.; Hertweck, C. Clostrindolin is an Antimycobacterial Pyrone Alkaloid from Clostridium beijerinckii. Org. Biomol. Chem. 2019, 17, 6119–6121. [Google Scholar] [CrossRef]
- Ezaki, M.; Muramatsu, H.; Takase, S.; Hashimoto, M.; Nagai, K. Naphthalecin, a Novel Antibiotic Produced by the Anaerobic Bacterium, Sporotalea colonica sp. nov. J. Antibiot. 2008, 61, 207–212. [Google Scholar] [CrossRef]
- Mihaylova-Garnizova, R.; Davidova, S.; Hodzhev, Y.; Satchanska, G. Antimicrobial Peptides Derived from Bacteria: Classification, Sources, and Mechanism of Action Against Multidrug-Resistant Bacteria. Int. J. Mol. Sci. 2024, 25, 10788. [Google Scholar] [CrossRef]
- Xuan, J.; Feng, W.; Wang, J.; Wang, R.; Zhang, B.; Bo, L.; Chen, Z.S.; Yang, H.; Sun, L. Antimicrobial Peptides for Combating Drug-Resistant Bacterial Infections. Drug Resist. Updates 2023, 68, 100954. [Google Scholar] [CrossRef]
- Singh, B.; Kumar, N.; Yadav, A.; Rohan; Bhandari, K. Harnessing the Power of Bacteriocins: A Comprehensive Review on Sources, Mechanisms, and Applications in Food Preservation and Safety. Curr. Microbiol. 2025, 82, 174. [Google Scholar] [CrossRef] [PubMed]
- Barber, J.M.; Robb, F.T.; Webster, J.R.; Woods, D.R. Bacteriocin Production by Clostridium acetobutylicum in an Industrial Fermentation Process. Appl. Environ. Microbiol. 1979, 37, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Li, A.W.; Verpoorte, J.A.; Lewis, R.G.; Mahony, D.E. Characterization of Bacteriocin 28 Produced by Clostridium perfringens. Can. J. Microbiol. 1982, 28, 860–873. [Google Scholar] [CrossRef] [PubMed]
- Ionesco, H.; Wolff, A. The Mode of Action of Bacteriocin N5 Purified from Clostridium perfringens. C. R. Acad. Hebd. Seances Acad. Sci. D 1975, 126, 343–356. [Google Scholar]
- Dineen, S.S.; Bradshaw, M.; Johnson, E.A. Cloning, Nucleotide Sequence, and Expression of the Gene Encoding the Bacteriocin Boticin B from Clostridium botulinum Strain 213B. Appl. Environ. Microbiol. 2000, 66, 5480–5483. [Google Scholar] [CrossRef]
- Lau, A.H.S.; Hawirko, R.Z.; Chow, C.T. Purification and Properties of Boticin P Produced by Clostridium botulinum. Can. J. Microbiol. 1974, 20, 385–390. [Google Scholar] [CrossRef]
- Wang, T.; Fu, J.; Xiao, X.; Wang, F.; Jin, M.; Wang, Y.; Zong, X. CBP22, a Novel Bacteriocin Isolated from Clostridium butyricum ZJU-F1, Protects Against LPS-Induced Intestinal Injury Through Maintaining the Tight Junction Complex. Mediat. Inflamm. 2021, 2021, 8032125. [Google Scholar] [CrossRef]
- Clarke, D.J.; Robson, R.M.; Morris, J.G. Purification of Two Clostridium Bacteriocins by Procedures Appropriate to Hydrophobic Proteins. Antimicrob. Agents Chemother. 1975, 7, 256–264. [Google Scholar] [CrossRef]
- Alejo Hernandez, M.A.; Villavicencio Sánchez, K.P.; Sánchez Morales, R.; Hernández-Magro Gil, K.G.; Moreno-Gutiérrez, D.S.; Sanchez-Rueda, E.G.; Teresa-Cruz, Y.; Choi, B.; Hernández García, A.; Romero-Rodríguez, A.; et al. Discovery of Antimicrobial Peptides Clostrisin and Cellulosin from Clostridium: Insights into Their Structures, Co-localized Biosynthetic Gene Clusters, and Antibiotic Activity. Beilstein J. Org. Chem. 2024, 20, 1800–1816. [Google Scholar] [CrossRef]
- Hongo, M.; Murata, A.; Ogata, S.; Kono, K.; Kato, F. Characterization of a Temperate Phage and Four Bacteriocins Produced by Nonpathogenic Clostridium Species. Agric. Biol. Chem. 1968, 32, 773–780. [Google Scholar] [CrossRef]
- Ogata, S.; Kato, F.; Hongo, M. Bacteriocins of Nonpathogenic Clostridium Species. IV. Study on Adsorption of Inducible Bacteriocin Clostocin O Toward Sensitive Bacteria. Agric. Biol. Chem. 1976, 40, 1093–1099. [Google Scholar] [CrossRef]
- Rizzacasa, M.; Ricca, M. Chemistry and Biology of Acyloin Natural Products. Synthesis 2023, 55, 2273–2284. [Google Scholar] [CrossRef]
- Wang, C.; Wambui, J.; Fernandez-Cantos, M.V.; Jurt, S.; Broos, J.; Stephan, R.; Kuipers, O.P. Heterologous Expression and Characterization of Estercin A, a Class II Lanthipeptide Derived from Clostridium estertheticum CF016, with Antimicrobial Activity against Clinically Relevant Pathogens. J. Nat. Prod. 2025, 88, 262–273. [Google Scholar] [CrossRef] [PubMed]
- Szadkowska, M.; Olewniczak, M.; Kloska, A.; Jankowska, E.; Kapusta, M.; Rybak, B.; Wyrzykowski, D.; Zmudzinska, W.; Gieldon, A.; Kocot, A.; et al. A Novel Cryptic Clostridial Peptide That Kills Bacteria by a Cell Membrane Permeabilization Mechanism. Microbiol. Spectr. 2022, 10, e01657-22. [Google Scholar] [CrossRef]
- Cooper, D.G.; Zajic, J.E.; Gerson, D.F.; Manninen, K.I. Isolation and Identification of Biosurfactants Produced During Anaerobic Growth of Clostridium pasteurianum. J. Ferment. Technol. 1980, 58, 83–86. [Google Scholar]
- Timbermont, L.; De Smet, L.; Van Nieuwerburgh, F.; Parreira, V.R.; Van Driessche, G.; Haesebrouck, F.; Ducatelle, R.; Prescott, J.; Deforce, D.; Devreese, B.; et al. Perfrin, a Novel Bacteriocin Associated with NetB Positive Clostridium perfringens Strains from Broilers with Necrotic Enteritis. Vet. Res. 2014, 45, 40. [Google Scholar] [CrossRef]
- Quratulain, S.; Qadeer, M.A.; Chaudhry, M.Y.; Kausar, A.R. Development and Characterization of Butanol–Resistant Strain of Clostridium acetobutylicum in Molasses Medium. Folia Microbiol. 1995, 40, 467–471. [Google Scholar] [CrossRef]
- Shabuer, M. Industrial Production of Acetone and Butanol by Fermentation—100 Years Later. FEMS Microbiol. Lett. 2016, 363, fnw134. [Google Scholar] [CrossRef]
- Martínez-Cuesta, M.C.; Kok, J.; Herranz, E.; Peláez, C.; Requena, T.; Buist, G. Requirement of Autolytic Activity for Bacteriocin-Induced Lysis. Appl. Environ. Microbiol. 2000, 66, 3174–3179. [Google Scholar] [CrossRef][Green Version]
- Granum, P.E.; Whitaker, J.R. Improved Method for Purification of Enterotoxin from Clostridium perfringens Type A. Appl. Environ. Microbiol. 1980, 39, 1120–1122. [Google Scholar] [CrossRef]
- Chen, J.S.; Hiu, S.F. Acetone–Butanol–Isopropanol Production by Clostridium beijerinckii (Synonym, Clostridium butylicum). Biotechnol. Lett. 1986, 8, 371–376. [Google Scholar] [CrossRef]
- Adler, B.A.; Kazakov, A.E.; Zhong, C.; Liu, H.; Kutter, E.; Lui, L.M.; Nielsen, T.N.; Carion, H.; Deutschbauer, A.M.; Mutalik, V.K.; et al. The Genetic Basis of Phage Susceptibility, Cross-Resistance and Host-Range in Salmonella. Microbiology 2022, 168, 001126. [Google Scholar] [CrossRef] [PubMed]
- Schieferdecker, S.; Shabuer, G.; Letzel, A.C.; Urbansky, B.; Ishida-Ito, M.; Ishida, K.; Cyrulies, M.; Dahse, H.M.; Pidot, S.; Hertweck, C.B. Biosynthesis of Diverse Antimicrobial and Antiproliferative Acyloins in Anaerobic Bacteria. ACS Chem. Biol. 2019, 14, 1490–1497. [Google Scholar] [CrossRef] [PubMed]
- Schwemmlein, N.; Pippel, J.; Gazdag, E.M.; Blankenfeldt, W. Crystal Structures of R-Type Bacteriocin Sheath and Tube Proteins CD1363 and CD1364 from Clostridium difficile in the Pre-assembled State. Front. Microbiol. 2018, 9, 1750. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.W.; Lillehoj, H.S. Role of Clostridium perfringens Necrotic Enteritis B-like Toxin in Disease Pathogenesis. Vaccines 2022, 10, 61. [Google Scholar] [CrossRef]
- Cheikhyoussef, A.; Chen, H.; Zhao, J.; Tang, J.; Zhang, H.; Chen, W. Bifidin I—A New Bacteriocin Produced by Bifidobacterium infantis BCRC 14602: Purification and Partial Amino Acid Sequence. Food Control 2010, 21, 746–753. [Google Scholar] [CrossRef]
- Javvadi, S.G.; Kujawska, M.; Papp, D.; Gontarczyk, A.M.; Jordan, A.; Lawson, M.A.; O’Neill, I.J.; Alcon-Giner, C.; Kiu, R.; Clarke, P.; et al. A Novel Bacteriocin Produced by Bifidobacterium longum subsp. infantis Has Dual Antimicrobial and Immunomodulatory Activity. bioRxiv 2022. [Google Scholar] [CrossRef]
- Kaewsrichan, J.; Douglas, C.W.; Nissen-Meyer, J.; Fimland, G.; Teanpaisan, R. Characterization of a Bacteriocin Produced by Prevotella nigrescens ATCC 25261. Lett. Appl. Microbiol. 2004, 39, 451–458. [Google Scholar] [CrossRef]
- Faye, T.; Langsrud, T.; Nes, I.F.; Holo, H. Biochemical and Genetic Characterization of Propionicin T1, a New Bacteriocin from Propionibacterium thoenii. Appl. Environ. Microbiol. 2000, 66, 4230–4236. [Google Scholar] [CrossRef]
- Sanchez-Gallardo, R.; O’Connor, P.M.; O’Neill, I.J.; McDonnell, B.; Lee, C.; Moore, R.L.; McAuliffe, F.M.; Cotter, P.D.; van Sinderen, D. Pseudocin 196, a Novel Lantibiotic Produced by Bifidobacterium pseudocatenulatum Elicits Antimicrobial Activity Against Clinically Relevant Pathogens. Gut Microbes 2024, 16, 2387139. [Google Scholar] [CrossRef]
- Dabard, J.; Bridonneau, C.; Phillipe, C.; Anglade, P.; Molle, D.; Nardi, M.; Ladiré, M.; Girardin, H.; Marcille, F.; Gomez, A.; et al. Ruminococcin A, a New Lantibiotic Produced by a Ruminococcus gnavus Strain Isolated from Human Feces. Appl. Environ. Microbiol. 2001, 67, 4111–4118. [Google Scholar] [CrossRef]
- Crost, E.H.; Ajandouz, E.H.; Villard, C.; Geraert, P.A.; Puigserver, A.; Fons, M. Ruminococcin C, a New Anti-Clostridium perfringens Bacteriocin Produced in the Gut by the Commensal Bacterium Ruminococcus gnavus E1. Biochimie 2011, 93, 1487–1494. [Google Scholar] [CrossRef]
- Kim, Y.T.; Kim, C.H.; Kwon, J.G.; Cho, J.H.; Shin, Y.S.; Kim, H.B.; Lee, J.H. In Vivo Trial of Bifidobacterium longum Revealed the Complex Network Correlations Between Gut Microbiota and Health Promotional Effects. Front. Microbiol. 2022, 13, 886934. [Google Scholar] [CrossRef]
- Nieto Lozano, J.C.; Meyer, J.N.; Sletten, K.; Peláez, C.; Nes, I.F. Purification and Amino Acid Sequence of a Bacteriocin Produced by Pediococcus acidilactici. J. Gen. Microbiol. 1992, 138, 1985–1990. [Google Scholar] [CrossRef]
- Drider, D.; Fimland, G.; Héchard, Y.; Mcmullen, L.M.; Prévost, H. The Continuing Story of Class IIa Bacteriocins. Microbiol. Mol. Biol. Rev. 2006, 70, 564–582. [Google Scholar] [CrossRef] [PubMed]
- Dasen, G.; Smutny, J.; Teuber, M.; Meile, L. Classification and Identification of Propionibacteria Based on Ribosomal RNA Genes and PCR. Syst. Appl. Microbiol. 1998, 21, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Karbalaei-Heidari, H.R.; Budisa, N. Combating Antimicrobial Resistance with New-To-Nature Lanthipeptides Created by Genetic Code Expansion. Front. Microbiol. 2020, 11, 590522. [Google Scholar] [CrossRef] [PubMed]
- Field, D.; Fernandez de Ullivarri, M.; Ross, R.P.; Hill, C. After a Century of Nisin Research—Where Are We Now? FEMS Microbiol. Rev. 2023, 47, fuad023. [Google Scholar] [CrossRef]
- Bierbaum, G.; Sahl, H.G. Lantibiotics: Mode of Action, Biosynthesis and Bioengineering. Curr. Pharm. Biotechnol. 2009, 10, 2–18. [Google Scholar] [CrossRef]
- Chakraborty, H.J.; Gangopadhyay, A.; Datta, A. Prediction and Characterisation of Lantibiotic Structures with Molecular Modelling and Molecular Dynamics Simulations. Sci. Rep. 2019, 9, 7169. [Google Scholar] [CrossRef] [PubMed]
- Pujol, A.; Crost, E.H.; Simon, G.; Barbe, V.; Vallenet, D.; Gomez, A.; Fons, M. Characterization and Distribution of the Gene Cluster Encoding RumC, an Anti-Clostridium perfringens Bacteriocin Produced in the Gut. FEMS Microbiol. Ecol. 2011, 78, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Balty, C.; Guillot, A.; Fradale, L.; Brewee, C.; Boulay, M.; Kubiak, X.; Benjdia, A.; Berteau, O. Ruminococcin C, an Anti-Clostridial Sactipeptide Produced by a Prominent Member of the Human Microbiota Ruminococcus gnavus. J. Biol. Chem. 2019, 294, 14512–14525. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.S.; Hansford, K.A.; Blaskovich, M.A.T.; Halai, R.; Cooper, M.A. Glycopeptide Antibiotics: Back to the Future. J. Antibiot. 2014, 67, 631–644. [Google Scholar] [CrossRef]
- Jakaria, S.M.; Budil, D.E.; Murtagh, J. Glycopeptide Antibiotic Drug Stability in Aqueous Solution. AAPS Open 2022, 8, 20. [Google Scholar] [CrossRef]
- Khairnar, S.; Das, A.; Oupicky, D.; Sadykov, M.R.; Romanova, S. Strategies to Overcome Antibiotic Resistance: Silver Nanoparticles and Vancomycin in Pathogen Eradication. RSC Pharm. 2025, 2, 455–479. [Google Scholar] [CrossRef]
- Blaskovich, M.A.T.; Hansford, K.A.; Butler, M.S.; Jia, Z.; Mark, A.E.; Cooper, M.A. Developments in Glycopeptide Antibiotics. ACS Infect. Dis. 2018, 4, 715–735. [Google Scholar] [CrossRef]
- Nakayama, H.; Kurokawa, K.; Lee, B.L. Lipoproteins in Bacteria: Structures and Biosynthetic Pathways. FEBS J. 2012, 279, 4247–4268. [Google Scholar] [CrossRef]
- Patel, S.; Ahmed, S.; Eswari, J.S. Therapeutic Cyclic Lipopeptides Mining from Microbes: Latest Strides and Hurdles. World J. Microbiol. Biotechnol. 2015, 31, 1177–1193. [Google Scholar] [CrossRef]
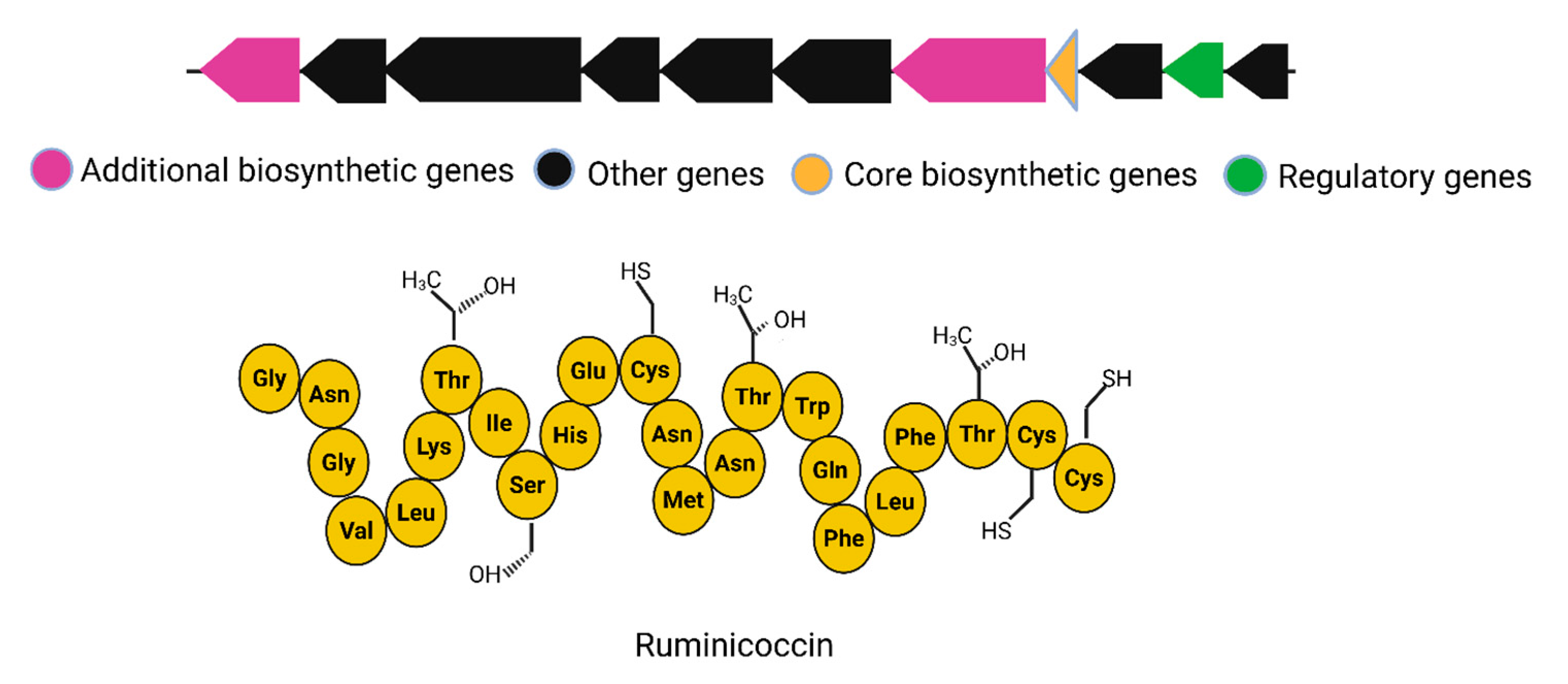
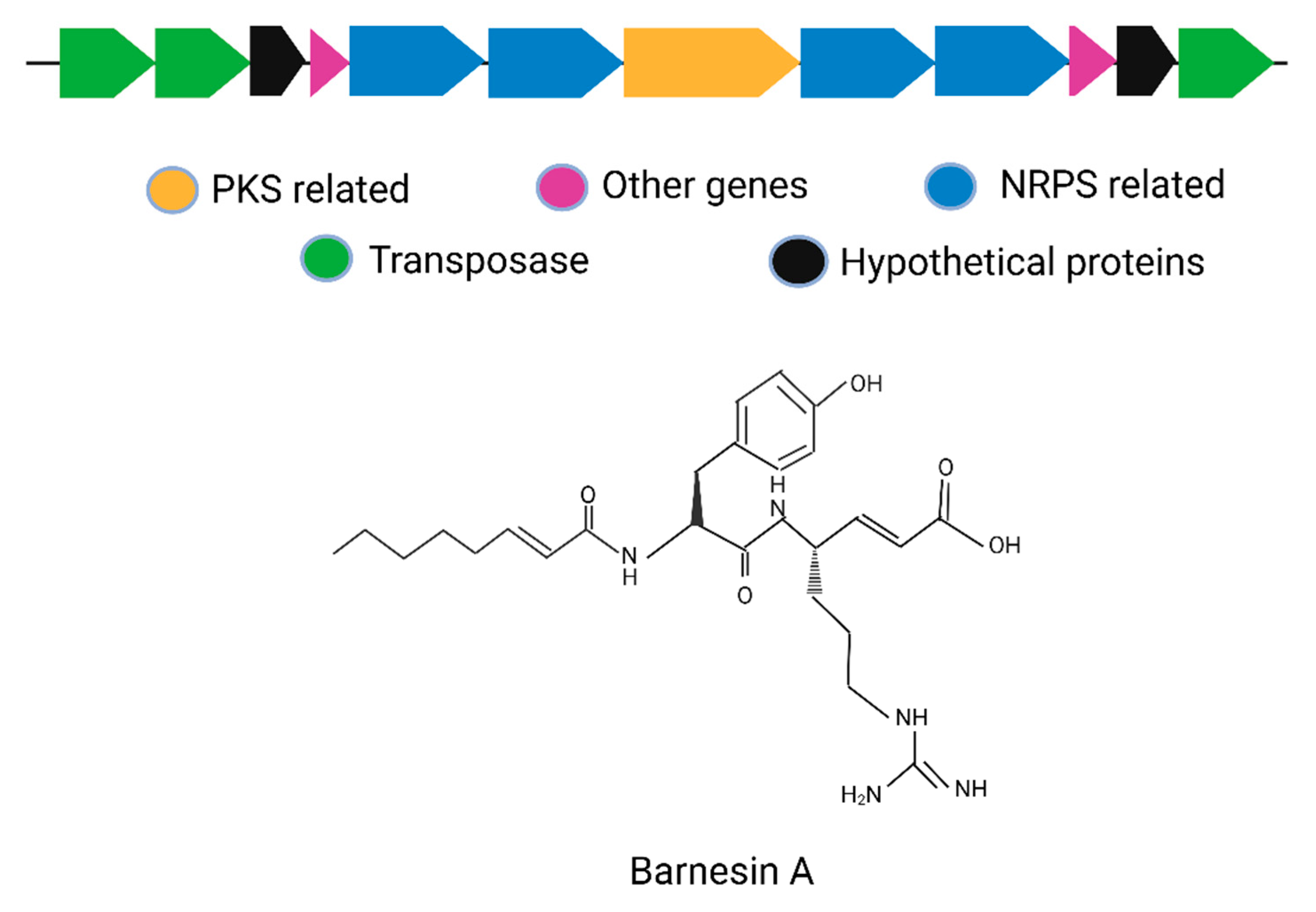
| S. No | Compound | Source | Class | Mol. Weight | Mode of Action | MIC Values | Activity Spectrum | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | Bacteriocin | C. acetobutylicum | Bacteriocin | Sedimentation coefficient is 6S (Sucrose gradient method) | Bactericidal not bacteriolytic | NR | Narrow spectrum (C. acetobutylicum and C. felsineum) | [43] |
| 2 | Bacteriocin 28 | C. perfringens | Bacteriocin | ~100 kDa | NR | NR | Narrow spectrum (Antibacterial) | [44] |
| 3 | Bacteriocin N5 | C. perfringens | Bacteriocin | 82 kDa | Bactericidal | NR | Narrow spectrum (Clostridium strains) | [45] |
| 4 | Boticin B | C. botulinum | Bacteriocin | 5.1 3 kDa or 4.0 kDa | Bactericidal | NR | Narrow spectrum (C. botulinum strains and related Clostridia) | [46] |
| 5 | Boticin P | C. botulinum | Bacteriocin | NR | NR | NR | Narrow spectrum (C. botulinum) | [47] |
| 6 | CBP22 | C. butyricum | Bacteriocin | 2.2 kDa | LPS | 128 µg/mL, 64 µg/mL for E. coli ATCC 25922, S. aureus ATCC 26923 | Broad spectrum (E. coli ATCC 25922, and S. aureus ATCC 26923) | [48] |
| 7 | Circularin and Closticin 574 | C. beijerinckii & C. tyrobutyricum | Cyclic antibacterial peptide & Class II Bacteriocin | 6.771 kDa and ~2.2 kDa | NR | NR | Narrow spectrum (C. tyrobutyricum, Lactococcus and Lactobacillus strains) | [31] |
| 8 | Closthioamide | C. cellulolyticum | Polythioamide Antibiotic | 0.69 kDa | Bactericidal | 0.5 µg/mL for MRSA, VRE & 9.0 µg/mL for E. coli | Broad spectrum (E. faecalis, Neisseria gonorrhoeae, and Staphylococcus aureus and E. coli) | [34] |
| 9 | Clostrubin | C. beijerinckii | Polyphenolic polyketide Antibiotic | 0.4 kDa | NR | 0.12 to 0.97 µΜ | Narrow spectrum (Mycobacterial strains, VRE, and MRSA) | [29] |
| 10 | Clostridocins | C. perfringens and C. butyricum | Bacteriocin | 32.5 kDa and 76 kDa | Lytic and non-lytic effect (C. pasteurianum) | NR | Narrow spectrum (Clostridium pasteurianum) | [49] |
| 11 | Clostrisin and cellulosin | C. cellulovorans. | Bacteriocin | 5.8 kDa and 9.1 kDa | Biostatic | 5.6 µg/mL and 4.8 µg/mL for S.epidermidis, and P. aeruginosa. | Narrow spectrum (multidrug- resistant S. epidermidis MIQ43 and Pseudomonas aeruginosa PA14) | [50] |
| 12 | Clostrocin A, B, C, D, E | Clostridium spp. | Bacteriocin | NR | Bactericidal | NR | Gram positive bacteria | [51] |
| 13 | Clostocin O | Clostridium spp. | Bacteriocin | NR | Bactericidal (Cell wall lysis) | NR | Narrow spectrum (closely related bacterial strains) | [52] |
| 14 | Clostrocyloin | C. beijerinckii | Bacteriocin | 0.25 kDa | NR | NR | Narrow spectrum (Sporobolomyces, M. vaccae, B. subtilis) | [53] |
| 15 | Clostrindolin | C. beijerinckii | Pyrone alkaloid | 0.24 kDa | NR | ~4 µg/mL | Narrow spectrum (M. vaccae) | [38] |
| 16 | Diffocins | C. difficile | R type Bacteriocin | ~200 kDa | Bactericidal | NR | Narrow spectrum (other strains of the same Species) | [30] |
| 17 | Estericin A | C. estertheticum | Class II lanthipeptide | 6.69 kDa | Bactericidal | 1 µg/mL for C. perfringens | Narrow spectrum (MRSA, C. perfringens, and C.tetani.) | [54] |
| 18 | Intestinalin (P30) | C. intestinale | Bacteriocin | 0.023 kDa | Cell membrane permeabilization | 0.2 µg/mL | Broad spectrum | [55] |
| 19 | Neutral Lipopeptide | C. pasteurianum | Lipopeptide | NR | NR | NR | NR | [56] |
| 20 | Perfrin | C. perfringens | Bacteriocin | 11.5 kDa | Bactericidal activity (Pore formation) | NA | Narrow spectrum (C. perfringens) | [57] |
| S. No | Compound | Source | Class | Mol. Weight | Mode of Action | MIC Values | Activity Spectrum | References |
|---|---|---|---|---|---|---|---|---|
| 1 | Barnesin | S. barnesii | Lipo-dipeptide | 0.48 kDa | Inhibition of cysteine protease | NR | Broad spectrum (Gram positive & Gram negative) | [11] |
| 1 | Bifidin I | B. bifidum NCDC 1452 | Bacteriocin | 2.8 kDa | NR | NR | Broad spectrum (E. coli and S. aureus) | [67] |
| 2 | Bifidococcin_664 | B. longum subsp. infantis | Bacteriocin | 14.9 kDa | NR | NR | Narrow spectrum (C. perfringens) | [68] |
| 3 | Naphthalecin | S. colonica | Polyketide antibiotic | 0.2 kDa | NR | NR | Gram positive bacteria | [39] |
| 4 | Nigrescin | P. nigrescens | Bacteriocin | 41 kDa | Bactericidal mode of action | 10.7 µg/mL for P. gingivalis | Narrow spectrum (Dental pathogens) | [69] |
| 5 | Propionicin T1 | P. thoenii | Bacteriocin | 7.1 kDa | Bactericidal mode of action | NR | Narrow spectrum (Propionibacterium strains) | [70] |
| 6 | Pseudocin 196 | B. pseudocatenulatum | Lantibiotic | 2.6 kDa | NR | 0.2 µm for Lactococcus cremoris HP | Narrow spectrum (Clostridium, Streptococcus, Lactococcus spp.) | [71] |
| 7 | Ruminicoccin | R. gnavus | sactipeptide | NR | NR | 0.8 to 50 µg/mL | Narrow spectrum (S. aureus VRE, nisin-resistant Bacillus subtilis and methicillin-resistant S. aureus (MRSA) | [72,73] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mishra, M.; Sharma, U.; Rawat, M.; Harshvardhan; Singh, S.S.; Korpole, S. Antimicrobial Compounds from Anaerobic Microorganisms: A Review of an Untapped Reservoir. Appl. Microbiol. 2025, 5, 68. https://doi.org/10.3390/applmicrobiol5030068
Mishra M, Sharma U, Rawat M, Harshvardhan, Singh SS, Korpole S. Antimicrobial Compounds from Anaerobic Microorganisms: A Review of an Untapped Reservoir. Applied Microbiology. 2025; 5(3):68. https://doi.org/10.3390/applmicrobiol5030068
Chicago/Turabian StyleMishra, Mamta, Upasana Sharma, Manisha Rawat, Harshvardhan, Shelley Sardul Singh, and Suresh Korpole. 2025. "Antimicrobial Compounds from Anaerobic Microorganisms: A Review of an Untapped Reservoir" Applied Microbiology 5, no. 3: 68. https://doi.org/10.3390/applmicrobiol5030068
APA StyleMishra, M., Sharma, U., Rawat, M., Harshvardhan, Singh, S. S., & Korpole, S. (2025). Antimicrobial Compounds from Anaerobic Microorganisms: A Review of an Untapped Reservoir. Applied Microbiology, 5(3), 68. https://doi.org/10.3390/applmicrobiol5030068