Quinolone and Colistin Resistance Genes in Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli of Diverse Phylogenetic Groups Isolated from Seafood in Mumbai, India
Abstract
1. Introduction
2. Materials and Methods
2.1. Detection of ESBL Phenotype in Escherichia coli Isolates
2.2. Quinolone Susceptibility Testing Using Disk Diffusion Assay
2.3. Screening of Colistin-Resistant Isolates Using Chromogenic Agar Medium
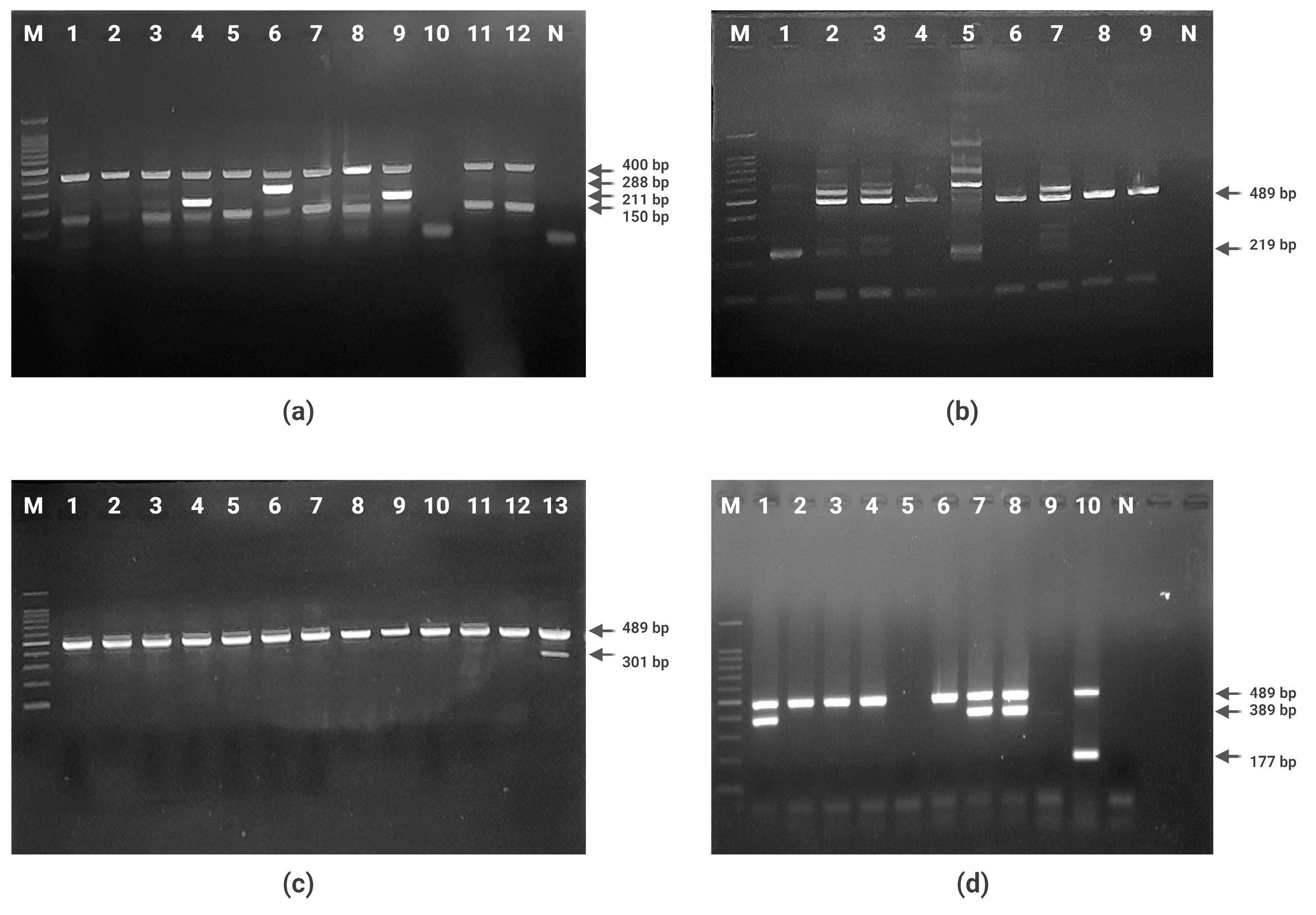
2.4. PCR Detection of Quinolone and Polymyxin Resistance Genes
2.5. Determination of Colistin Minimum Inhibitory Concentration (MIC) by Broth-Microdilution
2.6. Phylotyping of E. coli Isolates
3. Results
3.1. Quinolone and Colistin Resistance in ESBL-Producing E. coli and Their Genetic Determinants
3.2. Incidence of Colistin Resistance Gene mcr-1 and mcr-2 in E. coli
3.3. Phylogroup Evaluation of E. coli
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pitout, J.D.D. Multiresistant Enterobacteriaceae: New Threat of an Old Problem. Expert Rev. Anti Infect. Ther. 2008, 6, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Theuretzbacher, U. Global Antibacterial Resistance: The Never-Ending Story. J. Glob. Antimicrob. Resist. 2013, 1, 63–69. [Google Scholar] [CrossRef]
- Tewari, R.; Mitra, S.; Ganaie, F.; Das, S.; Chakraborty, A.; Venugopal, N.; Shome, R.; Rahman, H.; Shome, B.R. Dissemination and Characterisation of Escherichia coli Producing Extended-Spectrum β-Lactamases, AmpC β-Lactamases and Metallo-β-Lactamases from Livestock and Poultry in Northeast India: A Molecular Surveillance Approach. J. Glob. Antimicrob. Resist. 2019, 17, 209–215. [Google Scholar] [CrossRef]
- Varela, M.F.; Stephen, J.; Lekshmi, M.; Ojha, M.; Wenzel, N.; Sanford, L.M.; Hernandez, A.J.; Parvathi, A.; Kumar, S.H. Bacterial Resistance to Antimicrobial Agents. Antibiotics 2021, 10, 593. [Google Scholar] [CrossRef] [PubMed]
- Madec, J.-Y.; Haenni, M.; Nordmann, P.; Poirel, L. Extended-Spectrum β-Lactamase/AmpC- and Carbapenemase-Producing Enterobacteriaceae in Animals: A Threat for Humans? Clin. Microbiol. Infect. 2017, 23, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.T.T.; Alter, T.; Roesler, U.; Roschanski, N.; Huehn, S. Investigation of Extended-Spectrum and AmpC β-Lactamase-Producing Enterobacteriaceae from Retail Seafood in Berlin, Germany. J. Food Prot. 2018, 81, 1079–1086. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-Spectrum β-Lactamases: An Update on Their Characteristics, Epidemiology and Detection. JAC Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef]
- Price, L.B.; Johnson, J.R.; Aziz, M.; Clabots, C.; Johnston, B.; Tchesnokova, V.; Nordstrom, L.; Billig, M.; Chattopadhyay, S.; Stegger, M.; et al. The Epidemic of Extended-Spectrum-β-Lactamase-Producing Escherichia coli ST131 Is Driven by a Single Highly Pathogenic Subclone, H30-Rx. mBio 2013, 4, e00377-13. [Google Scholar] [CrossRef] [PubMed]
- Belas, A.; Marques, C.; Aboim, C.; Pomba, C. Emergence of Escherichia coli ST131 H30/H30-Rx Subclones in Companion Animals. J. Antimicrob. Chemother. 2019, 74, 266–269. [Google Scholar] [CrossRef]
- Pham, T.D.M.; Ziora, Z.M.; Blaskovich, M.A.T. Quinolone Antibiotics. Medchemcomm 2019, 10, 1719–1739. [Google Scholar] [CrossRef]
- Strahilevitz, J.; Jacoby, G.A.; Hooper, D.C.; Robicsek, A. Plasmid-Mediated Quinolone Resistance: A Multifaceted Threat. Clin. Microbiol. Rev. 2009, 22, 664–689. [Google Scholar] [CrossRef] [PubMed]
- Mirzaii, M.; Jamshidi, S.; Zamanzadeh, M.; Marashifard, M.; Malek Hosseini, S.A.A.; Haeili, M.; Jahanbin, F.; Mansouri, F.; Darban-Sarokhalil, D.; Khoramrooz, S.S. Determination of gyrA and parC Mutations and Prevalence of Plasmid-Mediated Quinolone Resistance Genes in Escherichia coli and Klebsiella pneumoniae Isolated from Patients with Urinary Tract Infection in Iran. J. Glob. Antimicrob. Resist. 2018, 13, 197–200. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Zhao, H. Quinolone Antibiotics: Resistance and Therapy. Infect. Drug Resist. 2023, 16, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L. Emergence of Plasmid-Mediated Resistance to Quinolones in Enterobacteriaceae. J. Antimicrob. Chemother. 2005, 56, 463–469. [Google Scholar] [CrossRef]
- Mirjalili, M.; Mirzaei, E.; Vazin, A. Pharmacological Agents for the Prevention of Colistin-Induced Nephrotoxicity. Eur. J. Med. Res. 2022, 27, 64. [Google Scholar] [CrossRef]
- Hussein, N.H.; Al-Kadmy, I.M.S.; Taha, B.M.; Hussein, J.D. Mobilized Colistin Resistance (Mcr) Genes from 1 to 10: A Comprehensive Review. Mol. Biol. Rep. 2021, 48, 2897–2907. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Smith, A.L. Emergence of Mobile Colistin Resistance Genes within Los Angeles County Wastewater. Environ. Sci. Technol. Lett. 2023, 10, 316–321. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, R.; Schwarz, S.; Wu, C.; Shen, J.; Walsh, T.R.; Wang, Y. Farm Animals and Aquaculture: Significant Reservoirs of Mobile Colistin Resistance Genes. Environ. Microbiol. 2020, 22, 2469–2484. [Google Scholar] [CrossRef]
- Singh, A.S.; Nayak, B.B.; Kumar, S.H. High Prevalence of Multiple Antibiotic-Resistant, Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli in Fresh Seafood Sold in Retail Markets of Mumbai, India. Vet. Sci. 2020, 7, 46. [Google Scholar] [CrossRef] [PubMed]
- CLSI Supplement M100; CLSI Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed. CLSI: Wayne, PA, USA, 2023.
- Gay, K.; Robicsek, A.; Strahilevitz, J.; Park, C.H.; Jacoby, G.; Barrett, T.J.; Medalla, F.; Chiller, T.M.; Hooper, D.C. Plasmid-Mediated Quinolone Resistance in Non-Typhi Serotypes of Salmonella enterica. Clin. Infect. Dis. 2006, 43, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Yamane, K.; Wachino, J.; Suzuki, S.; Arakawa, Y. Plasmid-Mediated qepA Gene among Escherichia coli Clinical Isolates from Japan. Antimicrob. Agents Chemother. 2008, 52, 1564–1566. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, G.A.; Walsh, K.E.; Mills, D.M.; Walker, V.J.; Oh, H.; Robicsek, A.; Hooper, D.C. qnrB, Another Plasmid-Mediated Gene for Quinolone Resistance. Antimicrob. Agents Chemother. 2006, 50, 1178–1182. [Google Scholar] [CrossRef] [PubMed]
- Rebelo, A.R.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A.; et al. Multiplex PCR for Detection of Plasmid-Mediated Colistin Resistance Determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for Surveillance Purposes. Eurosurveillance 2018, 23, 17-00672. [Google Scholar] [CrossRef] [PubMed]
- Onseedaeng, S.; Ratthawongjirakul, P. Rapid Detection of Genomic Mutations in gyrA and parC Genes of Escherichia coli by Multiplex Allele Specific Polymerase Chain Reaction. J. Clin. Lab. Anal. 2016, 30, 947–955. [Google Scholar] [CrossRef] [PubMed]
- CLSI Document M07-A10; CLSI Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. Approved Standard—10th ed. CLSI: Wayne, PA, USA, 2015.
- EUCAST European Committee on Antimicrobial Susceptibility: Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version; 2019. 2019.
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli Phylo-Typing Method Revisited: Improvement of Specificity and Detection of New Phylo-Groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Clermont, O.; Dixit, O.V.A.; Vangchhia, B.; Condamine, B.; Dion, S.; Bridier-Nahmias, A.; Denamur, E.; Gordon, D. Characterization and Rapid Identification of Phylogroup G in Escherichia coli, a Lineage with High Virulence and Antibiotic Resistance Potential. Environ. Microbiol. 2019, 21, 3107–3117. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.; Sung, K.; Kweon, O.; Khan, S.; Nawaz, S.; Steele, R. Characterisation of Novel Mutations Involved in Quinolone Resistance in Escherichia coli Isolated from Imported Shrimp. Int. J. Antimicrob. Agents 2015, 45, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Juraschek, K.; Deneke, C.; Schmoger, S.; Grobbel, M.; Malorny, B.; Käsbohrer, A.; Schwarz, S.; Meemken, D.; Hammerl, J.A. Phenotypic and Genotypic Properties of Fluoroquinolone-Resistant, Qnr-Carrying Escherichia coli Isolated from the German Food Chain in 2017. Microorganisms 2021, 9, 1308. [Google Scholar] [CrossRef] [PubMed]
- Belotindos, L.P.; Tsunoda, R.; Villanueva, M.A.; Nakajima, C.; Mingala, C.N.; Suzuki, Y. Characterisation of Plasmids Harbouring qnrA1, qnrS1, and qnrB4 in E. coli Isolated in the Philippines from Food-Producing Animals and Their Products. J. Glob. Antimicrob. Resist. 2022, 30, 38–46. [Google Scholar] [CrossRef]
- Jiang, H.-X.; Tang, D.; Liu, Y.-H.; Zhang, X.-H.; Zeng, Z.-L.; Xu, L.; Hawkey, P.M. Prevalence and Characteristics of β-Lactamase and Plasmid-Mediated Quinolone Resistance Genes in Escherichia coli Isolated from Farmed Fish in China. J. Antimicrob. Chemother. 2012, 67, 2350–2353. [Google Scholar] [CrossRef]
- Chandran, S.P.; Diwan, V.; Tamhankar, A.J.; Joseph, B.V.; Rosales-Klintz, S.; Mundayoor, S.; Lundborg, C.S.; Macaden, R. Detection of Carbapenem Resistance Genes and Cephalosporin, and Quinolone Resistance Genes along with oqxAB Gene in Escherichia coli in Hospital Wastewater: A Matter of Concern. J. Appl. Microbiol. 2014, 117, 984–995. [Google Scholar] [CrossRef]
- Shetty, S.S.; Deekshit, V.K.; Jazeela, K.; Vittal, R.; Rohit, A.; Chakraborty, A.; Karunasagar, I. Plasmid-Mediated Fluoroquinolone Resistance Associated with Extra-Intestinal Escherichia coli Isolates from Hospital Samples. Indian. J. Med. Res. 2019, 149, 192–198. [Google Scholar] [CrossRef]
- Ewers, C.; Göpel, L.; Prenger-Berninghoff, E.; Semmler, T.; Kerner, K.; Bauerfeind, R. Occurrence of Mcr-1 and Mcr-2 Colistin Resistance Genes in Porcine Escherichia coli Isolates (2010-2020) and Genomic Characterization of mcr-2-Positive E. coli. Front. Microbiol. 2022, 13, 1076315. [Google Scholar] [CrossRef]
- Mikhayel, M.; Leclercq, S.O.; Sarkis, D.K.; Doublet, B. Occurrence of the Colistin Resistance Gene mcr-1 and Additional Antibiotic Resistance Genes in ESBL/AmpC-Producing Escherichia coli from Poultry in Lebanon: A Nationwide Survey. Microbiol. Spectr. 2021, 9, e0002521. [Google Scholar] [CrossRef] [PubMed]
- Karim, M.R.; Zakaria, Z.; Hassan, L.; Faiz, N.M.; Ahmad, N.I. The Occurrence and Molecular Detection of mcr-1 and mcr-5 Genes in Enterobacteriaceae Isolated from Poultry and Poultry Meats in Malaysia. Front. Microbiol. 2023, 14, 1208314. [Google Scholar] [CrossRef]
- Ovejero, C.M.; Delgado-Blas, J.F.; Calero-Caceres, W.; Muniesa, M.; Gonzalez-Zorn, B. Spread of mcr-1-Carrying Enterobacteriaceae in Sewage Water from Spain. J. Antimicrob. Chemother. 2017, 72, 1050–1053. [Google Scholar] [CrossRef] [PubMed]
- Manohar, P.; Shanthini, T.; Ayyanar, R.; Bozdogan, B.; Wilson, A.; Tamhankar, A.J.; Nachimuthu, R.; Lopes, B.S. The Distribution of Carbapenem- and Colistin-Resistance in Gram-Negative Bacteria from the Tamil Nadu Region in India. J. Med. Microbiol. 2017, 66, 874–883. [Google Scholar] [CrossRef] [PubMed]
- Gandra, S.; Tseng, K.K.; Arora, A.; Bhowmik, B.; Robinson, M.L.; Panigrahi, B.; Laxminarayan, R.; Klein, E.Y. The Mortality Burden of Multidrug-Resistant Pathogens in India: A Retrospective, Observational Study. Clin. Infect. Dis. 2019, 69, 563–570. [Google Scholar] [CrossRef]
- Ghafur, A.; Shankar, C.; GnanaSoundari, P.; Venkatesan, M.; Mani, D.; Thirunarayanan, M.A.; Veeraraghavan, B. Detection of Chromosomal and Plasmid-Mediated Mechanisms of Colistin Resistance in Escherichia coli and Klebsiella pneumoniae from Indian Food Samples. J. Glob. Antimicrob. Resist. 2019, 16, 48–52. [Google Scholar] [CrossRef]
- Anyanwu, M.U.; Jaja, I.F.; Nwobi, O.C. Occurrence and Characteristics of Mobile Colistin Resistance (mcr) Gene-Containing Isolates from the Environment: A Review. Int. J. Environ. Res. Public Health 2020, 17, 1028. [Google Scholar] [CrossRef] [PubMed]
- Lemlem, M.; Aklilu, E.; Mohamed, M.; Kamaruzzaman, N.F.; Zakaria, Z.; Harun, A.; Devan, S.S.; Kamaruzaman, I.N.A.; Reduan, M.F.H.; Saravanan, M. Phenotypic and Genotypic Characterization of Colistin-Resistant Escherichia coli with mcr-4, mcr-5, mcr-6, and mcr-9 Genes from Broiler Chicken and Farm Environment. BMC Microbiol. 2023, 23, 392. [Google Scholar] [CrossRef] [PubMed]
- Delannoy, S.; Le Devendec, L.; Jouy, E.; Fach, P.; Drider, D.; Kempf, I. Characterization of Colistin-Resistant Escherichia coli Isolated from Diseased Pigs in France. Front. Microbiol. 2017, 8, 2278. [Google Scholar] [CrossRef] [PubMed]
- Rafique, M.; Potter, R.F.; Ferreiro, A.; Wallace, M.A.; Rahim, A.; Ali Malik, A.; Siddique, N.; Abbas, M.A.; D’Souza, A.W.; Burnham, C.-A.D.; et al. Genomic Characterization of Antibiotic Resistant Escherichia coli Isolated From Domestic Chickens in Pakistan. Front. Microbiol. 2019, 10, 3052. [Google Scholar] [CrossRef] [PubMed]
- Macori, G.; Nguyen, S.V.; Naithani, A.; Hurley, D.; Bai, L.; El Garch, F.; Woehrlé, F.; Miossec, C.; Roques, B.; O’Gaora, P.; et al. Characterisation of Early Positive Mcr-1 Resistance Gene and Plasmidome in Escherichia coli Pathogenic Strains Associated with Variable Phylogroups under Colistin Selection. Antibiotics 2021, 10, 1041. [Google Scholar] [CrossRef] [PubMed]
| Antibiotic | No. (%) | ||
|---|---|---|---|
| Resistant | Intermediate | Sensitive | |
| Nalidixic Acid (NA) | 132 (49.07%) | 75 (27.88%) | 62 (23.04%) |
| Ciprofloxacin (CIP) | 156 (57.99%) | 84 (31.22%) | 29 (10.78%) |
| Ofloxacin (OF) | 23 (8.55%) | 6 (2.23%) | 240 (89.21%) |
| Levofloxacin (LE) | 39 (14.49%) | 110 (40.89%) | 120 (44.60%) |
| Norfloxacin (NX) | 47 (17.47%) | 91 (33.82%) | 131 (48.67%) |
| Moxifloxacin (MO) | 198 (73.60%) | - | 71 (26.39%) |
| Gene | No. (%) Positive |
|---|---|
| qnrS | 145 (53.9%) |
| qnrB | 20 (7.43%) |
| qnrA | 0 |
| mcr-2 | 38 (14.12%) |
| mcr-1 | 0 |
| QRDR Mutation | No. (%) Positive |
|---|---|
| gyrA83 | 162 (68.93%) |
| gyrA87 | 166 (70.63%) |
| parC80 | 16 (68.93%) |
| parC84 | 186 (79.14%) |
| Phylogroup | No. of Isolates (%) | No. of Isolates Harboring One or More qnr Genes (%) | No. of Isolates Harboring mcr-2 Gene (%) |
|---|---|---|---|
| B1 | 125 (46.46%) | 66 (44%) | 27 (71.05%) |
| UN | 62 (23.04%) | 23 (15.33%) | 2 (5.26%) |
| A | 31 (11.52%) | 23 (15.33%) | 4 (10.52%) |
| D | 21 (7.80%) | 18 (12%) | 2 (5.26%) |
| C | 17 (6.31%) | 13 (8.66%) | 1 (2.63%) |
| B2 | 6 (2.23%) | 3 (2%) | 1 (2.63%) |
| F | 3 (1.11%) | 2 (1.33%) | 0 |
| E | 3 (1.11%) | 1 (0.66%) | 1 (2.63%) |
| G | 1(0.37%) | 1 (0.66%) | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhanush, C.K.; Lekshmi, M.; Girisha, S.K.; Nayak, B.B.; Kumar, S.H. Quinolone and Colistin Resistance Genes in Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli of Diverse Phylogenetic Groups Isolated from Seafood in Mumbai, India. Appl. Microbiol. 2025, 5, 3. https://doi.org/10.3390/applmicrobiol5010003
Dhanush CK, Lekshmi M, Girisha SK, Nayak BB, Kumar SH. Quinolone and Colistin Resistance Genes in Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli of Diverse Phylogenetic Groups Isolated from Seafood in Mumbai, India. Applied Microbiology. 2025; 5(1):3. https://doi.org/10.3390/applmicrobiol5010003
Chicago/Turabian StyleDhanush, Chandrashekar K., Manjusha Lekshmi, Shivani Kallappa Girisha, Binaya Bhusan Nayak, and Sanath H. Kumar. 2025. "Quinolone and Colistin Resistance Genes in Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli of Diverse Phylogenetic Groups Isolated from Seafood in Mumbai, India" Applied Microbiology 5, no. 1: 3. https://doi.org/10.3390/applmicrobiol5010003
APA StyleDhanush, C. K., Lekshmi, M., Girisha, S. K., Nayak, B. B., & Kumar, S. H. (2025). Quinolone and Colistin Resistance Genes in Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli of Diverse Phylogenetic Groups Isolated from Seafood in Mumbai, India. Applied Microbiology, 5(1), 3. https://doi.org/10.3390/applmicrobiol5010003






