Abstract
Although biofilms contribute to bacterial tolerance to desiccation and survival in low-moisture foods, the molecular mechanisms underlying biofilm formation have not been fully understood. This study created a mutant library from Salmonella Enteritidis using mini-Tn10 transposon mutagenesis. The biofilm-forming potential of acquired mutants was assessed before the genomic DNA of the mutants that formed significantly (p ≤ 0.05) less biofilm mass than their wildtype parent strain was extracted for deep DNA sequencing. The gene of each mutant interrupted by mini-Tn10 insertion was identified by aligning obtained sequencing data with the reference Genbank sequences using a BLAST search. Sixty-four mutant colonies were selected, and five mutants that formed the least amount of biofilm mass compared to the wildtype parent strain were selected for sequencing analysis. The results of the BLAST search revealed that the gene interrupted by mini-Tn10 in each mutant is responsible for the biosynthesis of aldehyde dehydrogenase (EutE), cysteine desulfurase (SufS or SufE), a transporter protein, porin OmpL, and a ribbon–helix–helix protein from the CopG family, respectively. Knock-off mutant construction is a possible approach to verify the potential of the identified genes to serve as targets of antimicrobial intervention to control Salmonella colonization on low-moisture foods and in their production environment.
1. Introduction
Foodborne Salmonella infection is an important public health issue, accounting for over 1.2 million cases, 23,000 hospitalizations, and 450 deaths each year in the United States [1]. Globally, nontyphoidal Salmonella causes ca. 150 million illnesses and ca. 60,000 deaths each year [2]. The symptoms of Salmonella infections vary from self-limited gastroenteritis to life-threatening systemic complications [3]. Outbreaks of foodborne Salmonella infection have historically been associated with animal products such as poultry, meat, or dairy products [3]. Recently, however, the pathogen has been implicated in outbreaks associated with fresh produce, including low-moisture products such as cereals, chocolates, seeds, peanuts, and almonds [4,5]. These foods have been erroneously considered to be safe and unable to support the growth of pathogenic microorganisms [4,5]. It is now well documented that pathogens like Salmonella can survive for days or even years under desiccation and in low-moisture foods [6].
The control of Salmonella in/on low-moisture foods is challenging because the pathogen has developed stronger cross-tolerance to multiple environmental stressors such as heat and chemical sanitizers compared to the pathogen in high-moisture foods [7]. Moreover, low-moisture foods have a relatively longer shelf life, and most of them are ready-to-eat products that do not require an additional killing step to eliminate the pathogen before consumption [8]. For this reason, once Salmonella is present in/on low-moisture food or in the production environment, it has a high probability of leading to long-lasting and widespread outbreaks.
Several cell-defensive strategies, including the synthesis of certain extracellular polymeric substances and the formation of biofilms, can protect Salmonella cells from desiccation [5]. Biofilm formation is a food safety concern unlimited to low-moisture foods. However, when bacterial cells with extraordinary biofilm-forming abilities are introduced to low-moisture food, they could persist much longer than those lacking such capabilities [9]. In particular, Salmonella cells able to synthesize extracellular polysaccharides, one of the major components of the biofilm matrix, have been reported to play an important role in survival under desiccation [10,11].
Although it is well documented that mutations in genes involved in the biosynthesis of curli, as well as cellulose and other exopolysaccharides, could affect cell initial attachment and biofilm formation [12,13,14], genetic determinants encoding many other cellular components that may have critical roles in biofilm formation remain unknown. To further understand the underlying molecular mechanisms of biofilm formation by Salmonella, mini-Tn10 transposon mutagenesis was used initially to create a library of S. enteritidis mutants with randomly interrupted genes, and subsequently, the biofilm-forming ability of the wildtype Salmonella strain and its mutants was assessed and compared. To identify the defective gene of the mutants with a reduced ability to form biofilms, the genomic DNA sequence of the mutants was compared, using a BLAST search with the reference sequences of S. enteritidis deposited in Genbank.
2. Materials and Methods
2.1. Materials and Culture Conditions
The wildtype Salmonella strains used, and their mutants created, in this study are listed in Table 1. Salmonella enterica subsp. enterica serotype Enteritidis PT 30 isolated from a raw almond-related outbreak [15] and Escherichia coli strain BW20767 [16] were used in this study. The S. enteritidis strain was the causative agent of the 168 laboratory-confirmed cases of Salmonella infection (157 in Canada, 11 in the United States) between October 2000 and July 2001. It is a known biofilm producer, but its biofilm-forming ability relative to Salmonella reference strains is unknown. The E. coli BW 20767 was obtained from Dr. Nikki Freed at Massey University. Cultures of S. enteritidis and E. coli preserved at −80 °C were inoculated on tryptic soy agar (TSA; BBL/Difco, Sparks, MD, USA) and incubated at 37 °C for 18 h. The resulting culture was transferred to fresh TSA plates two consecutive times and incubated under the conditions described above. When necessary, the growth media were supplemented with antibiotics, kanamycin at 50 μg/mL, ampicillin at 100 μg/mL, and/or nalidixic acid at 30 μg/mL (MP Biomedicals, Santa Ana, CA, USA).

Table 1.
Bacterial strains used in this study.
2.2. Construction of Salmonella enteritidis Mutants via Mini-Tn10 Transposon Mutagenesis
Mini-Tn10 transposon mutagenesis was performed using a previously published protocol with some modifications [16]. The E. coli BW20767 strain harboring a kanamycin-resistant marker (kanr) on the mini-Tn10 transposon, an ampicillin-resistant marker (ampr) on the suicidal plasmid pJA1, and a nalidixic acid-sensitive marker (nals) on the chromosome was used as the donor strain and the S. enteritidis almond outbreak strain (amps, kans, nalr) was used as the recipient strain. The donor and recipient were grown individually in tryptic soy broth (TSB) at 37 °C for 18 h, and the resulting cultures were mixed in a 1:1 (v/v) ratio. The mixture (100 µL) was spotted on TSA and incubated at 37 °C for 18 h. To activate the transposon, the resulting culture mixture was grown in TSB containing 1 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG; Fisher Scientific, Waltham, MA, USA) at 37 °C for 6 h. Following the incubation, the culture was streaked on TSA supplemented with kanamycin and nalidixic acid and incubated at 37 °C overnight. Cells that were resistant to kanamycin and nalidixic acid on TSA were randomly selected. To select S. enteritidis mutants generated by transposon mutagenesis, the susceptibility of cells to ampicillin on TSA was examined. Colonies that were resistant to kanamycin and nalidixic acid but sensitive to ampicillin were streaked on XLT4 agar and MacConkey agar. Black colonies on XLT4 agar with supplements that were colorless on MacConkey agar were randomly selected, and each colony was transferred to TSB amended with two antibiotics, kanamycin and nalidixic acid, and incubated at 37 °C overnight before being used in the biofilm study. The microbiological media used in this study were purchased from Becton Dickinson (Sparks, MD, USA).
2.3. Assessment of Biofilm-Forming Capability of S. enteritidis Mutants
The biofilm-forming ability of the mutants was assessed using the crystal violet binding assay [17,18]. In brief, mutants and their wildtype parent were grown for 18 h in Luria–Bertani no salt (LBNS) broth with or without kanamycin and nalidixic acid, respectively. Each cell suspension was diluted (1:40; v/v), and the resulting cell suspension (2 mL) was inoculated into a 24-well polystyrene tissue culture plate for biofilm development. The plates were incubated for 7 days at 25 °C. Following the incubation, the cultures in LBNS were withdrawn, and loosely attached cells were removed by rinsing with sterilized water three times. The biofilm mass was fixed with 95% ethanol (Koptec, King of Prussia, PA, USA) for 10 min and air-dried for 10 min at ambient temperature. The fixed biofilms were stained with 2% crystal violet (Fisher Scientific, Waltham, MA, USA) for 15 min, and the excess stain was then rinsed off with running tap water, followed by drying the plates for 1 h at room temperature. The crystal violet stain in stained biofilms was extracted with an ethanol and acetone mixture (80:20; VWR International, LLC, Randor, PA, USA). The absorbance of the ethanol–acetone solution was measured using a spectrophotometer (Thermo Fisher Scientific, Freemont, CA, USA) at a wavelength of 550 nm. The assay was replicated three times with duplicates each time. Significant differences in the absorbance values of crystal violet in the ethanol and acetone solution were analyzed using a one-way analysis of variance (ANOVA) and Fisher’s least significant difference test (p ≤ 0.05). Mutant colonies with a significantly lower amount of biofilm mass in comparison to their wildtype parent were selected for sequencing analysis.
2.4. Mini-Tn10 Transposon Insertion Sites in S. enteritidis Mutants
The genomic DNA of mutants that formed significantly less (p ≤ 0.05) biofilm mass compared to their wildtype parent was extracted. In brief, each selected mutant was cultivated in 1 mL of TSB containing kanamycin and nalidixic acid for 18 h. The resulting culture was rinsed twice with sterile distilled water after centrifuging for 2 min at 14,000× g. After final washing and discarding the supernatant, the cell pellet was resuspended in sterilized distilled water (0.5 mL). Lysis buffer (20 µL) containing 20% (w/v) SDS, 0.5 M EDTA, and 1 M Tris-Cl was added to the cell suspension, followed by agitation at ambient temperature for 2 h on a Clay Adams brand @ Nutator (BD Biosciences, Bedford, MA, USA). A mixture of 0.5 mL of phenol and chloroform at a 1:1 ratio (v/v) was added to the sample for DNA purification, and the sample was agitated on the Nutator for 30 min. The sample was then centrifuged for 2 min at 14,000× g, and the supernatant containing DNA was collected. The DNA was precipitated with 1 mL of 100% ethanol overnight at −20 °C and precipitated DNA was collected after centrifugation under the conditions described above and dried subsequently at room temperature. The DNA dissolved in sterilized distilled water (30 µL) was submitted for purification and whole-genome sequencing at CD Genomics (Shirley, NY, USA, https://www.cd-genomics.com, accessed on 20 April 2024). The chemicals used for DNA extraction were from Fisher Scientific unless specified. PilotEdit software (version 9.3.0; http://Pilotedit.com, accessed on 20 April 2024) was used to analyze the acquired sequencing data. DNA sequences containing the sequence of IS10, TTTTTACCAAAATCATTAGGGGATT, were compared to the E. coli mini-Tn10-based transposon cassette (accession no. AJ601386.1) and S. enteritidis (accession no. CP050716.1) using a BLAST search (https://blast.ncbi.nlm.nih.gov, accessed on 20 April 2024).
3. Results
3.1. Libraries of S. enteritidis Mutants
The kanamycin resistance gene (kanr) on the mini-Tn10 transposon carried by the suicidal plasmid pJA1 was successfully inserted into the chromosome of the S. enteritidis recipient (nalr, amps). Sixty-four mutant colonies were selected from TSA supplemented with kanamycin, as well as nalidixic acid, the antibiotic resistance marker on the recipient chromosome. Cells of the mutant colonies were later confirmed to be sensitive to ampicillin, the gene of which was located on the suicidal plasmid that delivered the mini-Tn10 to the recipient cells (Figure 1). The mutant colonies had the morphological characteristics of Salmonella on XLT4 and MacConkey agar plates (Figure 1).
Figure 1.
Cultures of a selected Salmonella mutant on tryptic soy agar supplemented with 100 μg/mL ampicillin (left) and XLT-4 agar (middle), as well as the cultures of the E. coli donor BW20767 [pink] and the Salmonella mutant [colorless] (right) on MacConkey agar.
3.2. Selection of Mutants with Reduced Ability to Form Biofilms
Figure 2 shows the A550 values of the solutions of crystal violet extracted from the biofilms formed by the wildtype and mutant Salmonella cultures. Among tested mutants, SE-L3, SE-L19, SE-L29, SE-S15, and SE-S26 formed significantly (p ≤ 0.05) less biofilm mass compared to their wildtype parent. However, the biofilm masses from the five mutants were not significantly (p > 0.05) different from one another.
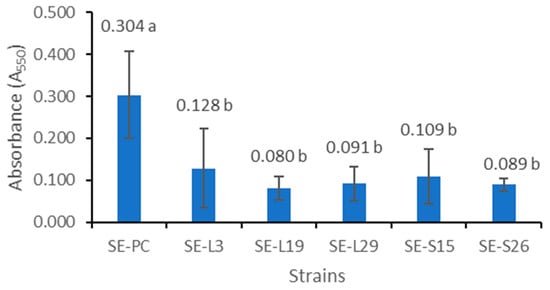
Figure 2.
Biofilm mass developed by wildtype and mutant S. enteritidis. The data represent the A550 values of the solutions of crystal violet extracted from biofilm mass. The error bars represent the standard deviations of the means. Means followed by different letters are significantly different (p ≤ 0.05). SE-PC: positive control from the wildtype parent strain.
3.3. Identification of the Mini-Tn10 Insertion Sites on the S. enteritidis Chromosome
The identified mini-Tn10 insertion sites in examined mutants are reported in Table 2. The genetic locations of mini-Tn10 insertion are shown in a circular chromosome map (Figure 3). In the SE-L3 mutant, the mini-Tn10 interrupted the eutE, a gene responsible for the biosynthesis of aldehyde dehydrogenase, while in SE-L19, SE-L29, SE-S15, and SE-S26, the transposon was found in the genes that encode for a transporter protein, outer membrane protein L, a ribbon–helix–helix protein from the CopG family, and cysteine desulfurase, respectively (Table 2).

Table 2.
Alignment of acquired DNA sequences, interrupted by mini-Tn10 in Salmonella mutants, with the reference Genbank sequences using a BLAST search *.
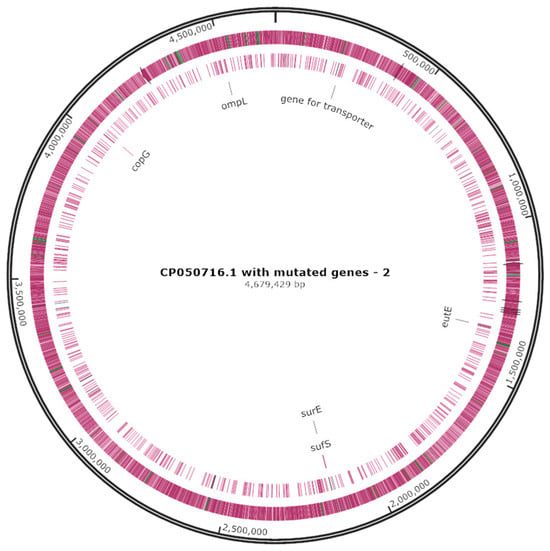
Figure 3.
Mini-Tn10 insertion locations in the SE-L3, SE-L19, SE-L29, SE-S15, and SE-S26 mutants on a circular chromosome map of S. enteritidis (accession no. CP050716.1).
In detail, bases 153 to 263 of the acquired DNA sequence from SE-L3 corresponded to bases 1347001 to 1347111 on an S. enteritidis genome (accession no. CP050716.1) and shared 100% identity and 0% gap with the genes encoding for the aldehyde dehydrogenase (EutE). In SE-L19, bases 151–262 of the sequencing data matched the sequence of a gene encoding for a transporter protein that corresponds to bases 229143 to 229254 on S. enteritidis genome CP050716.1 with 100% identity and 0% gap. The gene encoding for porin OmpL was interrupted by mini-Tn10 in the SE-L29 mutant. Bases 153 to 264 of the sequence obtained from SE-L29 shared 100% identity and 0% gap with the DNA sequence corresponding to bases 4505153 to 4505264 on S. enteritidis genome CP050716.1. In the SE-S15 mutant, bases 157–265 of the obtained sequence shared 99% identity and 0% gap with the gene encoding for a ribbon–helix–helix protein from the CopG family. In the SE-S26 mutant, bases 155–268 of the collected sequencing data corresponded to bases 2147129 to 2147242 of the reference S. enteritidis genome and matched sufS and sufE, genes that are involved in cysteine desulfuration.
Furthermore, bases 76–159, 74–150, 77–154, 80–163, and 79–156 of the acquired sequences from mutants SE-L3, L19, L29, S15, and S26, respectively, matched the E. coli kanamycin-resistant Tn10-based transposon cassette (accession no. AJ601386.1).
4. Discussion
The mini-Tn10 in the SE-L3 mutant interrupted a gene that encodes for aldehyde dehydrogenase, EutE, which is involved in ethanolamine degradation (Table 2). Ethanolamine is a prevalent compound on the cell membrane of the host intestine [19]. Intestinal-associated bacteria including Escherichia, Enterococcus, and Salmonella can metabolize ethanolamine as a carbon and nitrogen source, and the ability of cells to metabolize ethanolamine contributes to pathogenesis by outcompeting bacteria that cannot utilize it [20,21].
The genes required for ethanolamine utilization are clustered on the ethanolamine utilization operon (eut). EutBC is an enzyme of ethanolamine ammonia lyase that degrades ethanolamine into acetaldehyde (carbon source) and ammonia (nitrogen source). EutE, an aldehyde dehydrogenase, converts acetaldehyde into acetyl-CoA, which can be used in a myriad of metabolic reactions including the tricarboxylic acid (TCA) cycle. In addition, acetyl-CoA can be phosphorylated into acetyl phosphate by phosphate acetyltransferase, Pta, subsequently forming acetate by acetate kinase (AcaK) to generate ATP [20,22]. Thus, mutations in eutE can result in changes in the cellular concentration of acetaldehyde, acetyl-CoA, and downstream metabolites of acetyl-CoA such as acetyl phosphate in bacterial cells.
Acetyl phosphate has been shown to play important roles in the biosynthesis of flagella and curli, extracellular polymeric substances required for biofilm formation [22,23]. Acetyl phosphate transfers the phosphoryl group into the two-component signaling system, OmpR [24,25] and RcsB [26]. OmpR, a response regulator, is required for the regulation of the csgD promoter. CsgD is a biofilm regulator that is responsible for the biosynthesis of organelles involved in bacteria motility such as flagella and for the production of cellulose and curli [27]. RcsB is nevertheless an activator of colanic acid production, which is one of the major extracellular polymeric substances facilitating the maturation of biofilms [28]. It has been reported that the phosphorylation of RcsB acts as a key signal to switch planktonic cells to a sessile state in S. typhimurium [29].
Previous studies have shown that changes in the cellular level of acetyl-CoA through manipulation of the genes regulating acetyl-CoA metabolism affect cells’ ability to form biofilms [30,31]. For example, mutations in the acetyl-CoA positive regulatory genes, ackA (encoding for acetate kinase), pta (encoding for phosphotransacetylase), and ldhA (encoding for lactate dehydrogenase), caused the cells of E. coli to form more biofilms, while a mutation in the acetyl-CoA negative regulatory gene, pflA, decreased the cells’ ability to form biofilms [30]. In E. coli, the acetyl-CoA level was negatively regulated by PpiB, which belongs to the superfamily of peptidyl-prolyl cis/trans isomerase. Mutants with a defective ppiB produced a higher level of acetyl-CoA and produced more biofilm than the wildtype strain [32]. In the current study, the SE-L3 mutant with a defective gene encoding EutE might have been unable to convert acetaldehyde into acetyl-CoA, and consequentially, the cells of SE-L3 had a poorer ability to form biofilms.
In the SE-L19 and SE-L29 mutants, mini-Tn10 insertions disrupted the coding region of the genes encoding a transporter and porin OmpL, respectively. Bacteria have both passive and active transport systems. Passive transport allows passive diffusion of small molecules using channels such as porins in outer membranes, while active transport moves molecules against a concentration gradient, which requires ATP as an energy source [33,34]. Bacterial transporters are responsible for the uptake of essential nutrients including polysaccharides, amino acids, minerals, and vitamins, as well as the secretion of various molecules including toxins and antimicrobial agents out of the cell [35]. Thus, transporter proteins play critical roles in bacterial growth and antimicrobial resistance [36]. In addition to supporting survival against toxic compounds, the role of ABC transporters in biofilm development has been proposed in many microorganisms. For example, mutations in pstS, pstC, and pstA in the pst operon, encoding for the high-affinity phosphate-specific transporter in the ABC transporter family, decreased the amount of biofilm formed by bacteria such as Pseudomonas aureofaciens [36] and Proteus mirabilis [37]. The Pst system is required to repress the expression of phosphate (Pho) regulon; disruption of the Pst systems leads to constitutive expression of the Pho regulon [36,38]. Pho regulon expression has been demonstrated to inhibit biofilm formation by expression of RapA phosphodiesterase that lowers the level of c-di-GMP [30,31,32,33,34,35,36,37,38,39,40], a second messenger in an intracellular signaling system that controls bacterial lifestyle switching between sessile biofilm and planktonic cells [41]. In Streptococcus mutans, the deletion of pstS caused a reduction in extracellular polysaccharide production, which is necessary for adhesion and biofilm formation, and a reduced ability to attach to certain surfaces [38]. Similarly, the lapEBC-encoded ABC transporter plays important roles in the export of adhesion, LapA, and biofilm maturation in Pseudomonas fluorescens as the mutations within the lapEBC cluster caused the loss of detectable LapA on the cell surface and defects in irreversible attachment, thus forming a less matured biofilm structure compared to wildtype cells [42]. Collectively, these observations indicate that the mutations in the phosphate transporter and adhesin LapA transporter negatively regulate biofilm formation, possibly by activating the Pho regulon, which in turn decreases the cellular level of c-di-GMP and disrupts cell adhesion and colonization on surfaces, the events critical for the establishment of a biofilm community.
The outer membrane porin L (OmpL) is homologous to YshA [43], which is highly conserved and widely distributed among different Salmonella serovars [44]. The potential role of YshA in biofilm formation has been discussed [45]. In a previous study, the yshA-mutant cells of S. typhimurium accumulated less biofilm mass on a polyvinyl chloride plate than the wildtype cells in LB no salt broth [45]. The yshA is part of the yihU-yshA operon, which is required for the assembly and translocation of the O-antigen, one of the main constituents of the extracellular matrix of Salmonella [46]. Previous studies have shown that the O-antigen capsule is crucial for Salmonella attachment and biofilm formation on various surfaces [47,48].
The mass of biofilm was significantly lower when the gene encoding for a ribbon–helix–helix (RHH) protein from the CopG family was disrupted in the SE-L15 mutant (Figure 1). The RHH motif is a common structural motif of prokaryotic antitoxin proteins [49]. The link between the toxin–antitoxin system and biofilm formation has been explored. MazEF, for example, is one of the antitoxin–toxin systems in E. coli and many other pathogenic bacteria including S. enterica [50]. MazE, the antitoxin, is a putative transcriptional repressor of the CopG family [51]. It neutralizes MazF, a stable toxin that cleaves RNA to inhibit protein translation and interfere with vital cellular processes [52]. A significant increase in biofilm formation was observed in a Staphylococcus aureus strain lacking mazF expression [53]. Loss of MazF also caused increased expression of intercellular adhesion gene cluster (ica) and production of polysaccharide intercellular adhesin. The decrease in biofilm mass formed by the SE-S15 mutant with the knock-off gene encoding for the RHH protein of the CopG family could be due to an impaired RHH motif-containing antitoxin that releases the toxin, resulting in the inhibition of adhesion gene expression and subsequently biofilm formation.
The expression of sufS and sufE was interrupted in the SE-S26 mutant. SufS and SufE are part of the sulfur formation (SUF) system, involved in iron–sulfur (FeS) cluster biogenesis under iron starvation and oxidative stress in many bacteria [53,54]. The SUF system is encoded by the sufABCDSE operon, which encodes two protein complexes, SufSE and SufBCD, and one SufA protein [54]. SufS is a cysteine desulfurase that forms a complex with SufE, which has an unknown function. The SufSE complex mobilizes sulfur atoms from cysteine and provides sulfur atoms to the SufBCD complex to assemble an FeS cluster [54], which is a cofactor of several enzymes such as aconitase, succinate dehydrogenase, and glutamate synthase [54,55,56].
Aconitase is an enzyme that catalyzes the isomerization of citrate to isocitrate in the TCA cycle [55,57]. Mutating the gene encoding for aconitase resulted in a decrease in biofilm formation, possibly due to the interference of citrate flow in the TCA cycle [57]. Glutamate synthase is an FeS cluster-containing enzyme that connects the TCA cycle to nitrogen metabolism by converting 2-ketoglutarate, a TCA cycle intermediate, into glutamate, which contributes to ammonium assimilation and the biosynthesis of nitrogen-containing compounds such as amino acids [58]. B. subtilis cells with a mutation in gltA, the gene encoding for glutamate synthase, formed thin and flat pellicles and defective biofilms compared to the wildtype cells [58]. The biofilm defect in mutant cells could have resulted from an accumulation of citrate as evidenced by the restored biofilms in a gltA mutant with limited citrate synthase activity [58]. The relationship between citrate concentration in growth media and biofilm formation has been studied previously [59]. A high citrate concentration (2%) was bactericidal, while a low concentration (0.2%) facilitated biofilm formation [59]. Additionally, the accumulation of citrate in gltA mutant cells could cause iron shortage by chelating [58]. Iron shortage can induce a planktonic mode of growth in the liquid phase, while a high iron concentration stimulates cell aggregation and biofilm formation [60]. Thus, the insertion of mini-Tn10 into sufSE could likely fail to activate FeS assembly-dependent metabolic enzymes, aconitase, and glutamate synthase, subsequently interfering with the TCA cycle, resulting in an iron shortage due to the accumulation of TCA cycle intermediates and, consequently, defective biofilm formation.
Other than those identified in the current study, other Salmonella genes that play a role in biofilm formation have been identified in previous studies. Mutations in flgK and rfbA, encoding flagella and lipopolysaccharide production, respectively, in S. typhimurium DT104, resulted in decreased biofilm formation [45]. Solano et al. found that the synthesis of exopolysaccharide cellulose encoded by bcsABZC and bcsEFG operons was required for biofilm formation by S. enteritidis [13]. Barak et al. reported that mutations in the genes of S. enteritidis regulating curli, cellulose, and pili production reduced the ability of pathogenic cells to attach to plant tissues [12]. Chen and Wang found that the level of biofilm mass significantly decreased when cdg, trx2, and rtx were interrupted by mini-Tn10 insertion, while the interruption of fadI gene significantly increased the level of biofilm formation [61].
This study identified some of the genes that are involved in biofilm formation by S. enteritidis, namely those that are involved in the biosynthesis of aldehyde dehydrogenase (EutE), cysteine desulfurase (SufS or SufE), a transporter protein, porin OmpL, and a ribbon–helix–helix protein from the CopG family. The research could serve as a starting point for a series of future studies. Knock-off mutants could be constructed to verify the function of the genes. If successful, the proteins encoded by the genes characterized in the current study could serve as potential targets for the development of antimicrobial interventions to prevent pathogen contamination in low-moisture foods and their production environment.
Author Contributions
Conceptualization, J.C.; methodology, J.C. and S.L.; formal analysis, S.L.; resources, J.C.; data curation, S.L.; writing—original draft preparation, S.L.; writing—review and editing, J.C.; supervision, J.C.; project administration, J.C.; funding acquisition, J.C. All authors have read and agreed to the published version of the manuscript.
Funding
This material is based upon work that is supported by the National Institute of Food and Agriculture, U.S. Department of Agriculture, under award number 2014-67017-21705.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.-A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States-major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Center for Disease Control and Prevention. Salmonellosis, Nontyphoidal CDC Yellow Book. 2024. Available online: https://wwwnc.cdc.gov/travel/yellowbook/2024/infections-diseases/salmonellosis-nontyphoidal#:~:text=Epidemiology,travelers%20who%20return%20with%20diarrhea (accessed on 25 April 2024).
- Crum-Cianflone, N.F. Salmonellosis and the gastrointestinal tract: More than just peanut butter. Curr. Gastroenterol. Rep. 2008, 10, 424–431. [Google Scholar] [CrossRef]
- Beuchat, L.R.; Komitopolou, E.; Beckers, H.; Betts, R.P.; Bourdichon, F.; Fanning, S.; Joosten, H.M.; Ter Kuile, B.H. Low–water activity foods: Increased concern as vehicles of foodborne pathogens. J. Food Prot. 2013, 76, 150–172. [Google Scholar] [CrossRef]
- Finn, S.; Condell, O.; McClure, P.; Amézquita, A.; Fanning, S. Mechanisms of survival, responses and sources of Salmonella in low-moisture environments. Front. Microbiol. 2013, 4, 331. [Google Scholar] [CrossRef]
- Podolak, R.; Enache, E.; Stone, W.; Black, D.G.; Elliott, P.H. Sources and risk factors for contamination, survival, persistence, and heat resistance of Salmonella in low-moisture foods. J. Food Prot. 2010, 73, 1919–1936. [Google Scholar] [CrossRef]
- Gruzdev, N.; Pinto, R.; Sela, S. Effect of desiccation on tolerance of Salmonella enterica to multiple stresses. Appl. Environ. Microbiol. 2011, 77, 1667–1673. [Google Scholar] [CrossRef]
- Jayeola, V.; McClelland, M.; Porwollik, S.; Chu, W.; Farber, J.; Kathariou, S. Identification of novel genes mediating survival of Salmonella on low-moisture foods via transposon sequencing analysis. Front. Microbiol. 2020, 11, 726. [Google Scholar] [CrossRef]
- Iibuchi, R.; Hara-Kudo, Y.; Hasegawa, A.; Kumagai, S. Survival of Salmonella on a polypropylene surface under dry conditions in relation to biofilm-formation capability. J. Food Prot. 2010, 73, 1506–1510. [Google Scholar] [CrossRef]
- White, A.P.; Gibson, D.L.; Kim, W.; Kay, W.W.; Surette, M.G. Thin aggregative fimbriae and cellulose enhance long-term survival and persistence of Salmonella. J. Bacteriol. 2006, 188, 3219–3227. [Google Scholar] [CrossRef]
- Vestby, L.K.; Møretrø, T.; Ballance, S.; Langsrud, S.; Nesse, L.L. Survival potential of wild type cellulose deficient Salmonella from the feed industry. BMC Vet. Res. 2009, 5, 43. [Google Scholar] [CrossRef]
- Barak, J.D.; Gorski, L.; Naraghi-Arani, P.; Charkowski, A.O. Salmonella enterica virulence genes are required for bacterial attachment to plant tissue. Appl. Environ. Microbiol. 2005, 71, 5685–5691. [Google Scholar] [CrossRef]
- Solano, C.; García, B.; Valle, J.; Berasain, C.; Ghigo, J.-M.; Gamazo, C.; Lasa, I. Genetic analysis of Salmonella enteritidis biofilm formation: Critical role of cellulose. Mol. Microbiol. 2002, 43, 793–808. [Google Scholar] [CrossRef]
- O’Toole, G.; Kaplan, H.B.; Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000, 54, 49–79. [Google Scholar] [CrossRef]
- Isaacs, S.; Aramini, J.; Ciebin, B.; Farrar, J.A.; Ahmed, R.; Middleton, D.; Chandran, A.U.; Harris, L.J.; Howes, M.; Chan, E.; et al. An international outbreak of salmonellosis associated with raw almonds contaminated with a rare phage type of Salmonella Enteritidis. J. Food Prot. 2005, 68, 191–198. [Google Scholar] [CrossRef]
- Freed, N.E. Creation of a dense transposon insertion library using bacterial conjugation in enterobacterial strains such as Escherichia coli or Shigella flexneri. J. Vis. Exp. 2017, 12, e56216. [Google Scholar]
- Gazula, H.; Scherm, H.; Li, C.; Takeda, F.; Wang, P.; Chen, J. Ease of biofilm accumulation, and efficacy of sanitizing treatments in removing the biofilms formed, on coupons made of materials commonly used in blueberry packing environment. Food Control 2019, 104, 167–173. [Google Scholar] [CrossRef]
- Jain, S.; Chen, J. Attachment and biofilm formation by various serotypes of Salmonella as influenced by cellulose production and thin aggregative fimbriae biosynthesis. J. Food Prot. 2007, 70, 2473–2479. [Google Scholar] [CrossRef]
- Garsin, D.A. Ethanolamine utilization in bacterial pathogens: Roles and regulation. Nat. Rev. Microbiol. 2010, 8, 290–295. [Google Scholar] [CrossRef]
- Kaval, K.G.; Garsin, D.A. Ethanolamine utilization in bacteria. mBio 2018, 9, e00066-18. [Google Scholar] [CrossRef]
- Thiennimitr, P.; Winter, S.E.; Winter, M.G.; Xavier, M.N.; Tolstikov, V.; Huseby, D.L.; Sterzenbach, T.; Tsolis, R.M.; Roth, J.R.; Bäumler, A.J. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. USA 2011, 108, 17480–17485. [Google Scholar] [CrossRef]
- Kofoid, E.; Rappleye, C.; Stojiljkovic, I.; Roth, J. The 17-Gene Ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J. Bacteriol. 1999, 181, 5317–5329. [Google Scholar] [CrossRef]
- Prüß, B.M. Involvement of two-component signaling on bacterial motility and biofilm development. J. Bacteriol. 2017, 199, e00259-17. [Google Scholar] [CrossRef]
- Prüβ, B.M.; Wolfe, A.J. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol. Microbiol. 1994, 12, 973–984. [Google Scholar] [CrossRef]
- Shin, S.; Park, C. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J. Bacteriol. 1995, 177, 4696–4702. [Google Scholar] [CrossRef]
- Fredericks, C.E.; Shibata, S.; Aizawa, S.-I.; Reimann, S.A.; Wolfe, A.J. Acetyl phosphate-sensitive regulation of flagellar biogenesis and capsular biosynthesis depends on the Rcs phosphorelay. Mol. Microbiol. 2006, 61, 734–747. [Google Scholar] [CrossRef]
- Jubelin, G.; Vianney, A.; Beloin, C.; Ghigo, J.-M.; Lazzaroni, J.-C.; Lejeune, P.; Dorel, C. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 2005, 187, 2038–2049. [Google Scholar] [CrossRef]
- Prigent-Combaret, C.; Prensier, G.; Le Thi, T.T.; Vidal, O.; Lejeune, P.; Dorel, C. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: Role of flagella, curli and colanic acid. Environ. Microbiol. 2000, 2, 450–464. [Google Scholar] [CrossRef]
- Latasa, C.; García, B.; Echeverz, M.; Toledo-Arana, A.; Valle, J.; Campoy, S.; García-del Portillo, F.; Solano, C.; Lasa, I. Salmonella biofilm development depends on the phosphorylation status of RcsB. J. Bacteriol. 2012, 194, 3708–3722. [Google Scholar] [CrossRef]
- Mugabi, R.; Sandgren, D.; Born, M.; Leith, I.; Horne, S.M.; Prüβ, B.M. The role of activated acetate intermediates in the control of Escherichia coli biofilm amounts. WebmedCentral 2012, 3, 3577. [Google Scholar]
- Prüß, B.M.; Verma, K.; Samanta, P.; Sule, P.; Kumar, S.; Wu, J.; Christianson, D.; Horne, S.M.; Stafslien, S.J.; Wolfe, A.J.; et al. Environmental and genetic factors that contribute to Escherichia coli K-12 biofilm formation. Arch. Microbiol. 2010, 192, 715–728. [Google Scholar] [CrossRef]
- Skagia, A.; Zografou, C.; Vezyri, E.; Venieraki, A.; Katinakis, P.; Dimou, M. Cyclophilin PpiB is involved in motility and biofilm formation via its functional association with certain proteins. Genes Cells 2016, 21, 833–851. [Google Scholar] [CrossRef] [PubMed]
- Stillwell, W. Chapter 19-Membrane Transport. In An Introduction to Biological Membranes, 2nd ed.; Stillwell, W., Ed.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 423–451. [Google Scholar]
- Tarling, E.J.; de Aguiar Vallim, T.Q.; Edwards, P.A. Role of ABC transporters in lipid transport and human disease. Trends Endocrinol. Metab. 2013, 24, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.S.; Currie, M.J.; Wright, J.D.; Newton-Vesty, M.C.; North, R.A.; Mace, P.D.; Allison, J.R.; Dobson, R.C.J. Selective nutrient transport in bacteria: Multicomponent transporter systems reign supreme. Front. Mol. Biosci. 2021, 8, 578. [Google Scholar] [CrossRef] [PubMed]
- Monds, R.D.; Silby, M.W.; Mahanty, H.K. Expression of the Pho regulon negatively regulates biofilm formation by Pseudomonas aureofaciens PA147-2. Mol. Microbiol. 2001, 42, 415–426. [Google Scholar] [CrossRef] [PubMed]
- O’May, G.A.; Jacobsen, S.M.; Longwell, M.; Stoodley, P.; Mobley, H.L.T.; Shirtliff, M.E. The high-affinity phosphate transporter Pst in Proteus mirabilis HI4320 and its importance in biofilm formation. Microbiology 2009, 155, 1523–1535. [Google Scholar] [CrossRef] [PubMed]
- Luz, D.; Nepomuceno, R.; Spira, B.; Ferreira, R. The Pst system of Streptococcus mutans is important for phosphate transport and adhesion to abiotic surfaces. Mol. Oral. Microbiol. 2012, 27, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Monds, R.D.; Newell, P.D.; Gross, R.H.; O’Toole, G.A. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 2007, 63, 656–679. [Google Scholar] [CrossRef] [PubMed]
- Haddad, A.; Jensen, V.; Becker, T.; Häussler, S. The Pho regulon influences biofilm formation and type three secretion in Pseudomonas aeruginosa. Environ. Microbiol. Rep. 2009, 1, 488–494. [Google Scholar] [CrossRef]
- Valentini, M.; Filloux, A. Biofilms and cyclic di-GMP (c-di-GMP) signaling: Lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 2016, 291, 12547–12555. [Google Scholar] [CrossRef]
- Hinsa, S.M.; Espinosa-Urgel, M.; Ramos, J.L.; O’Toole, G.A. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 2003, 49, 905–918. [Google Scholar] [CrossRef]
- Freeman, T.C.; Landry, S.J.; Wimley, W.C. The prediction and characterization of YshA, an unknown outer-membrane protein from Salmonella typhimurium. Biochim. Biophys. Acta BBA-Biomembr. 2011, 1808, 287–297. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, Y.; Wan, C.; Xu, H.; Wei, H. Identification and characterization of OmpL as a potential vaccine candidate for immune-protection against salmonellosis in mice. Vaccine 2013, 31, 2930–2936. [Google Scholar] [CrossRef]
- Kim, S.H.; Wei, C.I. Molecular characterization of biofilm formation and attachment of Salmonella enterica serovar Typhimurium DT104 on food contact surfaces. J. Food Prot. 2009, 72, 1841–1847. [Google Scholar] [CrossRef]
- Gibson, D.L.; White, A.P.; Snyder, S.D.; Martin, S.; Heiss, C.; Azadi, P.; Surette, M.; Kay, W.W. Salmonella produces an O-antigen capsule regulated by AgfD and important for environmental persistence. J. Bacteriol. 2006, 188, 7722–7730. [Google Scholar] [CrossRef]
- Crawford, R.W.; Gibson, D.L.; Kay, W.W.; Gunn, J.S. Identification of a bile-induced exopolysaccharide required for Salmonella biofilm formation on gallstone surfaces. Infect. Immun. 2008, 76, 5341–5349. [Google Scholar] [CrossRef]
- Barak, J.D.; Jahn, C.E.; Gibson, D.L.; Charkowski, A.O. The role of cellulose and O-antigen capsule in the colonization of plants by Salmonella enterica. Mol. Plant-Microbe Interact. 2007, 20, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.T.; Espinosa, M.; Yeo, C.C. Keeping the wolves at bay: Antitoxins of prokaryotic type II toxin-antitoxin systems. Front. Mol. Biosci. 2016, 3, 9. [Google Scholar] [CrossRef]
- Yan, X.; Gurtler, J.B.; Fratamico, P.M.; Hu, J.; Juneja, V.K. Phylogenetic identification of bacterial MazF toxin protein motifs among probiotic strains and foodborne pathogens and potential implications of engineered probiotic intervention in food. Cell Biosci. 2012, 2, 39. [Google Scholar] [CrossRef] [PubMed]
- Mohammadzadeh, R.; Shivaee, A.; Ohadi, E.; Kalani, B.S. In silico insight into the dominant type II toxin–antitoxin systems and Clp proteases in Listeria monocytogenes and designation of derived peptides as a novel approach to interfere with this system. Int. J. Pept. Res. Ther. 2020, 26, 613–623. [Google Scholar] [CrossRef]
- Kolodkin-Gal, I.; Verdiger, R.; Shlosberg-Fedida, A.; Engelberg-Kulka, H. A Differential effect of E. coli toxin-antitoxin systems on cell death in liquid media and biofilm formation. PLoS ONE 2009, 4, e6785. [Google Scholar] [CrossRef]
- Ma, D.; Mandell, J.B.; Donegan, N.P.; Cheung, A.L.; Ma, W.; Rothenberger, S.; Shanks, R.M.Q.; Richardson, A.R.; Urish, K.L. The toxin-antitoxin MazEF drives Staphylococcus aureus biofilm formation, antibiotic tolerance, and chronic infection. mBio 2019, 10, e01658-19. [Google Scholar] [CrossRef] [PubMed]
- Loiseau, L.; Ollagnier-de-Choudens, S.; Nachin, L.; Fontecave, M.; Barras, F. Biogenesis of Fe-S cluster by the bacterial Suf system: SufS and SufE form a new type of cysteine desulfurase. J. Biol. Chem. 2003, 278, 38352–38359. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, A.G.; Netz, D.J.A.; Miethke, M.; Pierik, A.J.; Burghaus, O.; Peuckert, F.; Lill, R.; Marahiel, M.A. SufU Is an essential iron-sulfur cluster scaffold protein in Bacillus subtilis. J. Bacteriol. 2010, 192, 1643–1651. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C.A.; Al-Tameemi, H.M.; Mashruwala, A.A.; Rosario-Cruz, Z.; Chauhan, U.; Sause, W.E.; Torres, V.J.; Belden, W.J.; Boyd, J.M. The Suf iron-sulfur cluster biosynthetic system is essential in Staphylococcus aureus, and decreased Suf function results in global metabolic defects and reduced survival in human neutrophils. Infect. Immun. 2017, 85, e00100-17. [Google Scholar] [CrossRef] [PubMed]
- Shanks, R.M.Q.; Meehl, M.A.; Brothers, K.M.; Martinez, R.M.; Donegan, N.P.; Graber, M.L.; Cheung, A.L.; O’Toole, G.A. Genetic evidence for an alternative citrate-dependent biofilm formation pathway in Staphylococcus aureus that is dependent on fibronectin binding proteins and the GraRS two-component regulatory system. Infect. Immun. 2008, 76, 2469–2477. [Google Scholar] [CrossRef]
- Kimura, T.; Kobayashi, K. Role of glutamate synthase in biofilm formation by Bacillus subtilis. J. Bacteriol. 2020, 202, e00120-20. [Google Scholar] [CrossRef] [PubMed]
- Shanks, R.M.Q.; Sargent, J.L.; Martinez, R.M.; Graber, M.L.; O’Toole, G.A. Catheter lock solutions influence staphylococcal biofilm formation on abiotic surfaces. Nephrol. Dial. Transplant. 2006, 21, 2247–2255. [Google Scholar] [CrossRef] [PubMed]
- Berlutti, F.; Morea, C.; Battistoni, A.; Sarli, S.; Cipriani, P.; Superti, F.; Ammendolia, M.G.; Valenti, P. Iron availability influences aggregation, biofilm, adhesion and invasion of Pseudomonas aeruginosa and Burkholderia cenocepacia. Int. J. Immunopathol. Pharmacol. 2005, 18, 661–670. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Y. Genetic determinants of Salmonella enterica critical for attachment and biofilm formation. Int. J. Food Microbiol. 2020, 320, 108524. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).