Transcriptional Response of Salmonella enterica to Bacteriophage Treatments with Differential Multiplicities of Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteriophage Propagation and Titer Measurement
2.2. S. enterica Strain Storage Conditions
2.3. S. enterica Growth Assay
2.4. RNA Extraction, Reverse-Transcription and Real-Time Quantitative PCR (RT-qPCR)
2.5. Statistical Analysis
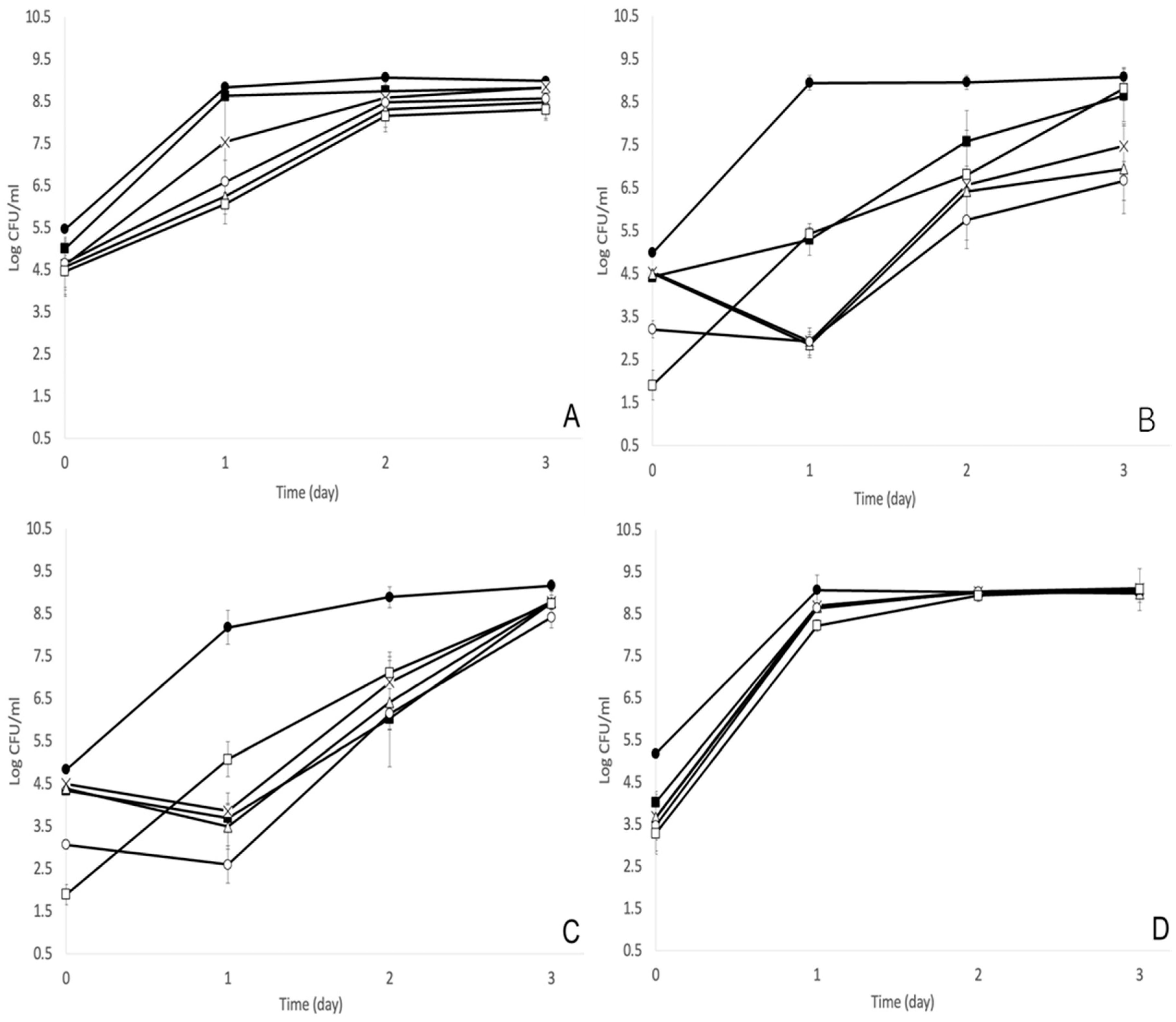
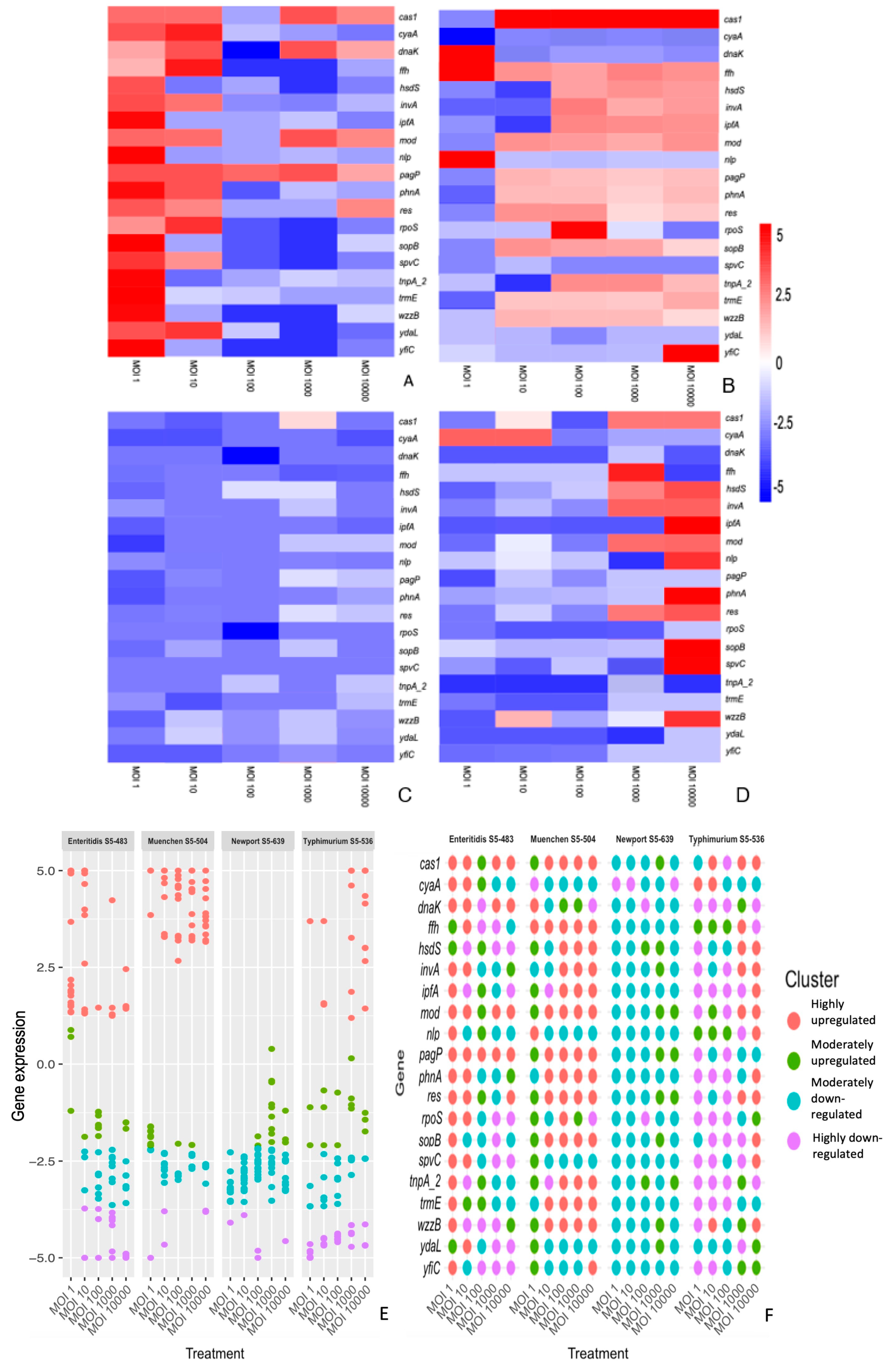
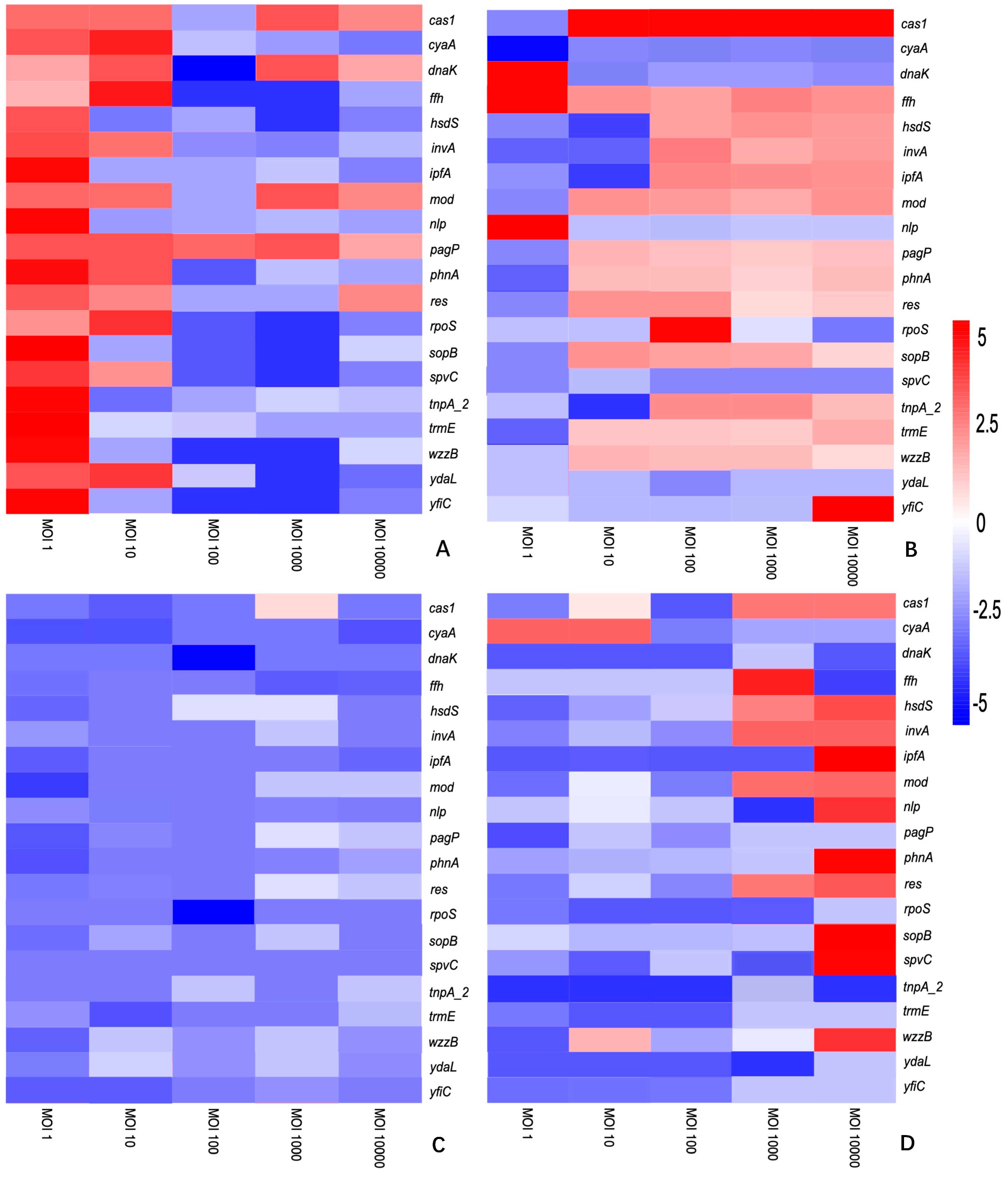
3. Results and Discussion
3.1. MOI of 1000 Was the Optimal Bacteriophage Treatment against S. enterica
3.2. The Differential Expression of Genes Is Dependent on the S. enterica Strain, Bacteriophage and Time of Treatment
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Centers for Disease Control and Prevention List of Selected Multistate Foodborne Outbreak Notices. Available online: https://www.cdc.gov/foodsafety/outbreaks/lists/outbreaks-list.html (accessed on 8 August 2023).
- Abdelhamid, A.G.; El-Dougdoug, N.K. Controlling Foodborne Pathogens with Natural Antimicrobials by Biological Control and Antivirulence Strategies. Heliyon 2020, 6, E05020. [Google Scholar] [CrossRef]
- Sahoo, M.; Panigrahi, C.; Aradwad, P. Management Strategies Emphasizing Advanced Food Processing Approaches to Mitigate Food Borne Zoonotic Pathogens in Food System. Food Front. 2022, 3, 641–665. [Google Scholar] [CrossRef]
- Carlson, A.; Skorbiansky, S.R. Strong Growth in Organic Market Slowed in 2021; U.S. Department of Agriculture: Washington, DC, USA, 2021.
- Połaska, M.; Sokołowska, B. Bacteriophages—A New Hope or a Huge Problem in the Food Industry. AIMS Microbiol. 2019, 5, 324–347. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, J.; Lv, M.; Shao, Z.; Hungwe, M.; Wang, J.; Bai, X.; Xie, J.; Wang, Y.; Geng, W. Metabolism Characteristics of Lactic Acid Bacteria and the Expanding Applications in Food Industry. Front. Bioeng. Biotechnol. 2021, 9, 612285. [Google Scholar] [CrossRef] [PubMed]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage Applications for Food Production and Processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef] [PubMed]
- Endersen, L.; Coffey, A. The Use of Bacteriophages for Food Safety. Curr. Opin. Food Sci. 2020, 36, 1–8. [Google Scholar] [CrossRef]
- Fernández, L.; Gutiérrez, D.; Rodríguez, A.; García, P. Application of Bacteriophages in the Agro-Food Sector: A Long Way Toward Approval. Front. Cell. Infect. Microbiol. 2018, 8, 296. [Google Scholar] [CrossRef] [PubMed]
- Sillankorva, S.M.; Oliveira, H.; Azeredo, J. Bacteriophages and Their Role in Food Safety. Int. J. Microbiol. 2012, 2012, 863945. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, M.; Martínez, S.; Tello, M.; Gallardo, M.J.; García, V. Use of Cocktail of Bacteriophage for Salmonella Typhimurium Control in Chicken Meat. Foods 2022, 11, 1164. [Google Scholar] [CrossRef]
- Wong, C.W.Y.; Delaquis, P.; Goodridge, L.; Lévesque, R.C.; Fong, K.; Wang, S. Inactivation of Salmonella enterica on Post-Harvest Cantaloupe and Lettuce by a Lytic Bacteriophage Cocktail. Curr. Res. Food Sci. 2020, 2, 25–32. [Google Scholar] [CrossRef]
- Kocharunchitt, C.; Ross, T.; McNeil, D.L. Use of Bacteriophages as Biocontrol Agents to Control Salmonella Associated with Seed Sprouts. Int. J. Food Microbiol. 2009, 128, 453–459. [Google Scholar] [CrossRef]
- Fong, K.; LaBossiere, B.; Switt, A.I.M.; Delaquis, P.; Goodridge, L.; Levesque, R.C.; Danyluk, M.D.; Wang, S. Characterization of Four Novel Bacteriophages Isolated from British Columbia for Control of Non-Typhoidal Salmonella in Vitro and on Sprouting Alfalfa Seeds. Front. Microbiol. 2017, 8, 2193. [Google Scholar] [CrossRef]
- Wong, C.W.Y.; Wang, S. Efficacy of Repeated Applications of Bacteriophages on Salmonella enterica-Infected Alfalfa Sprouts during Germination. Pathogens 2022, 11, 1156. [Google Scholar] [CrossRef]
- Fong, K.; Tremblay, D.M.; Delaquis, P.; Goodridge, L.; Levesque, R.C.; Moineau, S.; Suttle, C.A.; Wang, S. Diversity and Host Specificity Revealed by Biological Characterization and Whole Genome Sequencing of Bacteriophages Infecting Salmonella enterica. Viruses 2019, 11, 854. [Google Scholar] [CrossRef]
- Jackson, B.R.; Griffin, P.M.; Cole, D.; Walsh, K.A.; Chai, S.J. Outbreak-associated Salmonella enterica serotypes and food commodities, United States, 1998–2008. Emerg. Infect. Dis. 2013, 19, 1239–1244. [Google Scholar] [CrossRef]
- Hatfield, J.L.; Prueger, J.H. Temperature extremes: Effect on plant growth and development. Weather Clim. Extrem. 2015, 10, 4–10. [Google Scholar] [CrossRef]
- Motsa, M.M.; Slabbert, M.M.; van Averbeke, W.; Morey, L. Effect of light and temperature on seed germination of selected African leafy vegetables. S. Afr. J. Bot. 2015, 99, 29–35. [Google Scholar] [CrossRef]
- Pazzagli, M.; Malentacchi, F.; Simi, L.; Orlando, C.; Wyrich, R.; Günther, K.; Hartmann, C.C.; Verderio, P.; Pizzamiglio, S.; Ciniselli, C.M.; et al. SPIDIA-RNA: First External Quality Assessment for the Pre-Analytical Phase of Blood Samples Used for RNA Based Analyses. Methods 2013, 59, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Padhi, B.K.; Singh, M.; Rosales, M.; Pelletier, G.; Cakmak, S. A PCR-Based Quantitative Assay for the Evaluation of MRNA Integrity in Rat Samples. Biomol. Detect. Quantif. 2018, 15, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Fleige, S.; Pfaffl, M.W. RNA Integrity and the Effect on the Real-Time QRT-PCR Performance. Mol. Asp. Med. 2006, 27, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Nie, T.; Lin, F.; Connerton, I.F.; Lu, Z.; Zhou, S.; Hang, H. Resistance Mechanisms Adopted By a Salmonella Typhimurium Mutant Against Bacteriophage. Virus Res. 2019, 273, 197759. [Google Scholar] [CrossRef]
- Fernández, P.A.; Zabner, M.; Ortega, J.; Morgado, C.; Amaya, F.; Vera, G.; Rubilar, C.; Salas, B.; Cuevas, V.; Valenzuela, C.; et al. Novel Template Plasmids PCyaA’-Kan and PCyaA’-Cam for Generation of Unmarked Chromosomal CyaA’ Translational Fusion to T3SS Effectors in Salmonella. Microorganisms 2021, 9, 475. [Google Scholar] [CrossRef]
- Ramos-Morales, F.; Cardenal-Munoz, E.; Cordero-Alba, M.; Baison-Olmo, F. Generation and Use of Site-Directed Chromosomal CyaA′ Translational Fusions in Salmonella enterica. In Salmonella Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2015; Volume 2, pp. 93–104. [Google Scholar]
- Takaya, A.; Tomoyasu, T.; Matsui, H.; Yamamoto, T. The DnaK/DnaJ Chaperone Machinery of Salmonella enterica Serovar Typhimurium Is Essential for Invasion of Epithelial Cells and Survival within Macrophages, Leading to Systemic Infection. Infect. Immun. 2004, 72, 1364–1373. [Google Scholar] [CrossRef]
- Peschke, M.; Le Goff, M.; Koningstein, G.M.; Karyolaimos, A.; de Gier, J.W.; van Ulsen, P.; Luirink, J. SRP, FtsY, DnaK and YidC Are Required for the Biogenesis of the E. Coli Tail-Anchored Membrane Proteins DjlC and Flk. J. Mol. Biol. 2018, 430, 389–403. [Google Scholar] [CrossRef]
- Mohammed, B.T. Identification and Bioinformatic Analysis of InvA Gene of Salmonella in Free Range Chicken. Braz. J. Biol. 2022, 84, e263363. [Google Scholar] [CrossRef]
- Siddiky, N.A.; Sarker, M.S.; Khan, M.S.R.; Begum, R.; Kabir, M.E.; Karim, M.R.; Rahman, M.T.; Mahmud, A.; Samad, M.A. Virulence and Antimicrobial Resistance Profiles of Salmonella enterica Serovars Isolated from Chicken at Wet Markets in Dhaka, Bangladesh. Microorganisms 2021, 9, 952. [Google Scholar] [CrossRef] [PubMed]
- Yehia, H.M.; Fawzy Elkhadragy, M.; Al-Masoud1, A.A.; Al-Dagal1, M.M.; Al-Dagal, M. InvA Gene to Detect Salmonella enterica Serovar Typhimurium Supported by Serum Anti-Salmonella Antibodies and Protein Proles for Chicken Carcass Isolates. Res. Sq. 2020, 1–14. [Google Scholar] [CrossRef]
- Aviv, G.; Elpers, L.; Mikhlin, S.; Cohen, H.; Vitman Zilber, S.; Grassl, G.A.; Rahav, G.; Hensel, M.; Gal-Mor, O. The Plasmid-Encoded Ipf and Klf Fimbriae Display Different Expression and Varying Roles in the Virulence of Salmonella enterica Serovar Infantis in Mouse vs. Avian Hosts. PLoS Pathog. 2017, 13, e1006559. [Google Scholar] [CrossRef] [PubMed]
- Canals, R.; Hammarlöf, D.L.; Kröger, C.; Owen, S.V.; Yee Fong, W.; Zhu, X.; Wenner, N.; Carden, S.E.; Honeycutt, J.; Monack, M.; et al. Adding Function to the Genome of African Salmonella ST313. PLoS Biol. 2019, 17, e3000059. [Google Scholar] [CrossRef]
- Elfenbein, J.R.; Knodler, L.A.; Schaeffer, A.R.; Faber, F.; Bäumler, A.J.; Andrews-Polymenis, H.L. A Salmonella Regulator Modulates Intestinal Colonization and Use of Phosphonoacetic Acid. Front. Cell. Infect. Microbiol. 2017, 7, 69. [Google Scholar] [CrossRef]
- Kulakova, A.N.; Kulakov, L.A.; Akulenko, N.V.; Ksenzenko, V.N.; Hamilton, J.T.G.; Quinn, J.P. Structural and Functional Analysis of the Phosphonoacetate Hydrolase (PhnA) Gene Region in Pseudomonas Fluorescens 23F. J. Bacteriol. 2001, 183, 3268–3275. [Google Scholar] [CrossRef]
- Franco Meléndez, K.; Crenshaw, K.; Barrila, J.; Yang, J.; Gangaraju, S.; Davis, R.R.; Forsyth, R.J.; Ott, C.M.; Kader, R.; Curtiss, R.; et al. Role of RpoS in Regulating Stationary Phase Salmonella Typhimurium Pathogenesis-Related Stress Responses under Physiological Low Fluid Shear Force Conditions. mSphere 2022, 7, e00210-22. [Google Scholar] [CrossRef] [PubMed]
- Giacomodonato, M.N.; Sarnacki, S.H.; Llana, M.N.; Cerquetti, M.C. SopB Effector Protein of Salmonella Typhimurium Is Translocated in Mesenteric Lymph Nodes during Murine Salmonellosis. FEMS Microbiol. Lett. 2011, 317, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Xu, Q.; Cui, Y.; Yao, S.; Jin, S.; Zhang, Q.; Wen, Z.; Ruan, H.; Liang, X.; Chao, Y.; et al. Salmonella Effector SopB Reorganizes Cytoskeletal Vimentin to Maintain Replication Vacuoles for Efficient Infection. Nat. Commun. 2023, 14, 478. [Google Scholar] [CrossRef]
- Guiney, D.G.; Fierer, J. The Role of the Spv Genes in Salmonella Pathogenesis. Front. Microbiol. 2011, 2, 129. [Google Scholar] [CrossRef] [PubMed]
- Zuo, L.; Zhou, L.; Wu, C.; Wang, Y.; Li, Y.; Huang, R.; Wu, S. Salmonella spvC Gene Inhibits Pyroptosis and Intestinal Inflammation to Aggravate Systemic Infection in Mice. Front. Microbiol. 2020, 11, 562491. [Google Scholar] [CrossRef]
- Chang, H.K.; Mohseni, P.; Zylstra, G.J. Characterization and Regulation of the Genes for a Novel Anthranilate 1,2-Dioxygenase from Burkholderia Cepacia DBO1. J. Bacteriol. 2003, 185, 5871–5881. [Google Scholar] [CrossRef]
- Gall, A.R.; Datsenko, K.A.; Figueroa-Bossi, N.; Bossi, L.; Masuda, I.; Hou, Y.M.; Csonka, L.N. Mg2+ Regulates Transcription of MgtA in Salmonella Typhimurium via Translation of Proline Codons during Synthesis of the MgtL Peptide. Proc. Natl. Acad. Sci. USA 2016, 113, 15096–15101. [Google Scholar] [CrossRef]
- Thomason, M.K.; Fontaine, F.; De Lay, N.; Storz, G. A Small RNA That Regulates Motility and Biofilm Formation in Response to Changes in Nutrient Availability in Escherichia coli. Mol. Microbiol. 2012, 84, 17–35. [Google Scholar] [CrossRef]
- Golovina, A.Y.; Sergiev, P.V.; Golovin, A.V.; Serebryakova, M.V.; Demina, I.; Govorun, V.M.; Dontsova, O.A. The YfiC Gene of E. coli Encodes an Adenine-N6 Methyltransferase That Specifically Modifies A37 of TRNA1Val(Cmo5UAC). RNA 2009, 15, 1134–1141. [Google Scholar] [CrossRef]
- Mirzaei, M.K.; Nilsson, A.S. Isolation of Phages for Phage Therapy: A Comparison of Spot Tests and Efficiency of Plating Analyses for Determination of Host Range and Efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef]
- Abedon, S.T. Lysis from Without. Bacteriophage 2011, 1, 46. [Google Scholar] [CrossRef] [PubMed]
- Bull, J.J.; Christensen, K.A.; Scott, C.; Jack, B.R.; Crandall, C.J.; Krone, S.M. Phage-Bacterial Dynamics with Spatial Structure: Self Organization around Phage Sinks Can Promote Increased Cell Densities. Antibiotics 2018, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food & Drug Administration. GRAS Notice No. GRN 000435 [WWW Document]. 2021. Available online: https://www.fda.gov/media/151413/download (accessed on 25 September 2023).
- Rollins, M.C.F.; Chowdhury, S.; Carter, J.; Golden, S.M.; Wilkinson, R.A.; Bondy-Denomy, J.; Lander, G.C.; Wiedenheft, B. Cas1 and the Csy Complex Are Opposing Regulators of Cas2/3 Nuclease Activity. Proc. Natl. Acad. Sci. USA 2017, 114, E5113–E5121. [Google Scholar] [CrossRef] [PubMed]
- Dartois, W.; De Backer, O.; Colson, C. Sequence of the Salmonella Typhimurium StyLT 1 Restriction-Modification Genes: Homologies with EcoPI and EcoPS 5 Type-III R-M Systems and Presence of Helicase Domains. Gene 1993, 127, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Zaworski, J.; McClung, C.; Ruse, C.; Weigele, P.R.; Hendrix, R.W.; Ko, C.-C.; Edgar, R.; Hatfull, G.F.; Casjens, S.R.; Raleigh, E.A. Genome Analysis of Salmonella Enterica Serovar Typhimurium Bacteriophage L, Indicator for StySA (StyLT2III) Restriction-Modification System Action. G3 2021, 11, jkaa037. [Google Scholar] [CrossRef]
- Roer, L.; Hendriksen, R.S.; Leekitcharoenphon, P.; Lukjancenko, O.; Kaas, R.S.; Hasman, H.; Aarestrup, F.M. Is the Evolution of Salmonella enterica Subsp. Enterica Linked to Restriction-Modification Systems? mSystems 2016, 1, e00009-16. [Google Scholar] [CrossRef]
- Vasu, K.; Nagaraja, V. Diverse Functions of Restriction-Modification Systems in Addition to Cellular Defense. Microbiol. Mol. Biol. Rev. 2013, 77, 53–72. [Google Scholar] [CrossRef]




| Gene | Primer Direction | Sequence (5′ → 3′) |
|---|---|---|
| 16s | Forward | CAGAAGAAGCACCGGCTAAC |
| Reverse | GACTCAAGCCTGCCAGTTTC | |
| gapA | Forward | GGTGGTGACGTAGTGGCTGAA |
| Reverse | AGCGTTGGAAACGATGTCCTG | |
| rsrnC | Forward | GAAAAGCAGCCGCAGTTTAG |
| Reverse | CAGTTGGCTACCAACATCCA |
| Gene | Coding Protein | Function | Primer Direction | Sequence (5′ → 3′) | References |
|---|---|---|---|---|---|
| cas1 | CRISPR/Cas system-associated protein Cas1 | SOS response | Forward | GCAAAGCTGGCGTTAGATGA | [23] |
| Reverse | GATCCTTCAATACCGCGCAG | ||||
| cyaA | Adenylate cyclase | Metabolism and nutrient utilization | Forward | GTCCGCCGCTGCTTTTAC | [24,25] |
| Reverse | CGTCGTCCCATCCCCACT | ||||
| dnaK | Chaperon protein | Protein synthesis and folding | Forward | CGA TTA TGG ATG GAA CGC AGG | [26] |
| Reverse | GGC TGA CCA ACC AGA GTT | ||||
| ffh | Protein unit of signal recognition particle (SRP) | Protein synthesis and folding | Forward | TAA CAA GCG CCT GAC CCC | [27] |
| Reverse | AGA CCC GCC ATC AAT ACT | ||||
| hsdS | Type I restriction enzyme-specific protein | DNA modification and recombination | Forward | GTGTTCCTGTCCCACCTCTT | [23] |
| Reverse | TGGGATTTGCTCAAGACGTG | ||||
| invA | attachment/invasion protein | Pathogenicity/virulence | Forward | ACCGTGGTCCAGTTTATCGT | [23,28,29,30] |
| Reverse | GCTTTCCCTTTCCAGTACGC | ||||
| ipfA | Long polar fimbria | Pathogenicity/virulence | Forward | CTTCTGCTTTCGCTGCTGA | [29,31] |
| Reverse | CACAACTTCCTGGTTCTGG | ||||
| mod | Type III restriction–modification system methyltransferase | DNA modification and recombination | Forward | CCGAAACCGACCGCATTATT | [23] |
| Reverse | TTCATTCAGCGCCATTACCG | ||||
| nlp | ner-like regulatory protein | Cell envelope and membrane modification | Forward | ATGGATGTATGGAAAGCAAGC | [32] |
| Reverse | GCCAGCGTGGATGAACTC | ||||
| pagP | Lipid A palmitoyltransferase pagP | Cell envelope and membrane modification | Forward | GCG ACA GCC TGA GCA TTA T | [23] |
| Reverse | GGG ACT GAC CAA AAC CAA C | ||||
| phnA | Phosphonoacetate hydrolase (carbon-phosphorus bond cleavage enzyme) | Metabolism and nutrient utilization | Forward | CACTGCCCGCAATGTAAC | [25,33,34] |
| Reverse | CTTTGGTGCCGATTTTCA | ||||
| res | Type III restriction–modification system DNA helicase | DNA modification and recombination | Forward | ATTCGTGACAATAAGCCCGC | [23] |
| Reverse | CTACGCTCGAATTGCAGCAT | ||||
| rpoS | RNA polymerase sigma factor rpoS | Stress resistance, nutrient utilization, and biofilm formation | Forward | GAATCTGACGAACACGCTCA | [35] |
| Reverse | CCACGCAAGATGACGATATG | ||||
| sopB | Type III secretion system effector protein | Pathogenicity/virulence | Forward | GCTCGCCCGGAAATTATTGT | [23,36,37] |
| Reverse | GGTCCGCTTTAACTTTGGCT | ||||
| spvC | Salmonella plasmid virulence protein | Pathogenicity/virulence | Forward | ATTTGCCGGTGACAAGTTCC | [23,29,38,39] |
| Reverse | GGAGAAACGACGCACTGTAC | ||||
| tnpA_2 | Transposase | Transposition | Forward | CTTTACGAGCAGTTTGGGG | [40] |
| Reverse | CTGTATCTTCGCCGTGTTC | ||||
| trmE | tRNA methyltransferase | Protein synthesis and folding | Forward | CGTTGACCCTGCCGACAT | [41] |
| Reverse | CGAACCAGTGAGTGACCG | ||||
| wzzB | Polysaccharide antigen chain length regulator | Cell envelope and membrane modification | Forward | ACAGTTATGGCGTGGGAAGA | [23] |
| Reverse | GTTGAGCGCGTTGGTATAGG | ||||
| ydaL | Small MutS-related protein | Protein synthesis and folding | Forward | TCC TGA CGA CGG GCT TTC | [42] |
| Reverse | TTC CCA CTG CGT AAC | ||||
| yfiC | Recombinant protein | Metabolism and nutrient utilization | Forward | CTGGCTCCGTTCTTCGTC | [25,43] |
| Reverse | CACATTGTCATCCGTCCG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wong, C.W.Y.; Wang, S. Transcriptional Response of Salmonella enterica to Bacteriophage Treatments with Differential Multiplicities of Infection. Appl. Microbiol. 2024, 4, 390-405. https://doi.org/10.3390/applmicrobiol4010027
Wong CWY, Wang S. Transcriptional Response of Salmonella enterica to Bacteriophage Treatments with Differential Multiplicities of Infection. Applied Microbiology. 2024; 4(1):390-405. https://doi.org/10.3390/applmicrobiol4010027
Chicago/Turabian StyleWong, Catherine W. Y., and Siyun Wang. 2024. "Transcriptional Response of Salmonella enterica to Bacteriophage Treatments with Differential Multiplicities of Infection" Applied Microbiology 4, no. 1: 390-405. https://doi.org/10.3390/applmicrobiol4010027
APA StyleWong, C. W. Y., & Wang, S. (2024). Transcriptional Response of Salmonella enterica to Bacteriophage Treatments with Differential Multiplicities of Infection. Applied Microbiology, 4(1), 390-405. https://doi.org/10.3390/applmicrobiol4010027






