Abstract
The co-cultivation of sake yeast (AK25, K901, K1401, or K1801 strain) and the kuratsuki Bacillus A-10 and/or Priestia B-12 strains in koji solution was performed to demonstrate the effects of these two kuratsuki bacteria on sake taste. The results showed that the Brix and acidity patterns of sake preparations produced with and without these kuratsuki bacteria were very similar. This indicated that the addition of these kuratsuki bacteria did not inhibit ethanol fermentation or organic acid production by sake yeast. A taste recognition device showed that the effects of these kuratsuki bacteria on the saltiness and sourness of sake were greater than those on other taste properties. Astringency stimulation and saltiness of sake produced using the sake yeast K901 were increased by Bacillus A-10 and decreased by Priestia B-12. Except for these two cases, the taste intensities of sake preparations produced with the Bacillus A-10 and Priestia B-12 strains were very similar, but differed from those of sake produced with kuratsuki Kocuria. These results support our hypothesis that the flavor and taste of sake can be controlled by utilizing the interactions between kuratsuki bacteria and sake yeast. For crating the desired sake taste, a combination of kuratsuki bacteria and sake yeast should be considered.
1. Introduction
Sake, a traditional Japanese fermented alcoholic beverage, is produced from steamed polished rice, koji, and water, using koji mold (Aspergillus oryzae) and sake yeast (Saccharomyces cerevisiae). The koji mold and sake yeast convert rice starch into sugar and sugar into ethanol, respectively. Some lactic acid bacteria are used as fermentation starters, and this regards approximately 10% of all sake products. Bacteria other than lactic acid bacteria are not added during the sake production process. DNA analysis of foods and drinks obtained through processes involving fermentation reveals the type of microorganisms participating in the product-making process [,,,,,,,,,,,,]. DNA sequencing of bacteria participating in the sake production processes have shown that many bacteria are involved [,,,,]. Most of these bacteria are present by chance; however, some are introduced by necessity and are specific to each sake brewery [,,]. Bacteria that are introduced in the sake production process by necessity are known as kuratsuki bacteria []. The Japanese word “kuratsuki” corresponds to the term “sake brewery inhabiting”.
We isolated and identified the TGY1120_3 and TGY1127_2 strains belonging to the genus Kocuria as kuratsuki bacteria in sake from the Narimasa Sake Brewery in Toyama, Japan [,]. The TGY1120_3 and TGY1127_2 strains differ at the species level [], corresponding to Kocuria koreensis [] and K. uropygioeca [], respectively. In addition, we isolated and identified the strains A-10 and B-12 belonging to the genus Bacillus as kuratsuki bacteria in sake from the Shiraki-Tsunesuke Sake Brewery in Gifu, Japan []. In 2020, Gupta et al. taxonomically reconsidered the genus Bacillus [], and transferred some Bacillus species, such as Bacillus megaterium, to the new genus Priestia. Therefore, in this study, we sequenced the genomes of the strains A-10 and B-12 and classified them taxonomically.
The compositions of esters and organic acids produced by sake yeast strongly affect the flavor and taste of sake [,,,,,,,,,,]. Sake yeasts form a phylogenetic cluster different from that of beer and wine yeasts []. In addition, some chemical compounds are involved in interactions among microorganisms during sake production and affect the flavor and taste of sake [,,,,,]. Lactic acid bacteria used in the kimoto method interact with sake yeast, affecting its metabolism [,]. Previous reports showed that the interactions among microorganisms in the sake production process affect the flavor and taste of sake. Previously, we used kuratsuki Kocuria to study the interactions between kuratsuki bacteria and sake yeast during sake production [,,,]. First, co-cultivation experiments were performed using kuratsuki Kocuria and sake yeast in artificial media and koji solutions []. Although the number of viable cells of kuratsuki Kocuria gradually decreased, and the cells died during the 2-week culture period owing to ethanol production by sake yeast, the Kocuria TGY1127_2 strain survived longer than the TGY1120_3 strain [].
To confirm the effect of kuratsuki Kocuria on sake taste, we used the taste recognition device TS-5000Z (Intelligent Sensor Technology, Inc., Atsugi, Japan), which uses a receptor membrane composed of a lipid, a plasticizer, and polyvinyl chloride to detect taste-producing substances [,]. First, the taste recognition device showed that the addition of kuratsuki Kocuria to different koji solutions altered the taste of sake differently depending on the different koji solution []. Next, the change in taste with the addition of kuratsuki Kocuria differed depending on the sake yeast strain used []. Finally, the effect of kuratsuki Kocuria on sake taste differed for hydrogen- and non-hydrogen-treated kuratsuki Kocuria [], suggesting that different cell states may induce different microbial interactions. These results showed that when the sake production environment and the cell state of kuratsuki bacteria change, the interaction between kuratsuki bacteria and sake yeast also changes, affecting sake taste.
A sake test production was performed at an actual sake production company. Sake was produced using the Narimasa kuratsuki Kocuria strain TGY1127_2 at the Yoshinotomo Sake Brewery in Toyama, Japan. The Yoshinotomo Sake Brewery does not use Kocuria as a kuratsuki bacterium, because no Kocuria were detected in any samples from the Yoshinotomo Sake Brewery. The addition of kuratsuki Kocuria did not change the alcohol concentration, acidity, or amino acid content of sake. However, the taste of sake differed. Notably, 39 out of 41 individuals who were included in the taste test answered that sake produced with the addition of kuratsuki Kocuria tasted better than that produced without kuratsuki Kocuria [].
The results of the co-cultivation and sake test production showed that the addition of kuratsuki Kocuria strain TGY1127_2 changed the taste of sake and had a good effect on sake production. From the viewpoint of applied microbiology, the use of kuratsuki bacteria in sake production offers unprecedented possibilities for sake production. To the best of our knowledge, no bacteria other than lactic acid bacteria have been used for sake production. Therefore, data are available only for kuratsuki Kocuria. To create sake with a new taste using kuratsuki bacteria, more kuratsuki bacterial strains should be identified and characterized. In this study, we aimed to investigate the effects of the kuratsuki bacterial strains A-10 and B-12 on sake taste and found differences with respect to the effects of kuratsuki Kocuria strains.
2. Materials and Methods
2.1. Genome Sequencing of the kuratsuki Bacterial Strains A-10 and B-12
The genomes of the strains A-10 and B-12 were determined using both Nanopore (PromethION X5; Oxford, UK) and Illumina (MiSeq; San Diego, CA, USA) DNA sequencers. Hybrid assembly was performed using Trycycler version 0.5.3 []. Circulation analysis was performed using Circlator []. Illumina short-read mapping was performed using the Burrows–Wheeler aligner (BWA) version 0.7.12 [], and error correction was performed using Pilon version 1.23 [].
2.2. Cultivation of the Microorganisms
The sake yeast strains AK25, K901, K1401, and K1801, which are used in Japanese sake breweries [,,], and kuratsuki bacterial strains A-10 and B-12 [] were used in this study. Each strain was grown in TGY medium (5 g/L tryptone, 1 g/L glucose, and 3 g/L yeast extract) at 25 °C for 12 h prior to cultivation. As a control, 200 mL of water was added to 20 g of koji (Miyako koji, Isesou, Tokyo, Japan) and 1 mL of pre-cultivated cell suspensions of AK25 (4.11 109 cells/mL), K901 (6.43 107 cells/mL), K1401 (1.79 108 cells/mL), or K1801 (2.10 108 cells/mL). For co-cultivation, 200 mL of water was added to 20 g of koji, 1 mL of pre-cultivated yeast suspensions, and 1 mL of pre-cultivated A-10 (1.01 109 cells/mL) and/or B-12 (2.19 106 cells/mL) cell suspensions. Each mixed solution was incubated at 14 °C for 14 days.
2.3. Measurement of Brix and Acidity
Brix and acidity were measured each day using a PAL-BX/ACID digital refractometer (ATAGO, Tokyo, Japan). Each measurement was repeated three times. The median of the three values was chosen as the representative value.
2.4. Estimation of the Sake Taste
A TS-5000Z (Intelligent Sensor Technology, Inc., Atsugi, Japan) taste recognition device was used. Each taste sensor in this device has a different lipid membrane for taste estimation [,]. The strength of each taste is represented by the magnitude of its corresponding current value [,]. The taste intensity of sake with kuratsuki bacteria (A-10 or B-12 strains) minus that of sake without kuratsuki bacteria was calculated. The initial taste in terms of astringent stimulation, bitter miscellaneous taste, saltiness, sourness, and umami was measured using the sensors AE1, CO0, CT0, CA0, and AAE, respectively. The astringency, bitterness, and umami levels were measured using the intensity of the second measurement. The taste intensity for each yeast strain, with and without (control) kuratsuki bacteria, was measured. Each measurement was repeated four times. The obtained intensities were subjected to pairwise comparisons (intensity differences) for each sake yeast strain.
2.5. Statistical Analysis
Statistical analyses were performed using R software (R Project for Statistical Computing, http://www.R-project.org/ accessed on 1 January 2023). The Kolmogorov–Smirnov test was performed to compare the Brix and acidity change patterns. Analysis of variance (ANOVA) was performed using Bartlett’s test to compare the taste intensity differences with or without kuratsuki bacteria among different sake produced using different sake yeasts. Pairwise t-tests were performed using Bonferroni correction when the ANOVA showed a p-value < 0.05.
3. Results
3.1. Genome Characteristics
The complete genome sequences and eight contigs of strains A-10 and B-12 were obtained (Table 1). Taxonomic marker genes, such as ribosomal RNA-coding genes, showed that the strains A-10 and B-12 belong to the species Bacillus safensis and Priestia megaterium, respectively.

Table 1.
Summary features of the genomic sequences of the strains A-10 and B-12.
Bacillus A-10 contained four insertion sequence (IS) of three family transposases (Table 1). Two of the four transposases were identical. In contrast, the Priestia B-12 strain contained 20 transposases belonging to six different families (Table 1). Of the 20 transposases, 7, 6, 3, 2, 1, and 1 belonged to the IS4, IS110, IS21, IS1326, IS3, and IS466 families, respectively. Among the seven IS4 family of transposases, two and five were identical. Among the six IS110 family of transposases, two and four were identical. The IS3 family of transposase of Priestia B-12 differed from that of Bacillus A-10. Therefore, no transposase genes were exchanged between the Bacillus A-10 and Priestia B-12 strains.
3.2. Brix and Acidity Changes
The Brix and acidity patterns were very similar for sake preparations with and without kuratsuki bacteria (Figure 1). A significant difference (p < 0.05) was detected in only one case with the Kolmogorov–Smirnov tests. It was detected between the acidity of sake produced using only yeast K901 and that of sake produced using yeast K901, Bacillus A-10, and Priestia B-12 (Figure 1). This result was different from that obtained with the addition of the kuratsuki Kocuria TGY1127_2 strain in the sake production process []. The Brix and acidity of sake produced using only yeast AK25 were very different from those of sake produced using yeast AK25 and Kocuria TGY1127_2 []. However, this difference was not observed in this study.
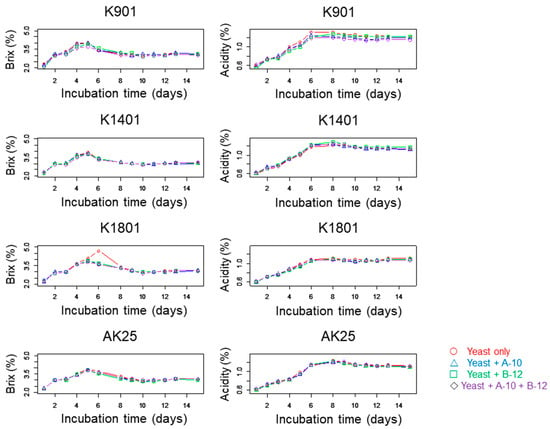
Figure 1.
Brix and acidity of sake produced with different strains of sake yeast (AK25, K901, K1401, or K1801) with or without kuratsuki Bacillus A-10 and/or Priestia B-12. The acidity of sake obtained using only yeast K901 was significantly (p < 0.05) different from that of sake obtained using yeast K901, Bacillus A-10, and Priestia B in the Kolmogorov–Smirnov tests. For the other sake productions, these values were not significantly different (p > 0.05).
3.3. Effects of kuratsuki Bacillus and Priestia on Sake Taste
The effects of kuratsuki Bacillus A-10 and Priestia B-12 were estimated using the TS-5000Z taste recognition device, and different taste intensities among different sake preparations produced using different sake yeast strains were detected by ANOVA. The ranges of the taste intensity differences were mostly between −1 and +1 (Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, Figure 7, Figures S1 and S2), i.e., much lower than those obtained for sake produced using kuratsuki Kocuria []. The effects of the addition of kuratsuki Bacillus A-10 and/or Priestia B-12 on saltiness and sourness were more significant than those on other tastes (Figure 4 and Figure 5). The largest taste intensity difference was detected in the sourness of sake produced using yeast K901, Bacillus A-10, and Priestia B-12 (Figure 5). This result is consistent with the fact that the acidity of sake produced with yeast K901 and that of sake produced with yeast K901, Bacillus A-10, and Priestia B-12 were significantly different (Figure 1).
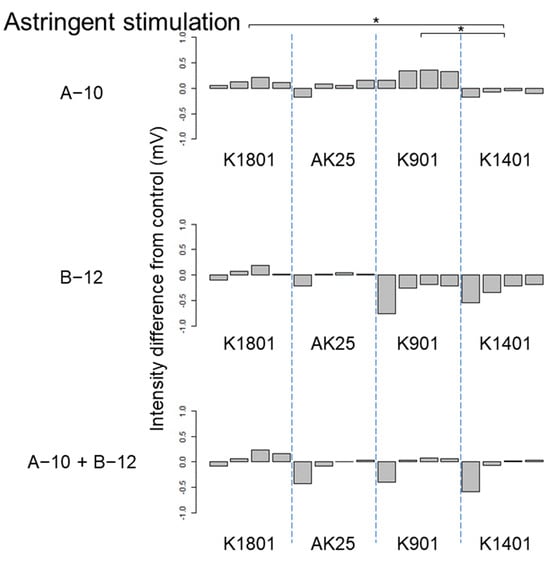
Figure 2.
Differences between the astringent taste intensities of sake produced using different sake yeasts (AK25, K901, K1401, and K1801) with or without kuratsuki Bacillus A-10 and/or Priestia B-12. The taste intensity for each yeast strain with and without the kuratsuki bacterium (control) was measured. Each measurement was repeated four times. Thus, four values of taste intensity for sake with kuratsuki bacterium and four values of taste intensity for sake without kuratsuki bacterium (control) were obtained and subjected to pairwise comparisons (intensity differences) for each sake yeast strain. Asterisk (*) indicates a significant difference (p < 0.05) in pairwise t-test.
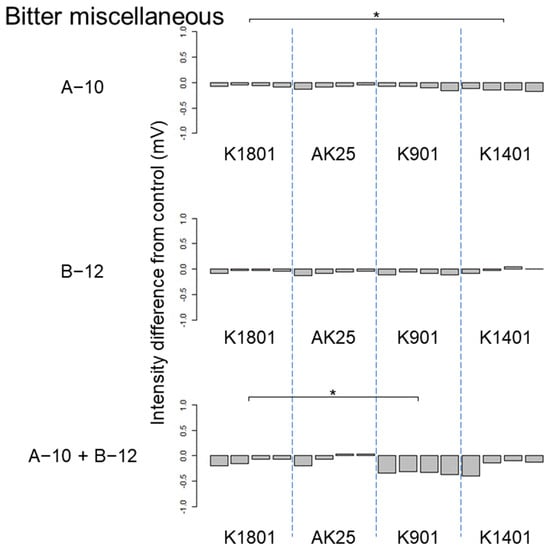
Figure 3.
Difference between the bitter miscellaneous taste intensities of sake produced using different sake yeasts (AK25, K901, K1401, and K1801) with or without kuratsuki Bacillus A-10 and/or Priestia B-12. The taste intensity for each yeast strain with and without the kuratsuki bacterium (control) was measured. Each measurement was repeated four times. Thus, four values of taste intensity for sake with kuratsuki bacterium and four values of taste intensity of sake without kuratsuki bacterium (control) were obtained and subjected to pairwise comparisons (intensity differences) for each sake yeast strain. Asterisk (*) indicates a significant difference (p < 0.05) in pairwise t-test.
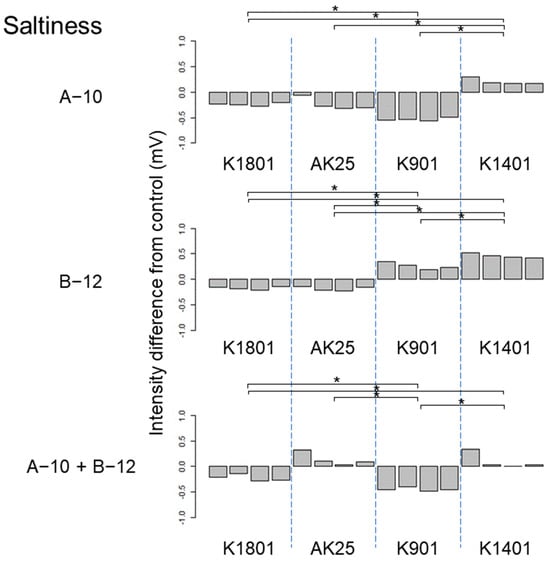
Figure 4.
Difference between the saltiness taste intensities of sake produced using different sake yeasts (AK25, K901, K1401, and K1801) with or without kuratsuki Bacillus A-10 and/or Priestia B-12. The taste intensity for each yeast strain with and without the kuratsuki bacterium (control) was measured. Each measurement was repeated four times. Thus, four values of taste intensity for sake with kuratsuki bacterium and four values of taste intensity for sake without kuratsuki bacterium (control) were obtained and subjected to pairwise comparisons (intensity differences) for each sake yeast strain. Asterisk (*) indicates a significant difference (p < 0.05) in pairwise t-test.
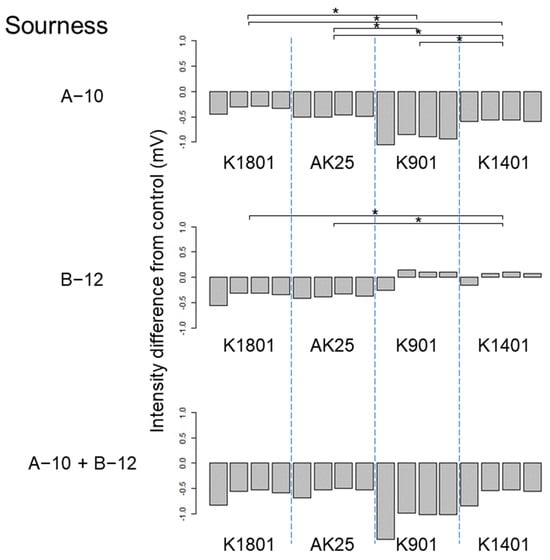
Figure 5.
Difference between the sourness taste intensities of sake produced using different sake yeasts (AK25, K901, K1401, and K1801) with or without kuratsuki Bacillus A-10 and/or Priestia B-12. The taste intensity for each yeast strain with and without the kuratsuki bacterium (control) was measured. Each measurement was repeated four times. Thus, four values of taste intensity for sake with kuratsuki bacterium and four values of taste intensity for sake without kuratsuki bacterium (control) were obtained and subjected to pairwise comparisons (intensity differences) for each sake yeast strain. Asterisk (*) indicates a significant difference (p < 0.05) in pairwise t-test.
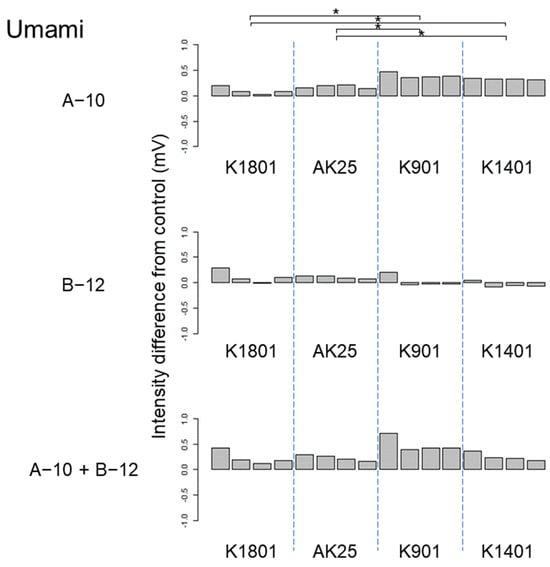
Figure 6.
Difference between the umami taste intensities of sake produced using different sake yeasts (AK25, K901, K1401, and K1801) with or without kuratsuki Bacillus A-10 and/or Priestia B-12. The taste intensity for each yeast strain with and without the kuratsuki bacterium (control) was measured. Each measurement was repeated four times. Thus, four values of taste intensity for sake with kuratsuki bacterium and four values of taste intensity for sake without kuratsuki bacterium (control) were obtained and subjected to pairwise comparisons (intensity differences) for each sake yeast strain. Asterisk (*) indicates a significant difference (p < 0.05) in pairwise t-test.
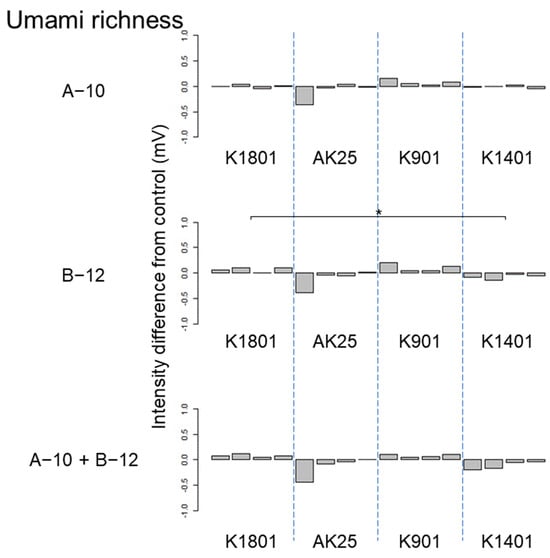
Figure 7.
Difference between the umami taste intensities of sake produced using different sake yeasts (AK25, K901, K1401, and K1801) with or without kuratsuki Bacillus A-10 and/or Priestia B-12. The taste intensity for each yeast strain with and without the kuratsuki bacterium (control) was measured. Each measurement was repeated four times. Thus, four values of taste intensity for sake with kuratsuki bacterium and four values of taste intensity for sake without kuratsuki bacterium (control) were obtained and subjected to pairwise comparisons (intensity differences) for each sake yeast strain. Asterisk (*) indicates a significant difference (p < 0.05) in pairwise t-test.
The results of ANOVA showed that the taste difference among four sake produced using different sake yeast strains was not significant (p > 0.05) in the following seven cases: astringency of sake produced using Bacillus A-10, Priestia B-12, and both Bacillus A-10 and Priestia B-12; astringent stimulation of sake produced using both Bacillus A-10 and Priestia B-12; bitter miscellaneous taste of sake produced using Bacillus B-12; umami taste of sake produced using Priestia B-12; and umami richness of sake produced using Bacillus A-10. Although differences were detected by ANOVA, different pairs were not identified by pairwise t-test in the following seven cases: astringent stimulation of sake produced using Priestia B-12; bitterness of sake produced using Bacillus A-10, Priestia B-12, and both Bacillus A-10 and Priestia B-12; sourness of sake produced using both Bacillus A-10 and Priestia B-12; umami taste of sake produced using both Bacillus A-10 and Priestia B-12; and umami richness of sake produced using both Bacillus A-10 and Priestia B-12. Therefore, the effects of the kuratsuki Bacillus A-10 and Priestia B-12 strains on sake taste were smaller than those of kuratsuki Kocuria TGY1127_2 strain [].
Although the Shiraki-Tunesuke kuratsuki bacteria were different at the genus level, the effects of their addition to the sake production process on sake taste were surprisingly similar. The difference between the effects of Bacillus A-10 and Priestia B-12 on sake taste was limited. The astringency stimulation of sake produced with sake yeast K901 was increased by Bacillus A-10 (Figure 2). However, Priestia B-12 decreased it (Figure 2). A similar pattern was detected for sake saltiness using yeast K901 (Figure 4). Except for these two cases, the changes in taste intensity induced by the strains A-10 and B-12 were similar. Therefore, sake yeast K901 interacted differently with the kuratsuki Bacillus A-10 and Priestia B-12 strains.
4. Discussion
Horizontal transfer of genetic information has been strongly suggested between different kuratsuki Kocuria strains (TGY1120_3 and TGY1127_2), because similar plasmids encoding identical transposase-coding gene were detected in the different Kocuria strains TGY1120_3 and TGY1127_2 []. In contrast, no plasmids were detected in the genomes of the kuratsuki Bacillus A-10 and Priestia B-12 strains. The Kocuria strains were kuratsuki bacteria from Narimasa but not kuratsuki bacteria from Shiraki-Tsunesuke. The Bacillus A-10 and Priestia B-21 strains were kuratsuki bacteria from Shiraki-Tsunesuke but not from Narimasa. This suggests that different sake production environments may influence differently genome evolution in kuratsuki bacteria.
The addition of Bacillus A-10 and/or Priestia B-12 to the sake production process using the sake yeasts AK25, K901, K1401, and K1801 did not significantly affect the Brix and acidity patterns (Figure 1). These results indicated that neither the kuratsuki Bacillus strain nor the Priestia B-12 strain inhibited ethanol fermentation and organic acid production by all four sake yeasts used in this study. Such characteristics can be expected for all kuratsuki bacteria used in sake production. If kuratsuki bacteria inhibit the metabolism of sake yeast, the characteristic flavor and taste of sake yeast may be lost.
Furthermore, the effects of Bacillus A-10 and/or Priestia B-12 addition to the sake production process on sake taste were limited (Figure 2, Figure 3, Figure 4, Figure 5, Figure 6 and Figure 7, Figures S1 and S2). The ranges of the taste intensity differences in this study were much smaller than those found for sake produced using kuratsuki Kocuria []. This suggests that kuratsuki Kocuria interacts more strongly with sake yeast than kuratsuki Bacillus or Priestia. The effects of kuratsuki Bacillus A-10 on sake taste were similar to those of kuratsuki Priestia B-12 in sake produced using the sake yeasts AK25, K1401, and K1801 (Figure 2, Figure 3, Figure 4, Figure 5, Figure 6 and Figure 7, Figures S1 and S2). However, the effects of these kuratsuki bacterial strains differed for sake produced using yeast K901 (Figure 2 and Figure 4). This indicated that a combination of kuratsuki bacteria and sake yeast should be considered when modifying sake taste. From the viewpoint of geographical indication, an increasing interest is recognized in autochthonous microorganisms related to fermented foods and drinks [,,]. It is necessary to perform additional research to investigate the interaction between different kuratsuki bacteria and sake yeasts in order to modify sake flavor and taste profiles.
When the effects of kuratsuki bacteria on sake taste were compared among sake preparations produced with only Bacillus A-10, only Priestia B-12, and both A-10 and B-12, the taste intensity of sake obtained with A-10 and B-12 was different from that of sake obtained with A-10 and that of sake obtained with B-12, in most cases (Figure 2, Figure 3, Figure 4, Figure 5, Figure 6 and Figure 7, Figures S1 and S2). This strongly suggests that the interaction between kuratsuki Bacillus A-10 and sake yeast is not independent of the interaction between kuratsuki Priestia B-12 and sake yeast. Therefore, microbial interactions may also occur between the Bacillus A-10 and Priestia B-12 strains.
5. Conclusions
Our aim was to use kuratsuki bacteria for sake production. The flavor and taste of sake can be controlled by utilizing the interactions between sake yeast and kuratsuki bacteria. To achieve this goal, kuratsuki bacteria should be collected from various sake breweries and characterized. To the best of our knowledge, this is the first study using kuratsuki bacteria for sake production. Currently, we have collected only four strains of kuratsuki bacteria from two different sake breweries and characterized only two strains, Kocuria TGY1120_3 and TGY1127_2, from one sake brewery.
In this study, we characterized the kuratsuki Bacillus A-10 and Priestia B-12 strains. This study elucidated differences in the effects of kuratsuki bacteria on sake taste. We were able to quantitatively show differences in the taste of sake and clearly show differences in the effects of different kuratsuki bacteria. Although all sake yeasts are Saccharomyces cerevisiae, kuratsuki bacteria are diverse and differ at the genus level. Surprisingly, the kuratsuki bacterial strains A-10 and B-12 differ at the genus level, although the taste intensity differences among sake preparations produced using the sake yeasts AK25, K1401, and K1801 and either kuratsuki Bacillus A-10 or kuratsuki Priestia B-12 were very small. To demonstrate kuratsuki bacterial diversification, additional kuratsuki bacteria should be collected and characterized.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/applmicrobiol4010011/s1. Figure S1: Difference between the astringency taste intensities of sake produced using different sake yeasts (AK25, K901, K1401, and K1801) with or without kuratsuki Bacillus A-10 and/or Priestia B-12. The taste intensity for each yeast strain with and without the kuratsuki bacterium (control) was measured. Each measurement was repeated four times. Thus, four values of taste intensity for sake with kuratsuki bacterium and four values of taste intensity for sake without kuratsuki bacterium (control) were obtained and subjected to pairwise comparisons (intensity differences) for each sake yeast strain. Figure S2: Difference between the bitter taste intensities of sake produced using different sake yeasts (AK25, K901, K1401, and K1801) with or without kuratsuki Bacillus A-10 and/or Priestia B-12. The taste intensity for each yeast strain with and without the kuratsuki bacterium (control) was measured. Each measurement was repeated four times. Thus, four values of taste intensity for sake with kuratsuki bacterium and four values of taste intensity for sake without kuratsuki bacterium (control) were obtained and subjected to pairwise comparisons (intensity differences) for each sake yeast strain.
Author Contributions
Conceptualization, H.N.; methodology, H.N.; formal analysis, K.K.; writing—original draft preparation, H.N.; supervision, H.N.; project administration, H.N.; funding acquisition, H.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by JSPS KAKENHI, grant number 21H02109 (to H.N.).
Data Availability Statement
Data are available upon request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Koyanagi, T.; Kiyohara, M.; Matsui, H.; Yamamoto, K.; Kondo, T.; Katayama, T.; Kumagai, H. Pyrosequencing survey of the microbial diversity of ‘narezushi’, an archetype of modern Japanese sushi. Lett. Appl. Microbiol. 2011, 53, 635–640. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Bamforth, C.W.; Mills, D.A. A review of molecular methods for microbial community profiling of beer and wine. J. Am. Soc. Brew. Chem. 2012, 70, 150–162. [Google Scholar] [CrossRef]
- Koyanagi, T.; Nakagawa, A.; Kiyohara, M.; Matsui, H.; Yamamoto, K.; Barla, F.; Take, H.; Katsuyama, Y.; Tsuji, A.; Shijimaya, M.; et al. Pyrosequencing analysis of microbiota in Kaburazushi, a traditional medieval sushi in Japan. Biosci. Biotechnol. Biochem. 2013, 77, 2125–2130. [Google Scholar] [CrossRef][Green Version]
- Kyung, K.H.; Pradas, E.M.; Kim, S.G.; Lee, Y.J.; Kim, K.H.; Choi, J.J.; Cho, J.H.; Chung, C.H.; Barrangou, R.; Breidt, F. Microbial ecology of watery kimuchi. J. Food Sci. 2015, 80, M1031–M1038. [Google Scholar] [CrossRef] [PubMed]
- Tamang, J.P.; Watanabe, K.; Holzapfel, W.H. Review: Diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 2016, 7, 377. [Google Scholar] [CrossRef]
- Wuyts, S.; Van Beeck, W.; Allonsius, C.N.; van den Broek, M.F.; Lebeer, S. Applications of plant-based fermented foods and their microbes. Curr. Opin. Biotechnol. 2020, 61, 45–52. [Google Scholar] [CrossRef]
- Whon, T.W.; Ahn, S.W.; Yang, S.; Kim, J.Y.; Kim, Y.B.; Kim, Y.; Hong, J.-M.; Jung, H.; Choi, Y.-E.; Lee, S.H.; et al. ODFM, an omics data resource from microorganisms associated with fermented foods. Sci. Data 2021, 8, 113. [Google Scholar] [CrossRef] [PubMed]
- Rodpai, R.; Sanpool, O.; Thanchomnang, T.; Wangwiwatsin, A.; Sadaow, L.; Phupiewkham, W.; Boonroumkaew, P.; Intapan, P.M.; Maleewong, W. Investigating the microbiota of fermented fish products (Pla-ra) from different communities of northeastern Thailand. PLoS ONE 2021, 16, e0245227. [Google Scholar] [CrossRef]
- Fraiture, M.-A.; Papazova, N.; Roosens, N.H.C. DNA walking strategy to identify unauthorized genetically modified bacteria in microbial fermentation products. Int. J. Food Microbiol. 2021, 337, 108913. [Google Scholar] [CrossRef]
- Walsh, A.M.; Leech, J.; Huttenhower, C.; Delhomme-Nguyen, H.; Crispie, F.; Chervaux, C.; Cotter, P.D. Integrated molecular approaches for fermented food microbiome research. FEMS Microbiol. Rev. 2023, 47, fuad001. [Google Scholar] [CrossRef]
- Le, M.-M.; Zhong, L.-W.; Ren, Z.-W.; An, M.-Q.; Long, Y.-H.; Ling, T.-J. Dynamic changes in the microbial community and metabolite profile during the pile fermentation process of Fuzhuan brink tea. J. Agric. Food Chem. 2023, 71, 19142–19153. [Google Scholar] [CrossRef] [PubMed]
- Fusco, V.; Fanelli, F.; Chieffi, D. Recent and advanced DNA-based technologies for the authentication of probiotic, protected designation of origin (PDO) and protected geographical indication (PGI) fermented foods and beverages. Foods 2023, 12, 3782. [Google Scholar] [CrossRef] [PubMed]
- Palmnäs-Bédard, M.; de Santa Izabel, A.; Dicksved, J.; Landberg, R. Characterization of the bacterial composition of 47 fermented foods in Sweden. Foods 2023, 12, 3827. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Ohta, M.; Lee, M.; Mills, D.A. Indigenous bacteria and fungi drive traditional kimoto sake fermentations. Appl. Environ. Microbiol. 2014, 80, 5522–5529. [Google Scholar] [CrossRef]
- Koyanagi, T.; Nakagawa, A.; Kiyohara, M.; Matsui, H.; Tsuji, A.; Barla, F.; Take, H.; Katsuyama, Y.; Tokuda, K.; Nakamura, S.; et al. Tracing microbiota changes in yamahai-moto, the traditional Japanese sake starter. Biosci. Biotechnol. Biochem. 2016, 80, 399–406. [Google Scholar] [CrossRef]
- Tsuji, A.; Kozawa, M.; Tokuda, K.; Enomoto, T.; Koyanagi, T. Robust domination of Lactobacillus sakei in microbiota during traditional Japanese sake starter yamahai-moto fermentation and the accompanying changes in metabolites. Curr. Microbiol. 2018, 75, 1498–1505. [Google Scholar] [CrossRef]
- Terasaki, M.; Nishida, H. Bacterial DNA diversity among clear and cloudy sakes, and sake-kasu. Open Bioinform. J. 2020, 13, 74–82. [Google Scholar] [CrossRef]
- Ito, K.; Niwa, R.; Kobayashi, K.; Nakagawa, T.; Hoshino, G.; Tsuchida, Y. A dark matter in sake brewing: Origin of microbes producing a Kimoto-style fermentation starter. Front. Microbiol. 2023, 14, 1112638. [Google Scholar] [CrossRef]
- Nishida, H. Sake brewing and bacteria inhabiting sake breweries. Front. Microbiol. 2021, 12, 602380. [Google Scholar] [CrossRef]
- Terasaki, M.; Kimura, Y.; Yamada, M.; Nishida, H. Genomic information of Kocuria isolates from sake brewing process. AIMS Microbiol. 2021, 7, 114–123. [Google Scholar] [CrossRef]
- Kanamoto, E.; Terashima, K.; Shiraki, Y.; Nishida, H. Diversity of Bacillus isolates from the sake brewing process at a sake brewery. Microorganisms 2021, 9, 1760. [Google Scholar] [CrossRef] [PubMed]
- Nishida, H. Kuratsuki bacteria and sake making. Biosci. Biotechnol. Biochem. 2023, zbad147. [Google Scholar] [CrossRef]
- Park, E.-J.; Roh, S.W.; Kim, M.-S.; Jung, M.-J.; Shin, K.-S.; Bae, J.-W. Kocuria koreensis, sp. nov., isolated from fermented seafood. Int. J. Syst. Evol. Microbiol. 2010, 60, 140–143. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Braun, M.S.; Wang, E.; Zimmermann, S.; Boutin, S.; Wink, M. Kocuria uropygioeca sp. nov. and Kocuria uropygialis sp. nov., isolated form the preen glands of Great Spotted Woodpeckers (Dendrocopos major). Syst. Appl. Microbiol. 2018, 41, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Patel, S.; Saini, N.; Chen, S. Robust demarcation of 17 distinct Bacillus species clades, proposed as novel Bacillaceae genera, by phylogenomics and comparative genomic analyses: Description of Robertmurraya kyonggiensis sp. nov. and proposal for an emended genus Bacillus limiting it only to the members of the Subtilis and Cereus clades of species. Int. J. Syst. Evol. Microbiol. 2020, 70, 5753–5798. [Google Scholar]
- Park, Y.C.; Shaffer, C.E.H.; Bennett, G.N. Microbial formation of esters. Appl. Microbiol. Biotechnol. 2009, 85, 13–25. [Google Scholar] [CrossRef]
- Saerens, S.M.G.; Delvaux, F.R.; Verstrepen, K.J.; Thevelein, J.M. Production and biological function of volatile esters in Saccharomyces cerevisiae. Microb. Biotechnol. 2010, 3, 165–177. [Google Scholar] [CrossRef]
- Procopio, S.; Qian, F.; Becker, T. Functional and regulation of yeast genes involved in higher alcohol and ester metabolism during beverage fermentation. Eur. Food Res. Technol. 2011, 233, 721–729. [Google Scholar] [CrossRef]
- Kitagaki, H.; Kitamoto, K. Breeding research on sake yeasts in Japan: History, recent technological advances, and future perspectives. Ann. Rev. Food Sci. Technol. 2013, 4, 215–235. [Google Scholar] [CrossRef]
- Dzialo, M.C.; Park, R.; Steensels, J.; Lievens, B.; Verstrepen, K.J. Physiology, ecology and industrial applications of aroma formation in yeast. FEMS Microbiol. Rev. 2017, 41, S95–S128. [Google Scholar] [CrossRef]
- Suto, M.; Kawashima, H. Compound specific carbon isotope analysis in sake by LC/IRMS and brewers’ alcohol proportion. Sci. Rep. 2019, 9, 17635. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Hu, W.; Xia, Y.; Mu, Z.; Tao, L.; Song, X.; Zhang, H.; Ni, B.; Ai, L. Flavor formation in Chinese rice wine (Huangjiu): Impacts of the flavor-active microorganisms, raw materials, and fermentation technology. Front. Microbiol. 2020, 11, 580247. [Google Scholar] [CrossRef] [PubMed]
- Suto, M.; Kawashima, H. Discrimination for sake brewing methods by compound specific isotope analysis and formation mechanism of organic acids in sake. Food Chem. 2022, 381, 132295. [Google Scholar] [CrossRef] [PubMed]
- Tekarslan-Sahin, S.H. Adaptive laboratory evolution of yeasts for aroma compound production. Fermentation 2022, 8, 372. [Google Scholar] [CrossRef]
- Yoshimoto, H.; Bogaki, T. Mechanisms of production and control of acetate esters in yeasts. J. Biosci. Bioeng. 2023, 136, 261–269. [Google Scholar] [CrossRef]
- Maruyama, H. Beer brewed with sake yeast strain has unique sake-like flavors. J. Am. Soc. Brew. Chem. 2023; in press. [Google Scholar] [CrossRef]
- Gonçalves, M.; Pontes, A.; Almeida, P.; Barbosa, R.; Serra, M.; Libkind, D.; Hutzler, M.; Gonçalves, P.; Sampaio, J.P. Distinct domestication trajectories in top-fermenting beer yeasts and wine yeasts. Curr. Biol. 2016, 26, 2750–2761. [Google Scholar] [CrossRef]
- Mizoguchi, H.; Ikeda, T.; Hara, S. Differences in the intracellular lipids of sake yeast in main mash seeded respectively with two kinds of seed mash: Kimoto and sokujo-moto. J. Ferment. Bioeng. 1995, 80, 586–591. [Google Scholar] [CrossRef]
- Sawada, K.; Sato, T.; Harajima, H.; Jayakody, L.N.; Hirata, M.; Yamashiro, M.; Tajima, M.; Mitsutake, S.; Nagao, K.; Tsuge, K.; et al. Glucosylceramide contained in koji mold-cultured cereal confers membrane and flavor modification and stress tolerance to Saccharomyces cerevisiae during coculture fermentation. Appl. Environ. Microbiol. 2015, 81, 3688–3698. [Google Scholar] [CrossRef]
- Huang, Z.-R.; Hong, J.-L.; Xu, J.-X.; Li, L.; Guo, W.-L.; Pan, Y.-Y.; Chen, S.-J.; Bai, W.-D.; Rao, P.-F.; Ni, L.; et al. Exploring core functional microbiota responsible for the production of volatile flavor during the traditional brewing of Wuyi Hong Qu glutinous rice wine. Food Microbiol. 2018, 76, 487–496. [Google Scholar] [CrossRef]
- Xu, J.-Z.; Zhang, Y.-Y.; Zhang, W.-G. Correlation between changes in flavor compounds and microbial community ecological succession in the liquid fermentation of rice wine. World J. Microbiol. Biotechnol. 2023, 40, 17. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Niwa, R.; Yamagishi, Y.; Kobayashi, K.; Tsuchida, Y.; Hoshino, G.; Nakagawa, T.; Watanabe, T. A unique case in which Kimoto-style fermentation was completed with Leuconostoc as the dominant genus without transitioning to Lactobacillus. J. Biosci. Bioeng. 2023, 135, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, H.; Watanabe, K.; Wakai, Y. Combination of four bacterial strains isolated from Yamahai-shubo in traditional Japanese sake brewing. Food Sci. Nutr. 2023, 11, 2990–3001. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, D.; Kumano, M.; Sugimoto, Y.; Ito, M.; Ohashi, M.; Sunada, K.; Takahashi, T.; Yamada, T.; Takagi, H. Metabolic switching of sake yeast by kimoto lactic acid bacteria through the [GAR+] non-genetic element. J. Biosci. Bioeng. 2018, 126, 624–629. [Google Scholar] [CrossRef]
- Watanabe, D.; Takagi, H. Yeast prion-based metabolic reprogramming induced by bacteria in fermented foods. FEMS Yeast Res. 2019, 19, foz061. [Google Scholar] [CrossRef]
- Terasaki, M.; Inoue, A.; Kanamoto, E.; Yoshida, S.; Yamada, M.; Toda, H.; Nishida, H. Co-cultivation of sake yeast and Kocuria isolates from the sake brewing process. FEMS Microbiol. Lett. 2021, 368, fnab053. [Google Scholar] [CrossRef]
- Yazaki, A.; Nishida, H. Effect of kuratsuki Kocuria on sake brewing in different koji conditions. FEMS Microbiol. Lett. 2023, 370, fnad020. [Google Scholar] [CrossRef]
- Yazaki, A.; Nishida, H. Effect of kuratsuki Kocuria on sake’s taste varies depending on the sake yeast strain used in sake brewing. Arch. Microbiol. 2023, 205, 290. [Google Scholar] [CrossRef]
- Saito, M.; Nishida, H. Molecular hydrogen treatment of sake yeast and kuratsuki bacteria affects sake taste. Fermentation 2023, 9, 516. [Google Scholar] [CrossRef]
- Toko, K. Taste sensor. Sens. Actuators B Chem. 2000, 64, 205–215. [Google Scholar] [CrossRef]
- Toko, K. Research and development of taste sensors as a novel analytical tool. Proc. Jpn. Acad. Ser. B 2023, 99, 173–189. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Cerdeira, L.T.; Hawkey, J.; Méric, G.; Vezina, B.; Wyres, K.L.; Holt, K.E. Trycycler: Consensus long-read assemblies for bacterial genomes. Genome Biol. 2021, 22, 266. [Google Scholar] [CrossRef]
- Hunt, M.; De Silva, N.; Otto, T.D.; Parkhill, J.; Keane, J.A.; Harris, S.R. Circlator: Automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015, 16, 294. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Ohya, Y.; Kashima, M. History, lineage and phenotypic differentiation of sake yeast. Biosci. Biotechnol. Biochem. 2019, 83, 1442–1448. [Google Scholar] [CrossRef]
- Zhang, K.; Wu, W.; Yan, Q. Research advances on sake rice, koji, and sake yeast: A review. Food Sci. Nutr. 2020, 8, 2995–3003. [Google Scholar] [CrossRef] [PubMed]
- Negoro, H.; Ishida, H. Development of sake yeast breeding and analysis of genes related to its various phenotypes. FEMS Yeast Res. 2022, 22, foac057. [Google Scholar] [CrossRef]
- Capozzi, V.; Spano, G. Food microbial biodiversity and “microbes of protected origin”. Front. Microbiol. 2011, 2, 237. [Google Scholar] [CrossRef]
- Capozzi, V.; Fragasso, M.; Russo, P. Microbiological safety and the management of microbial resources in artisanal foods and beverages: The need for a transdisciplinary assessment to conciliate actual trends and risks avoidance. Microorganisms 2020, 8, 306. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).