Genetic Analysis and Epidemiological Impact of SARS-CoV-2: A Multinational Study of 1000 Samples Using RT-PCR
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Sample Collection and Dataset
2.2. Design of qRT-PCR Primers Targeting E, N, and RdRp Genes
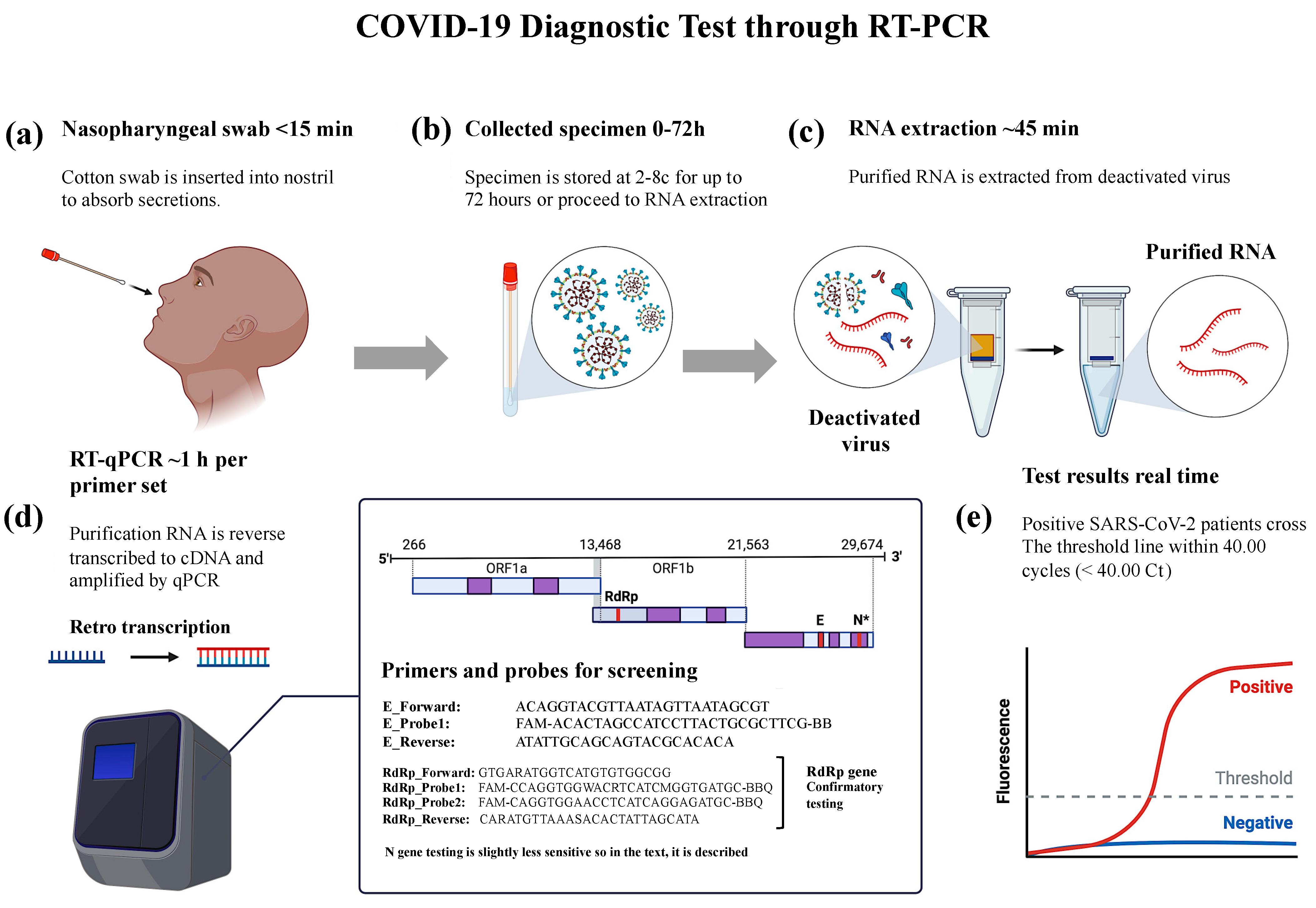
2.3. Detection of SARS-CoV-2 through RNA Isolation and RT-PCR
2.4. Statistical Analysis
3. Results
3.1. Effective Primers Designed and PCR Validation
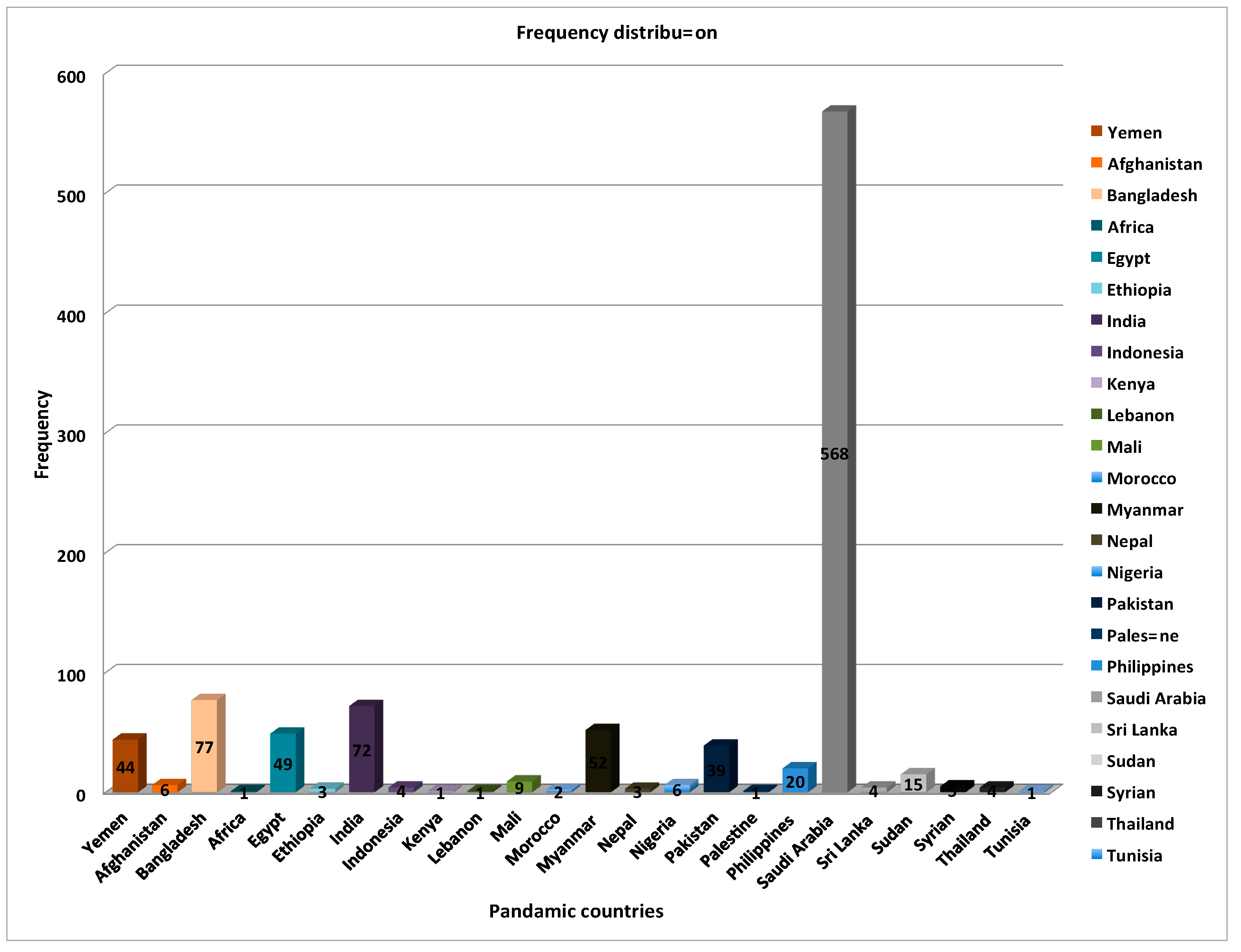
3.2. Wide-Ranging Characteristics of Patient Sample Distribution
3.3. SARS-CoV-2 Genes Detected via Quantitative RT-PCR
4. Discussions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Msemburi, W.; Karlinsky, A.; Knutson, V.; Aleshin-Guendel, S.; Chatterji, S.; Wakefield, J. The WHO estimates of excess mortality associated with the COVID-19 pandemic. Nature 2022, 613, 130–137. [Google Scholar] [CrossRef]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.; Lau, E.H.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [PubMed]
- Ceraolo, C.; Giorgi, F.M. Genomic variance of the 2019-nCoV coronavirus. J. Med. Virol. 2020, 92, 522–528. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, W.; Zhang, S.; Wei, P.; Zhang, L.; Chen, D.; Qiu, S.; Li, X.; Zhao, J.; Shi, Y.; et al. Two novel human coronavirus OC43 genotypes circulating in hospitalized children with pneumonia in China. Emerg. Microbes Infect. 2022, 11, 168–171. [Google Scholar] [CrossRef]
- Lai, A.; Bergna, A.; Caucci, S.; Clementi, N.; Vicenti, I.; Dragoni, F.; Cattelan, A.M.; Menzo, S.; Pan, A.; Callegaro, A.; et al. Molecular Tracing of SARS-CoV-2 in Italy in the First Three Months of the Epidemic. Viruses 2020, 12, 798. [Google Scholar] [CrossRef]
- Coburn, B.J.; Wagner, B.G.; Blower, S. Modeling influenza epidemics and pandemics: Insights into the future of swine flu (H1N1). BMC Med. 2009, 7, 30. [Google Scholar] [CrossRef]
- Cheng, Q.; Liu, Z.; Cheng, G.; Huang, J. Heterogeneity and effectiveness analysis of COVID-19 prevention and control in major cities in China through time-varying reproduction number estimation. Sci. Rep. 2020, 10, 21953. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. Author Correction: A new coronavirus associated with human respiratory disease in China. Nature 2020, 580, E7. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time, RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef]
- Nadareishvili, I.; Zhulina, A.; Tskitishvili, A.; Togonidze, G.; Bloom, D.E.; Lunze, K. The Approach to the COVID-19 Pandemic in Georgia-A Health Policy Analysis. Int. J. Public Health 2022, 67, 1604410. [Google Scholar] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Duffus, D.; Olifer, A. Introductory life science mathematics and quantitative neuroscience courses. CBE—Life Sci. Educ. 2010, 9, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Qahtani, M.; Afifi, I. COVID-19 Vaccine and Its Impacts on Dental Practice in Saudi Arabia. J. Umm Al-Qura Univ. Med. Sci. 2022, 8, 29–33. [Google Scholar] [CrossRef]
- Alkhamiss, A.S.; Alodheilah, A.; Alzaghran, R.; Alfurayh, K.; Alsultan, F.; Alshamlan, R.; Alharbi, K.; Alnafjan, A.; Alduraibi, A.; Alshomar, A.; et al. Impact of Exercise on COVID-19 Patients in Saudi Arabia. J. Umm Al-Qura Univ. Med. Sci. 2022, 8, 13–18. [Google Scholar] [CrossRef]
- Tayyib, N.; Abo-Zaid, M.A.; Ismail, A.H.; Mohammed, S.S.; Amin, A.H. COVID-19 Case Report: Circulating blood cells morphology in convalescent patient. J. Umm Al-Qura Univ. Med. Sci. 2022, 8, 25–28. [Google Scholar] [CrossRef]
- AlQarni, M.A.; Marghalani, A.A.; Al-Zahrani, A.M.; Nazer, T.S. Prevalence of Severe Acute Respiratory Syndrome Coronavirus 2 in Asymptomatic Surgical Pediatric Patients in Makkah, Saudi Arabia. J. Umm Al-Qura Univ. Med. Sci. 2022, 8, 51–54. [Google Scholar] [CrossRef]
- Lee, R.A.; Herigon, J.C.; Benedetti, A.; Pollock, N.R.; Denkinger, C.M. Performance of Saliva, Oropharyngeal Swabs, and Nasal Swabs for SARS-CoV-2 Molecular Detection: A Systematic Review and Meta-analysis. J. Clin. Microbiol. 2021, 59, 10–1128. [Google Scholar] [CrossRef]
- Harapan, H.; Itoh, N.; Yufika, A.; Winardi, W.; Keam, S.; Te, H.; Megawati, D.; Hayati, Z.; Wagner, A.L.; Mudatsir, M. Coronavirus disease 2019 (COVID-19): A literature review. J. Infect. Public Health 2020, 13, 667–673. [Google Scholar] [CrossRef]
- Tsang, N.N.Y.; So, H.C.; Ng, K.Y.; Cowling, B.J.; Leung, G.M.; Ip, D.K.M. Diagnostic performance of different sampling approaches for SARS-CoV-2 RT-PCR testing: A systematic review and meta-analysis. Lancet Infect. Dis. 2021, 21, 1233–1245. [Google Scholar] [CrossRef] [PubMed]
- Colton, H.; Ankcorn, M.; Yavuz, M.; Tovey, L.; Cope, A.; Raza, M.; Keeley, A.J.; Poller, B.; Parker, M.; de Silva, T.I.; et al. Improved sensitivity using a dual target, E and RdRp assay for the diagnosis of SARS-CoV-2 infection: Experience at a large NHS Foundation Trust in the UK. J. Infect. 2021, 82, 159–198. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, H.; Tabaraei, A.; Hosseini, S.M.; Khosravi, A.; Nikoo, H.R. Real-time PCR Ct value in SARS-CoV-2 detection: RdRp or N gene? Infection 2022, 50, 537–540. [Google Scholar] [CrossRef]
- Valadan, R.; Golchin, S.; Alizadeh-Navaei, R.; Haghshenas, M.; Zargari, M.; Mousavi, T.; Ghamati, M. Differential gene expression analysis of common target genes for the detection of SARS-CoV-2 using real time-PCR. AMB Express. 2022, 12, 112. [Google Scholar] [CrossRef] [PubMed]
- Asiri, A.M.; Alomary, S.A.; Alqahtani, S.A.; Adam, I.F.; Amer, S.A. Determinants of the Underlying Causes of Mortality during the First Wave of COVID-19 Pandemic in Saudi Arabia: A Descriptive National Study. Int. J. Environ. Res. Public Health 2021, 18, 12647. [Google Scholar] [CrossRef]
- Asdaq, S.M.B.; Rabbani, S.I.; Alshammari, M.K.; Alshammari, R.S.; Kamal, M.; Imran, M.; AlShammari, N.A.; Al Twallah, M.F.; Alshahrani, A.H. Burden of COVID-19: A preliminary analysis in the population of Saudi Arabia. PeerJ 2022, 10, e13219. [Google Scholar] [CrossRef] [PubMed]
- Alsayer, R.M.; Alsharif, H.M.; Al Baadani, A.M.; Kalam, K.A. Clinical and epidemiological characteristics of COVID-19 mortality in Saudi Arabia. Saudi Med. J. 2021, 42, 1083–1094. [Google Scholar] [CrossRef]
- Hung, I.F.; Lau, S.K.; Woo, P.C.; Yuen, K.Y. Viral loads in clinical specimens and SARS manifestations. Hong Kong Med. J. 2009, 15 (Suppl. S9), 20–22. [Google Scholar] [CrossRef]
- Hamdy, M.E.; El Deeb, A.H.; Hagag, N.M.; Shahein, M.A.; Alaidi, O.; Hussein, H.A. Interspecies transmission of SARS-CoV-2 with special emphasis on viral mutations and ACE-2 receptor homology roles. Int. J. Vet. Sci. Med. 2023, 11, 55–86. [Google Scholar] [CrossRef]
- Reynolds, D.L.; Simpson, E.B. Evaluation of ivermectin antiviral activity against avian infectious bronchitis virus using a chicken embryo model. Int. J. Vet. Sci. Med. 2022, 10, 19–24. [Google Scholar] [CrossRef]
- Moraga-Fernández, A.; Sánchez-Sánchez, M.; Queirós, J.; Lopes, A.M.; Vicente, J.; Pardavila, X.; Sereno-Cadierno, J.; Alves, P.C.; de la Fuente, J.; Fernández de Mera, I.G. A study of viral pathogens in bat species in the Iberian Peninsula: Identification of new coronavirus genetic variants. Int. J. Vet. Sci. Med. 2022, 10, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Ameen, L.; Assaggaf, H.; Alsafi, R.; Minshawi, F.; Alghamdi, S.; Alharbi, A.; Qashqari, F.; Makhdoom, H.; Refaat, B.; Alsaif, B.; et al. Analysis of the Clinical Characteristics of COVID-19 Patient Severity Amongst Saudi Hospital Admission in 2020. J. Umm Al-Qura Univ. Med. Sci. 2022, 8, 18–23. [Google Scholar] [CrossRef]
- Alqasim, A. COVID-19: Cardiopathy, Vasculopathy and Coagulopathy. J. Umm Al-Qura Univ. Med. Sci. 2021, 7, 23–27. [Google Scholar] [CrossRef]
- Almoayad, F.; Elhessewi, G.; Alhashem, A.; Mahboub, S.; Fiala, L. Commitment to COVID-19 Precautionary Measures in Saudi Arabia: An Application of the Health Belief Model. J. Umm Al-Qura Univ. Med. Sci. 2022, 8, 24–28. [Google Scholar] [CrossRef]


| Gene | Sequence (5′->3′) | Template Strand | Length | Start | Stop | Tm | GC% |
|---|---|---|---|---|---|---|---|
| Envelop (E) | ACTACTCTGGTGTGTGGTGC | Plus | 20 | 45 | 64 | 59.61 | 55 |
| ACTCGTTTAGGGAAAGGGTCT | Minus | 21 | 221 | 201 | 58.38 | 47.62 | |
| Nucleocapsid (N) | ACAGGTTACGGTGTTAGGCG | Plus | 20 | 337 | 356 | 60.04 | 55 |
| AGGAGTACCCGTTTTCGCTG | Minus | 20 | 633 | 614 | 60.04 | 55 | |
| RNA-dependent RNA polymerase (RdRp) | CACCTACACACCTCAGCGTT | Plus | 20 | 17,773 | 17,792 | 59.97 | 55 |
| GCACGAACGTGACGAATAGC | Minus | 20 | 17,959 | 17,940 | 59.98 | 55 |
| Measures | Groups | Frequency | Percent Frequency (%) |
|---|---|---|---|
| Age | 33.86 ± 0.499 | ||
| Age groups | >60 years | 931 | 94.2 |
| ≤60 years | 57 | 5.8 | |
| Gender | Female | 313 | 31.7 |
| Male | 675 | 68.3 | |
| E gene | Negative | 671 | 67.9 |
| Positive | 317 | 32.1 | |
| N gene | Negative | 551 | 55.8 |
| Positive | 437 | 44.2 | |
| RdRp gene | Negative | 619 | 62.7 |
| Positive | 369 | 37.3 |
| Measures | Groups | E Gene | χ2 | P (1-Sided) | OR | 95% CI | ||
|---|---|---|---|---|---|---|---|---|
| Positive | Negative | LOWER | UPPER | |||||
| Gender | Female | 217 | 96 | 0.420 | 0.283 | 1.100 | 0.824 | 1.469 |
| Male | 454 | 221 | ||||||
| Age Groups | >60 | 636 | 295 | 1.177 | 0.173 | 1.355 | 0.781 | 2.351 |
| ≤60 | 35 | 22 | ||||||
| N Gene | ||||||||
| Positive | Negative | |||||||
| Gender | Female | 177 | 136 | 0.113 | 0.395 | 1.047 | 0.799 | 1.372 |
| Male | 374 | 301 | ||||||
| Age Groups | >60 | 519 | 412 | 0.003 | 0.533 | 0.984 | 0.574 | 1.687 |
| ≤60 | 32 | 25 | ||||||
| RdRp Gene | ||||||||
| Positive | Negative | |||||||
| Gender | Female | 203 | 110 | 0.951 | 0.183 | 1.149 | 0.869 | 1.519 |
| Male | 416 | 259 | ||||||
| Age Groups | >60 | 586 | 345 | 0.585 | 0.264 | 1.235 | 0.718 | 2.125 |
| ≤60 | 33 | 24 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banser, T.; Abduljaleel, Z.; Alzabeedi, K.H.; Alzahrani, A.A.; Khogeer, A.A.; Qabbani, F.H.; Almutairi, A.T.; Melebari, S.; Shahzad, N. Genetic Analysis and Epidemiological Impact of SARS-CoV-2: A Multinational Study of 1000 Samples Using RT-PCR. Appl. Microbiol. 2024, 4, 133-146. https://doi.org/10.3390/applmicrobiol4010010
Banser T, Abduljaleel Z, Alzabeedi KH, Alzahrani AA, Khogeer AA, Qabbani FH, Almutairi AT, Melebari S, Shahzad N. Genetic Analysis and Epidemiological Impact of SARS-CoV-2: A Multinational Study of 1000 Samples Using RT-PCR. Applied Microbiology. 2024; 4(1):133-146. https://doi.org/10.3390/applmicrobiol4010010
Chicago/Turabian StyleBanser, Talib, Zainularifeen Abduljaleel, Kamal H. Alzabeedi, Adil A. Alzahrani, Asim Abdulaziz Khogeer, Fadel Hassan Qabbani, Ahmed T. Almutairi, Sami Melebari, and Naiyer Shahzad. 2024. "Genetic Analysis and Epidemiological Impact of SARS-CoV-2: A Multinational Study of 1000 Samples Using RT-PCR" Applied Microbiology 4, no. 1: 133-146. https://doi.org/10.3390/applmicrobiol4010010
APA StyleBanser, T., Abduljaleel, Z., Alzabeedi, K. H., Alzahrani, A. A., Khogeer, A. A., Qabbani, F. H., Almutairi, A. T., Melebari, S., & Shahzad, N. (2024). Genetic Analysis and Epidemiological Impact of SARS-CoV-2: A Multinational Study of 1000 Samples Using RT-PCR. Applied Microbiology, 4(1), 133-146. https://doi.org/10.3390/applmicrobiol4010010




