Antibiotic Resistance Patterns of Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa Isolated from Hospital Wastewater
Abstract
:1. Introduction
2. Materials and Methods
2.1. Description of the Study Area
2.2. Samples Collection
2.3. Isolation and Biochemical Identification
2.4. Phenotypic Antibiotic Susceptibility Testing
2.5. Genotypic Analysis
2.5.1. DNA Extraction
2.5.2. Molecular Identification of Bacterial Species
2.5.3. Identification of Resistance Genes
2.5.4. Sequencing and Phylogenetic Analysis
3. Results
3.1. Isolation and Biochemical Identification
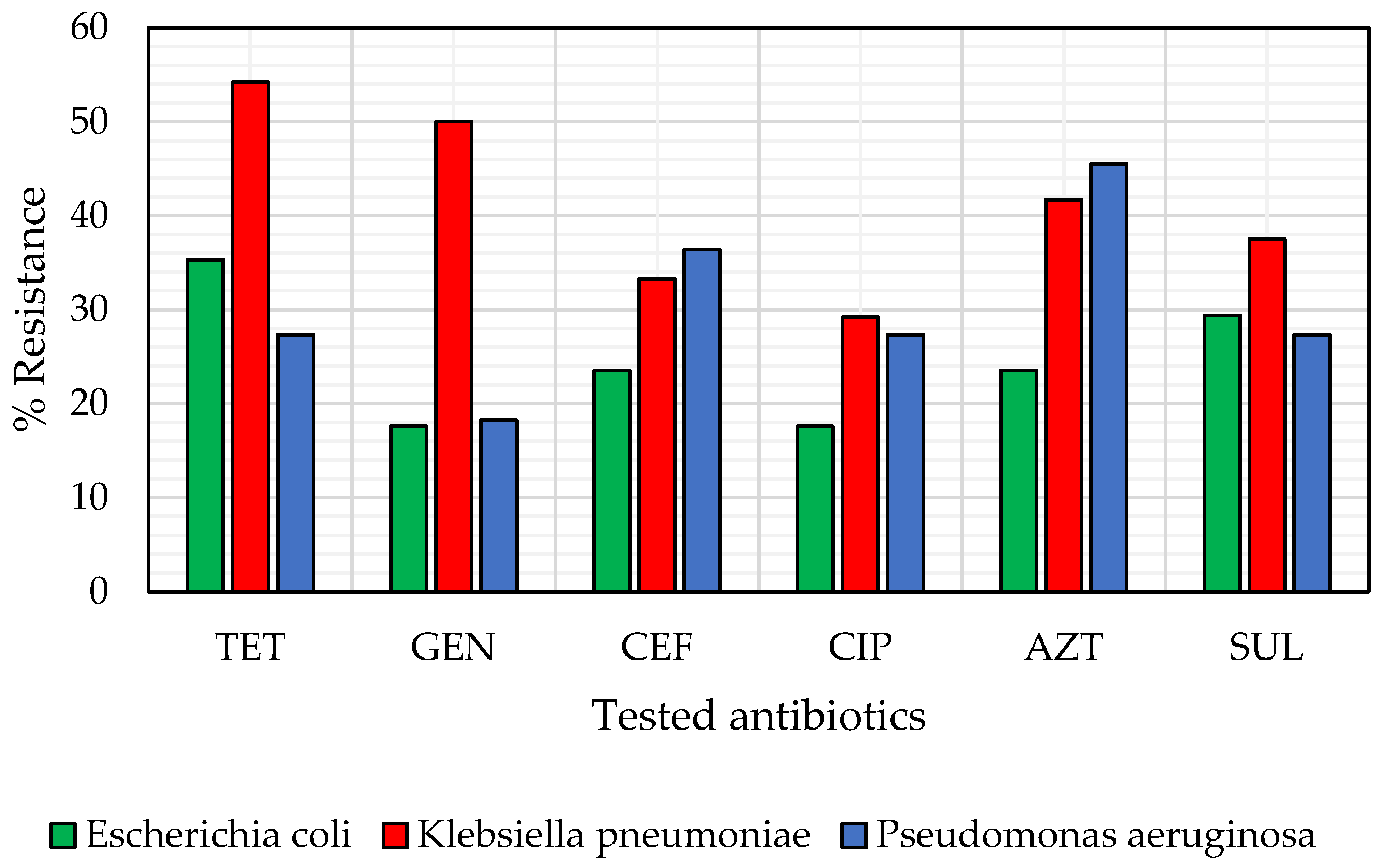
3.2. Phenotypic Antibiotic Susceptibility Testing
3.3. Molecular Identification of Bacterial Species
3.4. Identification of Resistance Genes
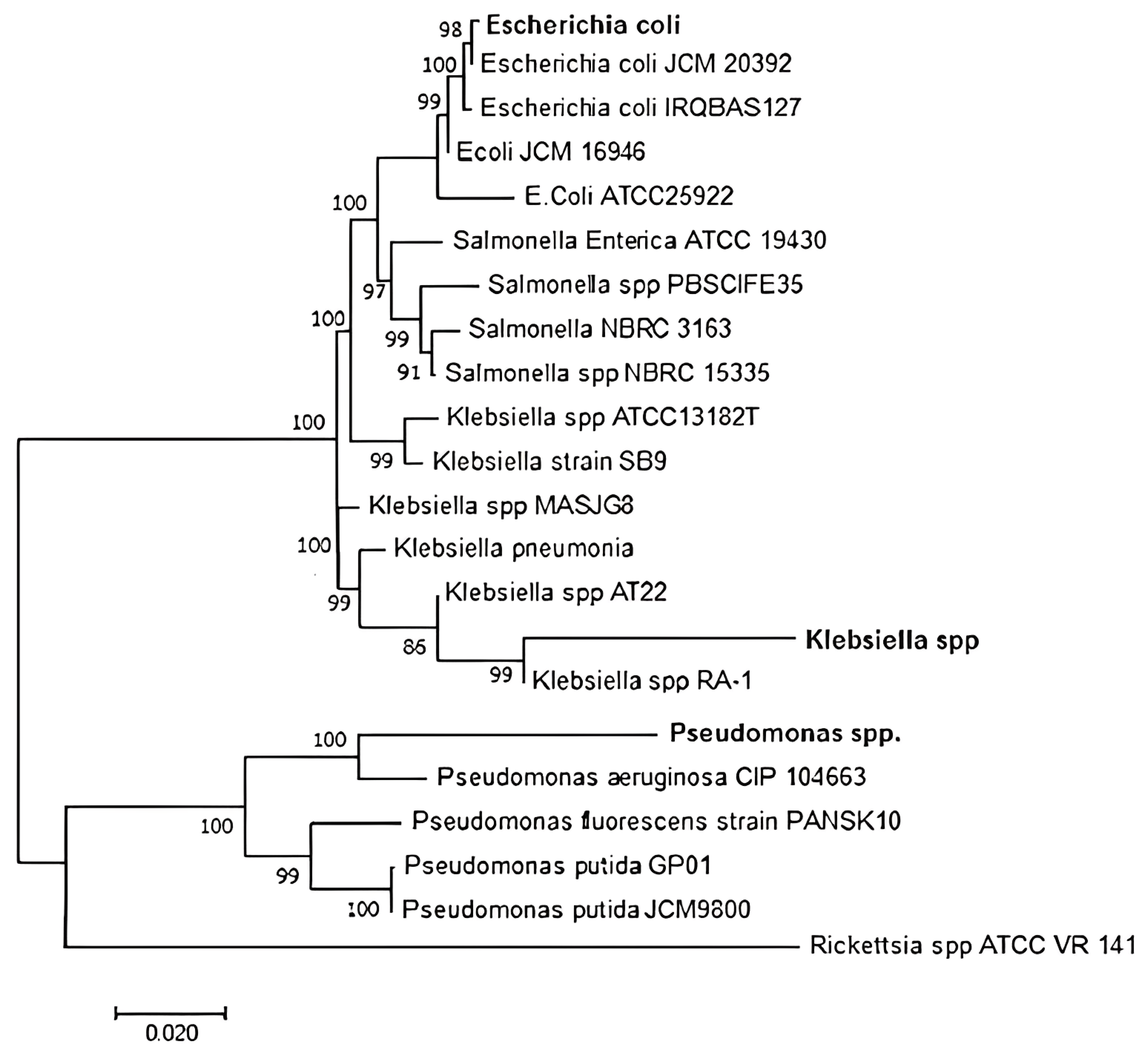
3.5. Sequencing and Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, M.; Jaiswal, S.; Sodhi, K.K.; Shree, P.; Singh, D.K.; Agrawal, P.K.; Shukla, P. Antibiotics bioremediation: Perspectives on its ecotoxicity and resistance. Environ. Int. 2019, 124, 448–461. [Google Scholar] [CrossRef]
- Ben, Y.; Fu, C.; Hu, M.; Liu, L.; Wong, M.H.; Zheng, C. Human health risk assessment of antibiotic resistance associated with antibiotic residues in the environment: A review. Environ. Res. 2019, 169, 483–493. [Google Scholar] [CrossRef]
- Le Page, G.; Gunnarsson, L.; Snape, J.; Tyler, C.R. Integrating human and environmental health in antibiotic risk assessment: A critical analysis of protection goals, species sensitivity and antimicrobial resistance. Environ. Int. 2017, 109, 155–169. [Google Scholar] [CrossRef]
- Amangelsin, Y.; Semenova, Y.; Dadar, M.; Aljofan, M.; Bjørklund, G. The Impact of Tetracycline Pollution on the Aquatic Environment and Removal Strategies. Antibiotics 2023, 12, 440. [Google Scholar] [CrossRef]
- Sabri, N.; van Holst, S.; Schmitt, H.; van der Zaan, B.; Gerritsen, H.; Rijnaarts, H.; Langenhoff, A. Fate of antibiotics and antibiotic resistance genes during conventional and additional treatment technologies in wastewater treatment plants. Sci. Total Environ. 2020, 741, 140199. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ryu, D.; Houtkooper, R.H.; Auwerx, J. Antibiotic use and abuse: A threat to mitochondria and chloroplasts with impact on research, health, and environment. BioEssays 2015, 37, 1045–1053. [Google Scholar] [CrossRef]
- Chen, J.; Cai, Y.; Deng, W.; Xing, S.; Liao, X. Transmission of tetracycline resistance genes and microbiomes from manure-borne black soldier fly larvae frass to rhizosphere soil and pakchoi endophytes. Front. Microbiol. 2022, 13, 1014910. [Google Scholar] [CrossRef]
- Hanna, N.; Tamhankar, A.J.; Lundborg, C.S. Antibiotic concentrations and antibiotic resistance in aquatic environments of the WHO Western Pacific and South-East Asia regions: A systematic review and probabilistic environmental hazard assessment. Lancet Planet. Health 2023, 7, e45–e54. [Google Scholar] [CrossRef]
- Qiu, J.; Chen, Y.; Feng, Y.; Li, X.; Xu, J.; Jiang, J. Adaptation of Rhizosphere Microbial Communities to Continuous Exposure to Multiple Residual Antibiotics in Vegetable Farms. Int. J. Environ. Res. Public Health 2023, 20, 3137. [Google Scholar] [CrossRef]
- Carvalho, I.T.; Santos, L. Antibiotics in the aquatic environments: A review of the European scenario. Environ. Int. 2016, 94, 736–757. [Google Scholar] [CrossRef]
- Konopka, J.K.; Chatterjee, P.; LaMontagne, C.; Brown, J. Environmental impacts of mass drug administration programs: Exposures, risks, and mitigation of antimicrobial resistance. Infect. Dis. Poverty 2022, 11, 78. [Google Scholar] [CrossRef]
- Dires, S.; Birhanu, T.; Ambelu, A.; Sahilu, G. Antibiotic resistant bacteria removal of subsurface flow constructed wetlands from hospital wastewater. J. Environ. Chem. Eng. 2018, 6, 4265–4272. [Google Scholar] [CrossRef]
- Kunhikannan, S.; Thomas, C.J.; Franks, A.E.; Mahadevaiah, S.; Kumar, S.; Petrovski, S. Environmental hotspots for antibiotic resistance genes. Microbiologyopen 2021, 10, e1197. [Google Scholar] [CrossRef]
- Khan, M.T.; Shah, I.A.; Ihsanullah, I.; Naushad, M.; Ali, S.; Shah, S.H.A.; Mohammad, A.W. Hospital wastewater as a source of environmental contamination: An overview of management practices, environmental risks, and treatment processes. J. Water Process Eng. 2021, 41, 101990. [Google Scholar] [CrossRef]
- Vieira, Y.; Pereira, H.A.; Leichtweis, J.; Mistura, C.M.; Foletto, E.L.; Oliveira, L.F.; Dotto, G.L. Effective treatment of hospital wastewater with high-concentration diclofenac and ibuprofen using a promising technology based on degradation reaction catalyzed by Fe0 under microwave irradiation. Sci. Total Environ. 2021, 783, 146991. [Google Scholar] [CrossRef] [PubMed]
- Ben, Y.; Hu, M.; Zhang, X.; Wu, S.; Wong, M.H.; Wang, M.; Andrews, C.B.; Zheng, C. Efficient detection and assessment of human exposure to trace antibiotic residues in drinking water. Water Res. 2020, 175, 115699. [Google Scholar] [CrossRef] [PubMed]
- Sosa-Hernández, J.E.; Rodas-Zuluaga, L.I.; López-Pacheco, I.Y.; Melchor-Martínez, E.M.; Aghalari, Z.; Limón, D.S.; Iqbal, H.M.; Parra-Saldívar, R. Sources of antibiotics pollutants in the aquatic environment under SARS-CoV-2 pandemic situation. Case Stud. Chem. Environ. Eng. 2021, 4, 100127. [Google Scholar] [CrossRef]
- Chang, Q.; Wang, W.; Regev-Yochay, G.; Lipsitch, M.; Hanage, W.P. Antibiotics in agriculture and the risk to human health: How worried should we be? Evol. Appl. 2015, 8, 240–247. [Google Scholar] [CrossRef] [Green Version]
- Ma, F.; Xu, S.; Tang, Z.; Li, Z.; Zhang, L. Use of antimicrobials in food animals and impact of transmission of antimicrobial resistance on humans. Biosaf. Health 2021, 3, 32–38. [Google Scholar] [CrossRef]
- Verraes, C.; Van Boxstael, S.; Van Meervenne, E.; Van Coillie, E.; Butaye, P.; Catry, B.; de Schaetzen, M.-A.; Van Huffel, X.; Imberechts, H.; Dierick, K.; et al. Antimicrobial resistance in the food chain: A review. Int. J. Environ. Res. Public Health 2013, 10, 2643–2669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Kong, L.; Gao, H.; Cheng, X.; Wang, X. A Review of Current Bacterial Resistance to Antibiotics in Food Animals. Front. Microbiol. 2022, 13, 822689. [Google Scholar] [CrossRef] [PubMed]
- Stanton, I.C.; Bethel, A.; Leonard, A.F.C.; Gaze, W.H.; Garside, R. Existing evidence on antibiotic resistance exposure and transmission to humans from the environment: A systematic map. Environ. Evid. 2022, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Lu, Y.; Zhang, J.; Liu, S.; Song, H. Constructed Wetland Revealed Efficient Sulfamethoxazole Removal but Enhanced the Spread. Molecules 2020, 25, 834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Shen, W.; Yan, L.; Wang, X.H.; Xu, H. Stepwise impact of urban wastewater treatment on the bacterial community structure, antibiotic contents, and prevalence of antimicrobial resistance. Environ. Pollut. 2017, 231, 1578–1585. [Google Scholar] [CrossRef] [PubMed]
- Monsalves, N.; Leiva, A.M.; Gómez, G.; Vidal, G. Antibiotic-Resistant Gene Behavior in Constructed Wetlands Treating Sewage: A Critical Review. Sustainability 2022, 14, 8524. [Google Scholar] [CrossRef]
- Li, S.; Zhang, C.; Li, F.; Hua, T.; Zhou, Q.; Ho, S.H. Technologies towards antibiotic resistance genes (ARGs) removal from aquatic environment: A critical review. J. Hazard. Mater. 2021, 411, 125148. [Google Scholar] [CrossRef]
- Kappell, A.D.; Kimbell, L.K.; Seib, M.D.; Carey, D.E.; Choi, M.J.; Kalayil, T.; Fujimoto, M.; Zitomer, D.H.; McNamara, P.J. Removal of antibiotic resistance genes in an anaerobic membrane bioreactor treating primary clarifier effluent at 20 °C. Environ. Sci. Water Res. Technol. 2018, 4, 1783–1793. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Harb, M.; Zarei-Baygi, A.; Stadler, L.B.; Smith, A.L. Comparative Analysis of Intracellular and Extracellular Antibiotic Resistance Gene Abundance in Anaerobic Membrane Bioreactor Effluent. bioRxiv 2019, 702076. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Deng, G.T.; Kou, K.I. Asymptotic behavior of fractional Laplacians in the half space. Appl. Math. Comput. 2015, 254, 125–132. [Google Scholar] [CrossRef]
- Stange, C.; Sidhu, J.P.S.; Toze, S.; Tiehm, A. Comparative removal of antibiotic resistance genes during chlorination, ozonation, and UV treatment. Int. J. Hyg. Environ. Health 2019, 222, 541–548. [Google Scholar] [CrossRef]
- Chen, P.; Yu, X.; Zhang, J.; Wang, Y. New and traditional methods for antibiotic resistance genes removal: Constructed wetland technology and photocatalysis technology. Front. Microbiol. 2023, 13, 1110793. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhuang, Y.; Geng, J.; Ren, H.; Xu, K.; Ding, L. Reduction of antibiotic resistance genes in municipal wastewater effluent by advanced oxidation processes. Sci. Total Environ. 2016, 550, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Ghernaout, D.; Elboughdiri, N. Is Not It Time to Stop Using Chlorine for Treating Water? OALib 2020, 7, 1106007. [Google Scholar] [CrossRef]
- Liu, L.; Liu, Y.H.; Wang, Z.; Liu, C.X.; Huang, X.; Zhu, G.F. Behavior of tetracycline and sulfamethazine with corresponding resistance genes from swine wastewater in pilot-scale constructed wetlands. J. Hazard. Mater. 2011, 278, 304–310. [Google Scholar] [CrossRef]
- Vymazal, J. Plants used in constructed wetlands with horizontal subsurface flow: A review. Hydrobiologia 2011, 674, 133–156. [Google Scholar] [CrossRef]
- Song, H.-L.; Zhang, S.; Guo, J.; Yang, Y.-L.; Zhang, L.-M.; Li, H.; Yang, X.-L.; Liu, X. Vertical up-flow constructed wetlands exhibited efficient antibiotic removal but induced antibiotic resistance genes in effluent. Chemosphere 2018, 203, 434–441. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Liu, C.; Li, K.; Su, J.; Zhu, G.; Liu, L. Performance of vertical up-flow constructed wetlands on swine wastewater containing tetracyclines and tet genes. Water Res. 2015, 70, 109–117. [Google Scholar] [CrossRef]
- Karlowsky, J.A.; Jones, M.E.; Draghi, D.C.; Thornsberry, C.; Sahm, D.F.; Volturo, G.A. Prevalence and antimicrobial susceptibilities of bacteria isolated from blood cultures of hospitalized patients in the United States in 2002. Ann. Clin. Microbiol. Antimicrob. 2004, 3, 7. [Google Scholar] [CrossRef] [Green Version]
- Bassetti, M.; Magnè, F.; Giacobbe, D.R.; Bini, L.; Vena, A. New antibiotics for Gram-negative pneumonia. Eur. Respir. Rev. 2022, 31, 166. [Google Scholar] [CrossRef]
- Ahmed, N.J. The Most Predominant Gram Negative Bacteria in a Public Hospital. J. Pharm. Res. Int. 2021, 33, 39–45. [Google Scholar] [CrossRef]
- Santos, J.V.d.O.; Júnior, S.D.d.C.; Medeiros, S.M.d.F.R.d.S.; Cavalcanti, I.D.L.; de Souza, J.B.; Coriolano, D.L.; da Silva, W.R.C.; Alves, M.H.M.E.; Cavalcanti, I.M.F. Panorama of Bacterial Infections Caused by Epidemic Resistant Strains. Curr. Microbiol. 2022, 79, 175. [Google Scholar] [CrossRef]
- Mkude, I.T.; Saria, J. Assessment of waste stabilization ponds (WSP) efficiency on wastewater treatment for agriculture reuse and other activities a case of Dodoma Municipality, Tanzania. Ethiop. J. Environ. Stud. Manag. 2014, 7, 298–304. [Google Scholar] [CrossRef]
- Desta, A.F.; Assefa, F.; Leta, S.; Stomeo, F.; Wamalwa, M.; Njahira, M.; Appolinaire, D. Microbial community structure and diversity in an integrated system of anaerobic-aerobic reactors and a constructed wetland for the treatment of tannery wastewater in Modjo, Ethiopia. PLoS ONE 2014, 9, e115576. [Google Scholar] [CrossRef] [PubMed]
- Imfeld, G.; Aragonés, C.E.; Fetzer, I.; Mészáros, É.; Zeiger, S.; Nijenhuis, I.; Nikolausz, M.; Delerce, S.; Richnow, H.H. Characterization of microbial communities in the aqueous phase of a constructed model wetland treating 1,2-dichloroethene-contaminated groundwater. FEMS Microbiol. Ecol. 2010, 72, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Mara, D.; Horan, N. Handbook of Water and Wastewater Microbiology; Elsevier: Amsterdam, The Netherlands, 2003. [Google Scholar] [CrossRef]
- Reasoner, D.J. Microbiology: Detection of bacterial pathogens and their occurrence. J. Water Pollut. Control. Fed. 1982, 54, 946–980. [Google Scholar]
- Jemal, M.; Tinshku, F.; Nigussie, Y.; Kefyalew, B.; Alemu, C.; Belay, M.; Belachew, T.; Ayelegn, B. Trend Analysis of Multidrug-Resistant Bacterial Pathogens Causing Neonatal Sepsis at University of Gondar Comprehensive Specialized Hospital, Northwest Ethiopia: A Retrospective Study. Int. J. Microbiol. 2021, 2021, 9992994. [Google Scholar] [CrossRef]
- Abbott, S.L.; Cheung, W.K.W.; Janda, J.M. The genus Aeromonas: Biochemical characteristics, atypical reactions, and phenotypic identification schemes. J. Clin. Microbiol. 2003, 41, 2348–2357. [Google Scholar] [CrossRef] [Green Version]
- El Deen, A.E.N.; Dorgham, M.; Hassan, A.H.M.; Hakim, A.S. Studies on Aeromonas hydrophila in cultured Oreochromis niloticus at Kafr El Sheikh Governorate, Egypt with reference to histopathological alterations in some. World J. Fish Mar. Sci. 2014, 3, 233–240. [Google Scholar] [CrossRef]
- Marijani, E. Prevalence and Antimicrobial Resistance of Bacteria Isolated from Marine and Freshwater Fish in Tanzania. Int. J. Microbiol. 2022, 2022, 4652326. [Google Scholar] [CrossRef]
- Some, S.; Mondal, R.; Mitra, D.; Jain, D.; Verma, D.; Das, S. Microbial pollution of water with special reference to coliform bacteria and their nexus with environment. Energy Nexus 2021, 1, 100008. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- Hoorzook, K.B.; Barnard, T.G. Culture independent DNA extraction method for bacterial cells concentrated from water. MethodsX 2022, 9, 101653. [Google Scholar] [CrossRef]
- Zaman, R.U.; Bushra, A.; Pospo, T.A.; Runa, M.A.; Tasnuva, S.; Parvin, M.S.; Islam, T. Detection of antimicrobial resistance genes in Lactobacillus spp. from poultry probiotic products and their horizontal transfer among Escherichia coli. Vet. Anim. Sci. 2023, 20, 100292. [Google Scholar] [CrossRef] [PubMed]
- Dashti, A.A.; Jadaon, M.M.; Abdulsamad, A.M.; Dashti, H.M. Heat treatment of bacteria: A simple method of DNA extraction for molecular techniques. Kuwait Med. J. 2009, 41, 117–122. [Google Scholar]
- Chentouf, H.F.; Rahli, F.; Benmechernene, Z.; Barros-Velazquez, J. 16S rRNA gene sequencing and MALDI TOF mass spectroscopy identification of Leuconostoc mesenteroides isolated from Algerian raw camel milk. J. Genet. Eng. Biotechnol. 2023, 21, 51. [Google Scholar] [CrossRef]
- Clarridge, J.E. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 2004, 17, 840–862. [Google Scholar] [CrossRef] [Green Version]
- Carriero, M.M.; Maia, A.A.M.; Sousa, R.L.M.; Henrique-Silva, F. Characterization of a new strain of Aeromonas dhakensis isolated from diseased pacu fish (Piaractus mesopotamicus) in Brazil. J. Fish Dis. 2016, 39, 1285–1295. [Google Scholar] [CrossRef]
- Srinivasan, R.; Karaoz, U.; Volegova, M.; MacKichan, J.; Kato-Maeda, M.; Miller, S.; Nadarajan, R.; Brodie, E.L.; Lynch, S.V. Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens. PLoS ONE 2015, 10, e0117617. [Google Scholar] [CrossRef]
- Fauzi, N.N.F.N.M.; Hamdan, R.H.; Mohamed, M.; Ismail, A.; Zin, A.A.M.; Mohamad, N.F.A. Prevalence, antibiotic susceptibility, and presence of drug resistance genes in Aeromonas spp. isolated from freshwater fish in Kelantan and Terengganu states, Malaysia. Vet. World 2021, 14, 2064–2072. [Google Scholar] [CrossRef] [PubMed]
- Agusi, E.; Kabantiyok, D.; Mkpuma, N.; Atai, R. Multidrug Resistant Escherichia Coli Isolates Expresses Several Virulence Genes in Poultry Samples from Jos, Nigeria. BMJ 2022, 1–15. [Google Scholar] [CrossRef]
- Kayode, A.J.; Okoh, A.I. Assessment of multidrug-resistant Listeria monocytogenes in milk and milk product and One Health perspective. PLoS ONE 2022, 17, e270993. [Google Scholar] [CrossRef]
- Schwartz, T.; Kohnen, W.; Jansen, B.; Obst, U. Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol. Ecol. 2003, 43, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Yang, X.; Jiao, S.; Zhang, J.; Ye, B.; Gao, S. Sulfonamide-resistant bacteria and their resistance genes in soils fertilized with manures from Jiangsu province, Southeastern China. PLoS ONE 2014, 9, e112626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tofteland, S.; Haldorsen, B.; Dahl, K.H.; Simonsen, G.S.; Steinbakk, M.; Walsh, T.R.; Sundsfjord, A. Effects of phenotype and genotype on methods for detection of extended-spectrum-β-lactamase-producing clinical isolates of Escherichia coli and Klebsiella pneumoniae in Norway. J. Clin. Microbiol. 2007, 45, 199–205. [Google Scholar] [CrossRef] [Green Version]
- Kallau, N.H.G.; Wibawan, I.W.T.; Lukman, D.W.; Sudarwanto, M.B. Detection of multi-drug resistant (MDR) Escherichia coli and tet gene prevalence at a pig farm in Kupang, Indonesia. J. Adv. Vet. Anim. Res. 2018, 5, 388–396. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Saitou, N.; Nei, M. ESCALA CIWA-AR Escala CIWA-Ar(Clinical Institute Withdrawal Assesment for Alcohol) Evaluación del Síndrome de Abstinencia Alcohólica. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar] [CrossRef]
- Addae-Nuku, D.S.; Kotey, F.C.; Dayie, N.T.; Osei, M.-M.; Tette, E.M.; Debrah, P.; Donkor, E.S. Multidrug-Resistant Bacteria in Hospital Wastewater of the Korle Bu Teaching Hospital in Accra, Ghana. Environ. Health Insights 2022, 16, 11786302221130613. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Yadav, B.; Tyagi, R.D. Microbiology of hospital wastewater. In Current Developments in Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2020; p. 14. [Google Scholar] [CrossRef]
- Yan, W.; Wang, N.; Wei, D.; Liang, C.; Chen, X.; Liu, L.; Shi, J. Bacterial community compositions and nitrogen metabolism function in a cattle farm wastewater treatment plant revealed by Illumina high-throughput sequencing. Environ. Sci. Pollut. Res. 2021, 28, 40895–40907. [Google Scholar] [CrossRef]
- Asfaw, T.; Negash, L.; Kahsay, A.; Weldu, Y. Antibiotic Resistant Bacteria from Treated and Untreated Hospital Wastewater at Ayder Referral Hospital, Mekelle, North Ethiopia. Adv. Microbiol. 2017, 7, 871–886. [Google Scholar] [CrossRef] [Green Version]
- Oyeleke, S.B.; Istifanus, N. The microbiological effects of hospital wastes on the environment. Afr. J. Biotechnol. 2009, 8, 6253–6257. [Google Scholar] [CrossRef] [Green Version]
- Reygaert, W. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018, 4, 482–501. [Google Scholar] [CrossRef]
- Lépesová, K.; Olejníková, P.; Mackuľak, T.; Tichý, J.; Birošová, L. Annual changes in the occurrence of antibiotic-resistant coliform bacteria and enterococci in municipal wastewater. Environ. Sci. Pollut. Res. 2019, 26, 18470–18483. [Google Scholar] [CrossRef]
- Mutuku, C.; Gazdag, Z.; Melegh, S. Occurrence of antibiotics and bacterial resistance genes in wastewater: Resistance mechanisms and antimicrobial resistance control approaches. World J. Microbiol. Biotechnol. 2022, 38, 152. [Google Scholar] [CrossRef] [PubMed]
- Grehs, B.W.N.; Linton, M.A.O.; Clasen, B.; de Oliveira Silveira, A.; Carissimi, E. Antibiotic resistance in wastewater treatment plants: Understanding the problem and future perspectives. Arch. Microbiol. 2021, 203, 1009–1020. [Google Scholar] [CrossRef]
- Breijyeh, Z.; Jubeh, B.; Karaman, R. Resistance of Gram-Negative Bacteria to Current Antibacterial Agents and Approaches to Resolve It. Molecules 2020, 25, 1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, V.; Datta, P. Next-generation strategy for treating drug resistant bacteria: Antibiotic hybrids. Indian J. Med. Res. 2019, 149, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Maria, R. Antibiotic resistance: What is so special about multidrug-resistant Gram-negative bacteria? GMS Hyg. Infect. Control 2017, 12, 1–24. [Google Scholar]
- Rozman, U.; Duh, D.; Cimerman, M.; Turk, S.Š. Hospital wastewater effluent: Hot spot for antibiotic resistant bacteria. J. Water Sanit. Hyg. Dev. 2020, 10, 171–178. [Google Scholar] [CrossRef]
- Verlicchi, P.; Al Aukidy, M.; Zambello, E. What have we learned fromworldwide experiences on the management and treatment of hospital effluent?—An overview and a discussion on perspectives. Sci. Total Environ. J. 2015, 514, 467–491. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.S.; Guo, W.Q.; Wu, Q.L.; Ren, N.Q.; Chang, J.S. Electro-peroxone pretreatment for enhanced simulated hospital wastewater treatment and antibiotic resistance genes reduction. Environ. Int. 2018, 115, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Ovung, A.; Bhattacharyya, J. Sulfonamide drugs: Structure, antibacterial property, toxicity, and biophysical interactions. Biophys. Rev. 2021, 13, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Thawng, C.N.; Lee, K.; Wellington, E.M.H.; Cha, C.J. A novel sulfonamide resistance mechanism by two-component flavin-dependent monooxygenase system in sulfonamide-degrading actinobacteria. Environ. Int. 2019, 127, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Balsalobre, L.; Blanco, A.; Alarcón, T. Beta-lactams. In Antibiotic Drug Resistance; Springer International Publishing: Berlin/Heidelberg, Germany, 2019; pp. 57–72. [Google Scholar] [CrossRef]
- Worthington, R.J.; Melander, C. Overcoming resistance to β-Lactam antibiotics. J. Org. Chem. 2013, 78, 4207–4213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bush, K.; Bradford, P.A. β-Lactams and β-Lactamase Inhibitors: An Overview. Cold Spring Harb. Perspect. Med. 2016, 8, 22. [Google Scholar] [CrossRef]
- Waśko, I.; Kozińska, A.; Kotlarska, E.; Baraniak, A. Clinically Relevant β-Lactam Resistance Genes in Wastewater Treatment Plants. Int. J. Environ. Res. Public Health 2022, 19, 21. [Google Scholar] [CrossRef]
- Lin, X.; Ruan, J.; Huang, L.; Zhao, J.; Xu, Y. Comparison of the elimination effectiveness of tetracycline and AmpC β-lactamase resistance genes in a municipal wastewater treatment plant using four parallel processes. Ecotoxicology 2021, 30, 1586–1597. [Google Scholar] [CrossRef]





| Primer Name | Sequence | Expected Band (bp) | Annealing Temp. | Reference |
|---|---|---|---|---|
| Sul1 F Sul1 R | CGGCGTGGGCTACCTGAACG GCCGATCGCGTGAAGGTTCCG | 450 bp | 55 °C | [64] |
| Sul2 F Sul2 R | GCGCTCAAGGCAGATGGCATT GCGTTTGATACCGGCACCCGT | 625 bp | 58 °C | [64] |
| bla SHV F bla SHV R | ATGCGTTATATTCGCCTGTG AGCGTTGGCCAGTGCTCGATC | 862 bp | 58 °C | [65] |
| bla CTXM F bla CTXM R | SCSATGTGCAGYACCAGTAA CCCGCRATATGRTTGGTGGTGGTG | 554 bp | 58 °C | [66] |
| bla TEM F bla TEM R | ATGAGTATTCMCATTTCCG CCMTGCTTMTCAGTGAGG | 858 bp | 50 °C | [65] |
| 16s rDNA F 16s rDNA R | AGAGTTTGATTCATGGCTCAG TACGGYTACCTTGTTACGACTT | 1500 bp | 58 °C | [64] |
| Bacterial ssp. | Number (N) | Sul1 (n) | Sul2 (n) | SHV (n) | CTXM (n) | TEM (n) | MDR Genes (%) |
|---|---|---|---|---|---|---|---|
| Pseudomonas ssp. | 4 | 3 | 0 | 3 | 0 | 1 | 35 |
| Klebsiella ssp. | 4 | 1 | 1 | 1 | 5 | 0 | 40 |
| E. coli ssp. | 4 | 0 | 1 | 1 | 0 | 2 | 20 |
| Resistance genes (%) | 33 | 17 | 42 | 42 | 25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karungamye, P.; Rugaika, A.; Mtei, K.; Machunda, R. Antibiotic Resistance Patterns of Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa Isolated from Hospital Wastewater. Appl. Microbiol. 2023, 3, 867-882. https://doi.org/10.3390/applmicrobiol3030060
Karungamye P, Rugaika A, Mtei K, Machunda R. Antibiotic Resistance Patterns of Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa Isolated from Hospital Wastewater. Applied Microbiology. 2023; 3(3):867-882. https://doi.org/10.3390/applmicrobiol3030060
Chicago/Turabian StyleKarungamye, Petro, Anita Rugaika, Kelvin Mtei, and Revocatus Machunda. 2023. "Antibiotic Resistance Patterns of Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa Isolated from Hospital Wastewater" Applied Microbiology 3, no. 3: 867-882. https://doi.org/10.3390/applmicrobiol3030060
APA StyleKarungamye, P., Rugaika, A., Mtei, K., & Machunda, R. (2023). Antibiotic Resistance Patterns of Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa Isolated from Hospital Wastewater. Applied Microbiology, 3(3), 867-882. https://doi.org/10.3390/applmicrobiol3030060






