Comparison of the Gill Microbiome of Retail Oysters from Two Geographical Locations Exhibited Distinct Microbial Signatures: A Pilot Study for Potential Future Applications for Monitoring Authenticity of Their Origins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Sample Processing and DNA Extraction
2.3. Microbiome Analysis
2.4. Data Analysis
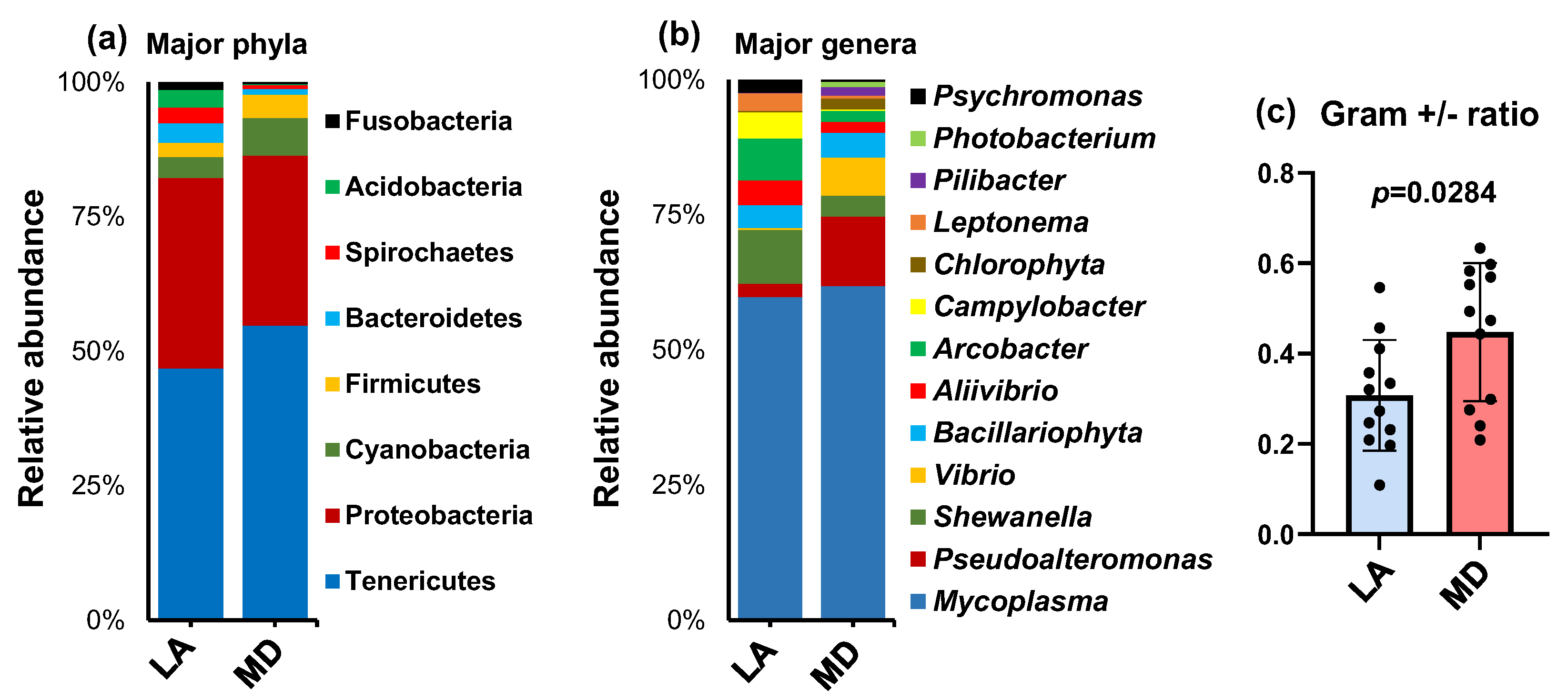
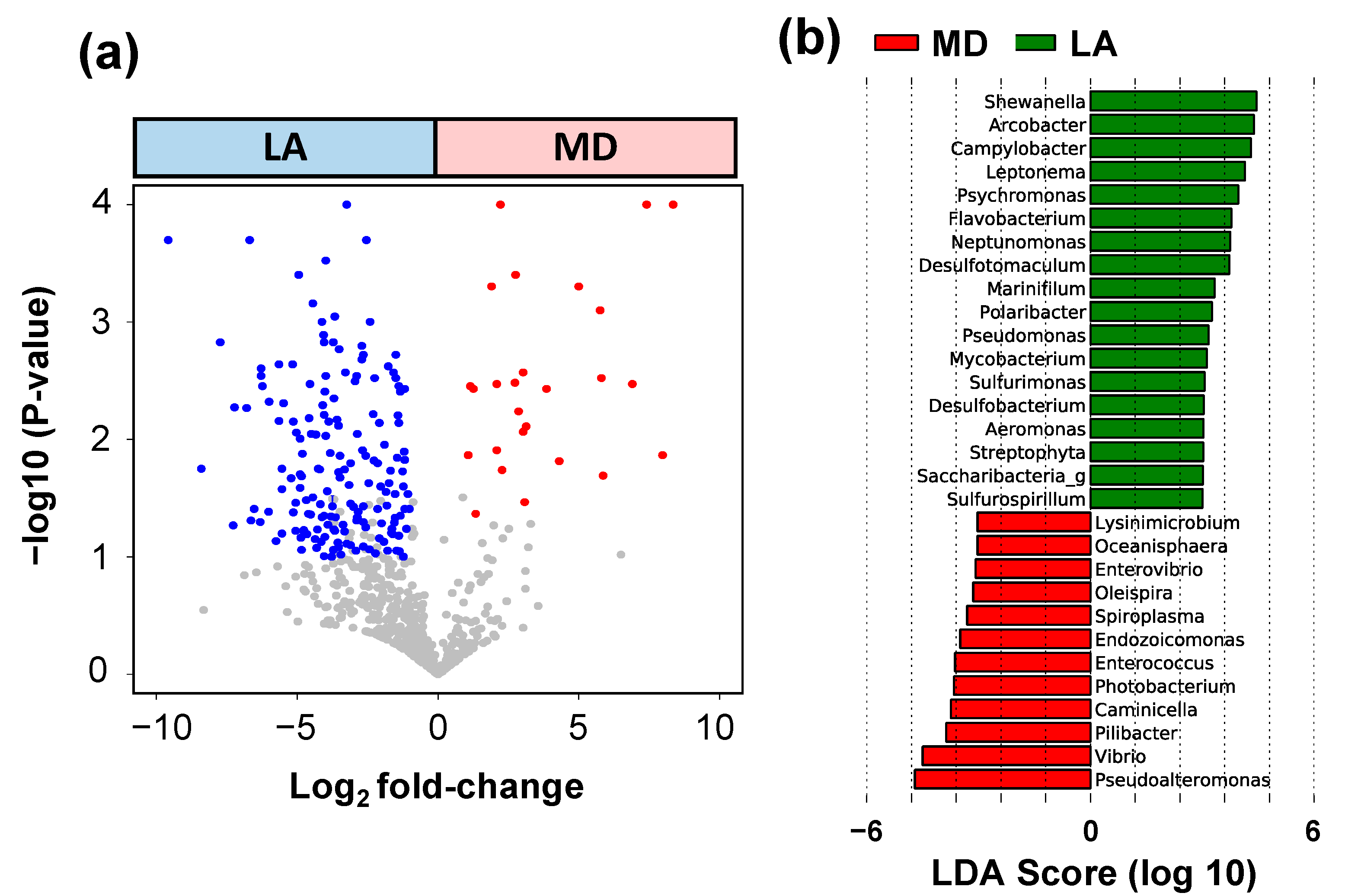
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- NOAA Seafood Import Monitoring Program|NOAA Fisheries. Available online: https://www.fisheries.noaa.gov/international/seafood-import-monitoring-program (accessed on 7 August 2020).
- NOAA Landings. Available online: https://www.fisheries.noaa.gov/foss/f?p=215:200:7602721600498::NO::: (accessed on 20 September 2022).
- Warner, K.; Timme, W.; Lowell, B.; Stiles, M.L. Persistent Seafood Fraud Found in South Florida. Oceana 2012, 1–20. Available online: https://oceana.org/reports/persistent-seafood-fraud-found-south-florida/ (accessed on 10 November 2022).
- Luque, G.M.; Donlan, C.J. The Characterization of Seafood Mislabeling: A Global Meta-Analysis. Biol. Conserv. 2019, 236, 556–570. [Google Scholar] [CrossRef]
- Blue Points: The Most Bastardized Name in Seafood and Why You Should Care|Pangea Shellfish Company. Available online: https://www.pangeashellfish.com/blog/blue-points-the-most-bastardized-name-in-seafood (accessed on 25 October 2022).
- Code of Federal Regulations. ECFR:: 7 CFR Part 60—Country of Origin Labeling for Fish and Shellfish. 2003. Available online: https://www.govinfo.gov/content/pkg/CFR-2012-title7-vol3/xml/CFR-2012-title7-vol3-part60.xml (accessed on 10 November 2022).
- FDA Single Laboratory Validated Method for DNA-Barcoding for the Species Identification of Fish. Available online: https://www.fda.gov/food/dna-based-seafood-identification/single-laboratory-validated-method-dna-barcoding-species-identification-fish (accessed on 30 May 2022).
- Sharma, L.; Nagpal, R.; Jackson, C.R.; Patel, D.; Singh, P. Antibiotic-Resistant Bacteria and Gut Microbiome Communities Associated with Wild-Caught Shrimp from the United States versus Imported Farm-Raised Retail Shrimp. Sci. Rep. 2021, 11, 3356. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Suresh Kumar, K.; Shin, K.-H. Applicability of Stable C and N Isotope Analysis in Inferring the Geographical Origin and Authentication of Commercial Fish (Mackerel, Yellow Croaker and Pollock). Food Chem. 2015, 172, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Black, C.; Chevallier, O.P.; Elliott, C.T. The Current and Potential Applications of Ambient Mass Spectrometry in Detecting Food Fraud. TrAC Trends Anal. Chem. 2016, 82, 268–278. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Ford, S.E.; Bushek, D.; Powell, E.N.; Bao, Z.; Guo, X. Effective Population Sizes of Eastern Oyster Crassostrea virginica (Gmelin) Populations in Delaware Bay, USA. J. Mar. Res. 2012, 70, 357–379. [Google Scholar] [CrossRef]
- Thongda, W.; Zhao, H.; Zhang, D.; Jescovitch, L.N.; Liu, M.; Guo, X.; Schrandt, M.; Powers, S.P.; Peatman, E. Development of SNP Panels as a New Tool to Assess the Genetic Diversity, Population Structure, and Parentage Analysis of the Eastern Oyster (Crassostrea virginica). Mar. Biotechnol. 2018, 20, 385–395. [Google Scholar] [CrossRef]
- Chin Chin, T.; Adibah, A.B.; Danial Hariz, Z.A.; Siti Azizah, M.N. Detection of Mislabelled Seafood Products in Malaysia by DNA Barcoding: Improving Transparency in Food Market. Food Control 2016, 64, 247–256. [Google Scholar] [CrossRef]
- Pacific Oyster Crassostrea Gigas. Available online: http://anchorenvironmental.co.za/sites/default/files/2017-11/BRBA%20C.%20gigas.pdf (accessed on 30 July 2022).
- Stevick, R.J.; Post, A.F.; Gómez-Chiarri, M. Functional Plasticity in Oyster Gut Microbiomes along a Eutrophication Gradient in an Urbanized Estuary. Anim. Microbiome 2021, 3, 5. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration (FDA). Bacteriological Analytical Manual (BAM) R59: Phosphate-Buffered Saline (PBS), PH 7.4. 2020. Available online: https://www.fda.gov/food/laboratory-methods-food/bam-r59-phosphate-buffered-saline-pbs-ph-74 (accessed on 10 November 2022).
- Nagpal, R.; Indugu, N.; Singh, P. Distinct Gut Microbiota Signatures in Mice Treated with Commonly Used Food Preservatives. Microorganisms 2021, 9, 2311. [Google Scholar] [CrossRef]
- Nagpal, R.; Neth, B.J.; Wang, S.; Craft, S.; Yadav, H. Modified Mediterranean-Ketogenic Diet Modulates Gut Microbiome and Short-Chain Fatty Acids in Association with Alzheimer’s Disease Markers in Subjects with Mild Cognitive Impairment. EBioMedicine 2019, 47, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Earth Microbiome Project (EMP). 16S Illumina Amplicon Protocol: Earth Microbiome Project. 2019. Available online: https://earthmicrobiome.org/protocols-and-standards/16s/ (accessed on 10 November 2022).
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-High-Throughput Microbial Community Analysis on the Illumina HiSeq and MiSeq Platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Saccon, T.D.; Nagpal, R.; Yadav, H.; Cavalcante, M.B.; Nunes, A.D.D.C.; Schneider, A.; Gesing, A.; Hughes, B.; Yousefzadeh, M.; Tchkonia, T.; et al. Senolytic Combination of Dasatinib and Quercetin Alleviates Intestinal Senescence and Inflammation and Modulates the Gut Microbiome in Aged Mice. J. Gerontol A Biol. Sci. Med. Sci. 2021, 76, 1895–1905. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for Comprehensive Statistical, Functional, and Meta-Analysis of Microbiome Data. Nat. Protocols 2020, 15, 799–821. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Kashulin, A.; Seredkina, N.; Sørum, H. Cold-Water Vibriosis. The Current Status of Knowledge. J. Fish Dis. 2017, 40, 119–126. [Google Scholar] [CrossRef]
- Kittinger, J.N.; Teh, L.C.L.; Allison, E.H.; Bennett, N.J.; Crowder, L.B.; Finkbeiner, E.M.; Hicks, C.; Scarton, C.G.; Nakamura, K.; Ota, Y.; et al. Committing to Socially Responsible Seafood. Science 2017, 356, 912–913. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Teixeira, J.S.; Ner, S.; Ma, K.V.; Petronella, N.; Banerjee, S.; Ronholm, J. Exploring the Potential of the Microbiome as a Marker of the Geographic Origin of Fresh Seafood. Front. Microbiol. 2020, 11, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Sommer, F.; Bäckhed, F. The Gut Microbiota—Masters of Host Development and Physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, Z.T.; Dufault-Thompson, K.; Russo, K.T.; Scro, A.K.; Smolowitz, R.M.; Gomez-Chiarri, M.; Zhang, Y. Microbiome Analysis Reveals Diversity and Function of Mollicutes Associated with the Eastern Oyster, Crassostrea Virginica. mSphere 2021, 6, e00227-21. [Google Scholar] [CrossRef] [PubMed]
- Pierce, M.L.; Ward, J.E. Gut Microbiomes of the Eastern Oyster (Crassostrea Virginica) and the Blue Mussel (Mytilus Edulis): Temporal Variation and the Influence of Marine Aggregate-Associated Microbial Communities. mSphere 2019, 4, e00730-19. [Google Scholar] [CrossRef] [Green Version]
- De Schryver, P.; Vadstein, O. Ecological Theory as a Foundation to Control Pathogenic Invasion in Aquaculture. ISME J. 2014, 8, 2360–2368. [Google Scholar] [CrossRef] [Green Version]
- King, G.M.; Judd, C.; Kuske, C.R.; Smith, C. Analysis of Stomach and Gut Microbiomes of the Eastern Oyster (Crassostrea Virginica) from Coastal Louisiana, USA. PLoS ONE 2012, 7, e51475. [Google Scholar] [CrossRef] [Green Version]
- Kong, N.; Han, S.; Fu, Q.; Yu, Z.; Wang, L.; Song, L. Impact of Ocean Acidification on the Intestinal Microflora of the Pacific Oyster Crassostrea Gigas. Aquaculture 2022, 546, 737365. [Google Scholar] [CrossRef]
- King, W.L.; Jenkins, C.; Go, J.; Siboni, N.; Seymour, J.R.; Labbate, M. Characterisation of the Pacific Oyster Microbiome During a Summer Mortality Event. Microb. Ecol. 2019, 77, 502–512. [Google Scholar] [CrossRef] [Green Version]
- Chauhan, A.; Wafula, D.; Lewis, D.E.; Pathak, A. Metagenomic Assessment of the Eastern Oyster-Associated Microbiota. Genome Announc. 2014, 2, e01083-14. [Google Scholar] [CrossRef] [Green Version]
- Arfken, A.; Song, B.; Bowman, J.S.; Piehler, M. Denitrification Potential of the Eastern Oyster Microbiome Using a 16S rRNA Gene Based Metabolic Inference Approach. PLOS ONE 2017, 12, e0185071. [Google Scholar] [CrossRef] [Green Version]
- Milan, M.; Maroso, F.; Dalla Rovere, G.; Carraro, L.; Ferraresso, S.; Patarnello, T.; Bargelloni, L.; Cardazzo, B.; Fariselli, P. Tracing Seafood at High Spatial Resolution Using NGS-Generated Data and Machine Learning: Comparing Microbiome versus SNPs. Food Chem. 2019, 286, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, T.; Marcelino, J.; Ricardo, F.; Soares, A.M.V.M.; Calado, R. Bacterial Communities 16S rDNA Fingerprinting as a Potential Tracing Tool for Cultured Seabass Dicentrarchus Labrax. Sci. Rep. 2017, 7, 11862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lokesh, J.; Kiron, V. Transition from Freshwater to Seawater Reshapes the Skin-Associated Microbiota of Atlantic Salmon. Sci. Rep. 2016, 6, 19707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [Green Version]
- Bevins, C.L.; Salzman, N.H. The Potter’s Wheel: The Host’s Role in Sculpting Its Microbiota. Cell Mol. Life Sci. 2011, 68, 3675–3685. [Google Scholar] [CrossRef] [Green Version]
- Ley, R.E.; Peterson, D.A.; Gordon, J.I. Ecological and Evolutionary Forces Shaping Microbial Diversity in the Human Intestine. Cell 2006, 124, 837–848. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations (FAO). Bivalve Depuration: Fundamental and Practical Aspects; FAO: Rome, Italy, 2008; Available online: https://www.fao.org/3/i0201e/i0201e.pdf (accessed on 10 November 2022).


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, P.; Williams, D.; Velez, F.J.; Nagpal, R. Comparison of the Gill Microbiome of Retail Oysters from Two Geographical Locations Exhibited Distinct Microbial Signatures: A Pilot Study for Potential Future Applications for Monitoring Authenticity of Their Origins. Appl. Microbiol. 2023, 3, 1-10. https://doi.org/10.3390/applmicrobiol3010001
Singh P, Williams D, Velez FJ, Nagpal R. Comparison of the Gill Microbiome of Retail Oysters from Two Geographical Locations Exhibited Distinct Microbial Signatures: A Pilot Study for Potential Future Applications for Monitoring Authenticity of Their Origins. Applied Microbiology. 2023; 3(1):1-10. https://doi.org/10.3390/applmicrobiol3010001
Chicago/Turabian StyleSingh, Prashant, David Williams, Frank J. Velez, and Ravinder Nagpal. 2023. "Comparison of the Gill Microbiome of Retail Oysters from Two Geographical Locations Exhibited Distinct Microbial Signatures: A Pilot Study for Potential Future Applications for Monitoring Authenticity of Their Origins" Applied Microbiology 3, no. 1: 1-10. https://doi.org/10.3390/applmicrobiol3010001
APA StyleSingh, P., Williams, D., Velez, F. J., & Nagpal, R. (2023). Comparison of the Gill Microbiome of Retail Oysters from Two Geographical Locations Exhibited Distinct Microbial Signatures: A Pilot Study for Potential Future Applications for Monitoring Authenticity of Their Origins. Applied Microbiology, 3(1), 1-10. https://doi.org/10.3390/applmicrobiol3010001






