Selection of Listeria monocytogenes InlA-Binding Peptides Using Phage Display—Novel Compounds for Diagnostic Applications?
Abstract
1. Introduction
2. Materials and Methods
2.1. Growth of Bacterial Strains
2.2. Protein Purification
2.3. Selection of Phages with Affinity to Internalin A
2.4. Phage-Enzyme-Linked Immunosorbent Assay (ELISA) Binding Assay
2.5. Isolation of Phage DNA and DNA Sequencing
2.6. Peptide Synthesis
2.7. Detection of Peptide Binding to Internalin A
2.8. Detection of Peptide Binding to Listeria Monocytogenes Using ELISA
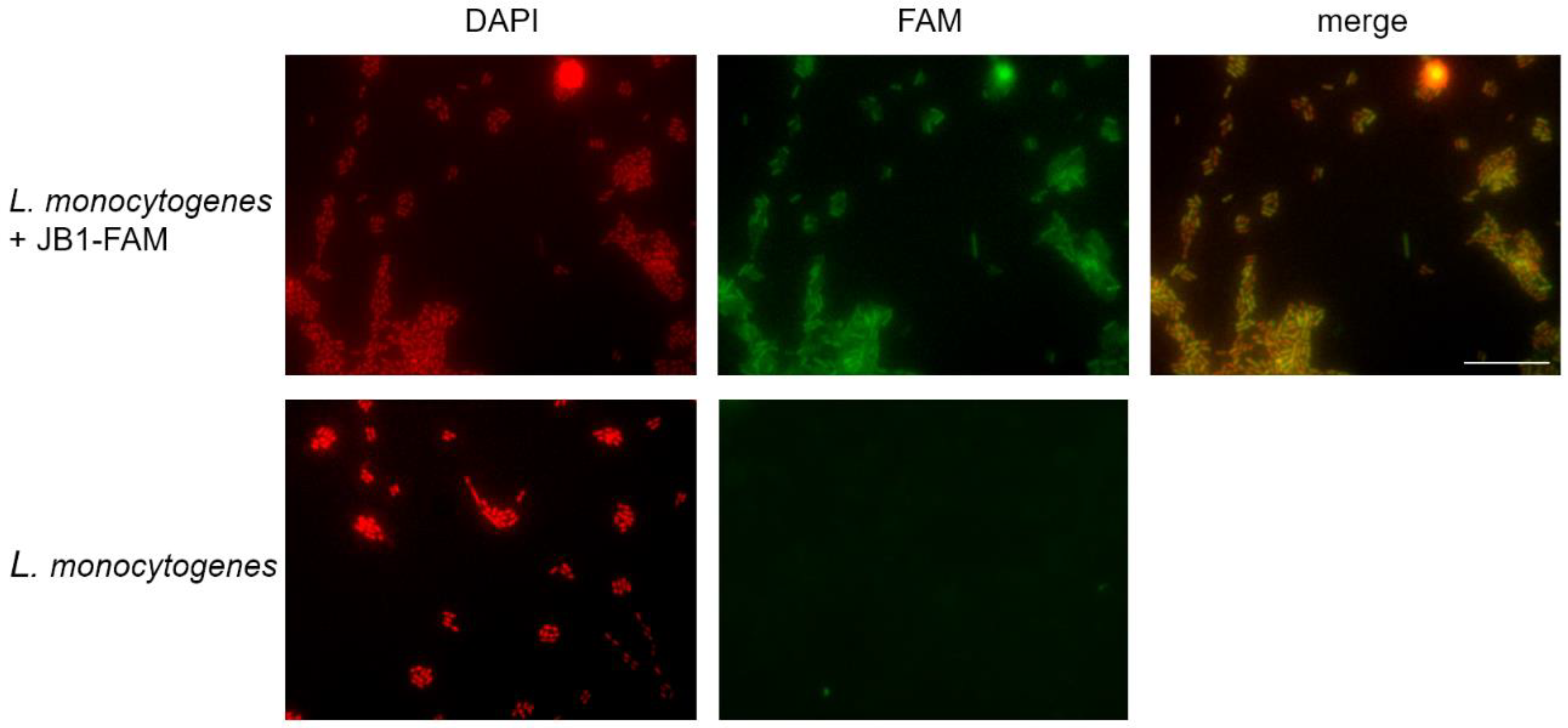
2.9. Detection of Peptide Binding to Listeria Monocytogenes Using Fluorescence Microscopy
2.10. Detection of E-cad GST-InlA Interaction
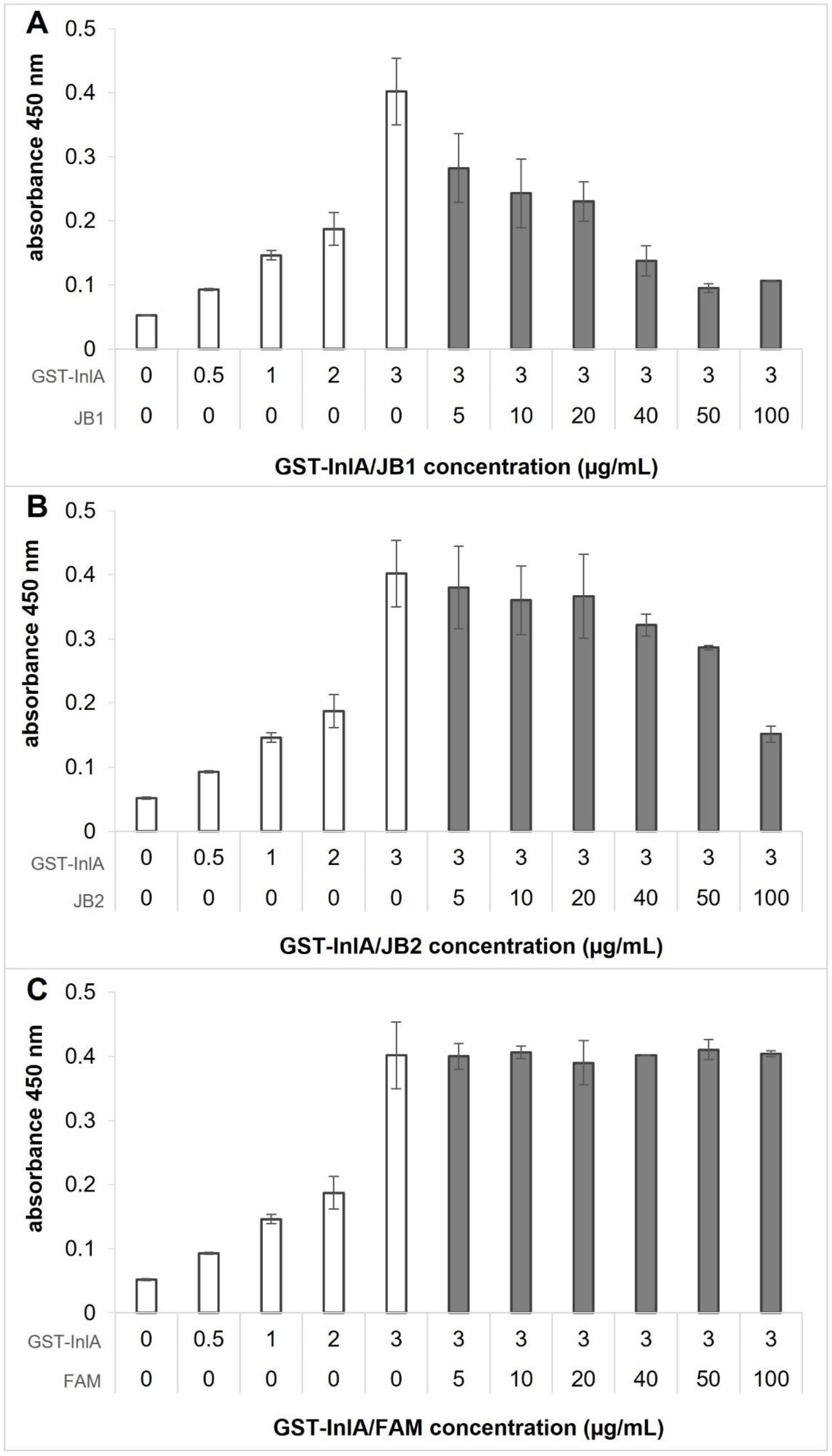
2.11. Inhibition of the E-cad GST-InlA Interaction Using InlA-Binding Peptides
3. Results
3.1. Biopanning of a Phage Display Library against InlA of L. monocytogenes Resulted in Selection of Five Peptides
3.2. Selected Peptides Interact with InlA of L. monocytogenes
3.3. JB1 and JB2 Demonstrate Binding to L. monocytogenes
3.4. JB1 and JB2 Compete with E-cad for Binding to InlA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, Q.; Gooneratne, R.; Hussain, M.A. Listeria monocytogenes in Fresh Produce: Outbreaks, Prevalence and Contamination Levels. Foods 2017, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Carpentier, B.; Cerf, O. Review—Persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 2011, 145, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Doran, K.S.; Banerjee, A.; Disson, O.; Lecuit, M. Concepts and mechanisms: Crossing host barriers. Cold Spring Harb. Perspect. Med. 2013, 3, a013433. [Google Scholar] [CrossRef] [PubMed]
- Lecuit, M. Human listeriosis and animal models. Microbes Infect. 2007, 9, 1216–1225. [Google Scholar] [CrossRef] [PubMed]
- Hof, H. History and epidemiology of listeriosis. FEMS Immunol. Med. Microbiol. 2003, 35, 199–202. [Google Scholar] [CrossRef]
- Cabanes, D.; Dehoux, P.; Dussurget, O.; Frangeul, L.; Cossart, P. Surface proteins and pathogenis potential of Listeria monocytogenes. Trends Microbiol. 2002, 10, 238–245. [Google Scholar] [CrossRef]
- Glaser, P.; Frangeul, L.; Buchrieser, C.; Rusniok, C.; Amend, A.; Baquero, F.; Berche, P.; Bloecker, H.; Brandt, P.; Chakraborty, T.; et al. Comparative genomics of Listeria species. Science 2001, 294, 849–852. [Google Scholar] [CrossRef]
- Disson, O.; Lecuit, M. In vitro and in vivo models to study human listeriosis: Mind the gap. Microbes Infect. 2013, 15, 971–980. [Google Scholar] [CrossRef]
- Tilney, L.G.; Portnoy, D.A. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell Biol. 1989, 109, 1597–1608. [Google Scholar] [CrossRef]
- Gaillard, J.L.; Berche, P.; Frehel, C.; Gouin, E.; Cossart, P. Entry of L. monocytogenes into cells is mediated y internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 1991, 65, 1127–1141. [Google Scholar] [CrossRef]
- Lecuit, M.; Ohayon, H.; Braun, L.; Mengaud, J.; Cossart, P. Internalin of Listeria monocytogenes with an intact leucine-rich repeat region is sufficient to promote internalization. Infect. Immun. 1997, 65, 5309–5319. [Google Scholar] [CrossRef] [PubMed]
- Mengaud, J.; Ohayon, H.; Gounon, P.; Mege, R.M.; Cossart, P. E-cadherin is the receptor for internalin, a surfece protein required for entry of L. monocytogenes into epithelial cells. Cell 1996, 84, 923–932. [Google Scholar] [CrossRef]
- van Roy, F.; Berx, G. The cell-cell adhesion molecule E-cadherin. Cell Mol. Life Sci. 2008, 65, 3756–3788. [Google Scholar] [CrossRef] [PubMed]
- Lecuit, M.; Dramsi, S.; Gottardi, C.; Fedor-Chaiken, M.; Gumbiner, B.; Cossart, P. A single amino acid in E-cadherin responsible for host specificity towords the hman pathogen Listeria monocytogenes. EMBO J. 1999, 18, 3956–3963. [Google Scholar] [CrossRef] [PubMed]
- Schubert, W.D.; Urbanke, C.; Ziehm, T.; Beier, V.; Machner, M.P.; Domann, E.; Wehland, J.; Chakraborty, T.; Heinz, D.W. Structure of internalin, a major invasion protein of Listeria monocytogenes, in complex with its human receptor E-cadherin. Cell 2002, 111, 825–836. [Google Scholar] [CrossRef]
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef]
- Krumpe, L.R.; Mori, T. Potential of phage-displayed peptide library technology to identify functional targeting peptides. Expert Opin. Drug Discov. 2007, 2, 525–537. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Sorokulova, I.B.; Vodyanoy, V.J.; Simonian, A.L. Lytic phage as a specific and selective probe for detection of Staphylococcus aureus—A surface plasmon resonance spectroscopic study. Biosens. Bioelectron. 2007, 22, 948–955. [Google Scholar] [CrossRef]
- Edgar, R.; McKinstry, M.; Hwang, J.; Oppenheim, A.B.; Fekete, R.A.; Giulian, G.; Merril, C.; Nagashima, K.; Adhya, S. High-sensitivity bacterial detection using biotin-tagged phage and quantum-dot nanocomplexes. Proc. Natl. Acad. Sci. USA 2006, 103, 4841–4845. [Google Scholar] [CrossRef]
- Goldman, E.R.; Anderson, G.P.; Liu, J.L.; Delehanty, J.B.; Sherwood, L.J.; Osborn, L.E.; Cummins, L.B.; Hayhurst, A. Facile generation of heat-stable antiviral and antitoxin single domain antibodies from a semisynthetic llama library. Anal. Chem. 2006, 78, 8245–8255. [Google Scholar] [CrossRef]
- Olsen, E.V.; Pathirana, S.T.; Samoylov, A.M.; Barbaree, J.M.; Chin, B.A.; Neely, W.C.; Vodyanoy, V. Specific and selective biosensor for Salmonella and its detection in the environment. J. Microbiol. Methods 2003, 53, 273–285. [Google Scholar] [CrossRef]
- Rao, S.S.; Mohan, K.V.K.; Gao, Y.; Atreya, C.D. Identification and evaluation of a novel peptide binding to the cell surface of Staphylococcus aureus. Microbiol. Res. 2013, 168, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Sorokulova, I.B.; Olsen, E.V.; Chen, I.-H.; Fiebor, B.; Barbaree, J.M.; Vodyanoy, V.J.; Chin, B.A.; Petrenko, V.A. Landscape phage probes for Salmonella typhimurium. J. Microbiol. Methods 2005, 63, 55–72. [Google Scholar] [CrossRef]
- Kenzel, J.; Schlegelmilch, L.; Funke, S.A. Phage display selection of specific ligands for Listeria monocytogenes—novel tools for diagnostic of therapeutic purposes. J. Microbiol. Biotechnol. 2018, 7, 31–38. [Google Scholar]
- Moreira, G.M.S.G.; Gronow, S.; Dübel, S.; Mendonça, M.; Moreira, Â.N.; Conceição, F.R.; Hust, M. Phage Display-Derived Monoclonal Antibodies Against Internalins A and B Allow Specific Detection of Listeria monocytogenes. Front. Public Health 2022, 10, 712657. [Google Scholar] [CrossRef] [PubMed]
- Behravesh, C.B.; Jones, T.F.; Vugia, D.J.; Long, C.; Marcus, R.; Smith, K.; Thomas, S.; Zansky, S.; Fullerton, K.E.; Henao, O.L.; et al. Deaths Associated with Bacterial Pathogens Transmitted Commonly Through Food: Foodborne Diseases Active Surveillance Network (FoodNet), 1996–2005. J. Infect. Dis. 2011, 204, 263–267. [Google Scholar] [CrossRef]
- Aureli, P.; Ferrini, A.M.; Mannoni, V.; Hodzic, S.; Wedell-Weergaard, C.; Oliva, B. Susceptibility of Listeria monocytogenes isolated from food in Italy to antibiotics. Int. J. Food Microbiol. 2003, 83, 325–330. [Google Scholar] [CrossRef]
- Li, L.; Olsen, R.H.; Ye, L.; Wang, W.; Shi, L.; Yan, H.; Meng, H. Characterization of Antimicrobial Resistance of Listeria monocytogenes Strains Isolated from a Pork Processing Plant and Its Respective Meat Markets in Southern China. Foodborne Pathog. Dis. 2016, 13, 262–268. [Google Scholar] [CrossRef]
- Li, Q.; Sherwood, J.S.; Logue, C.M. Antimicrobial resistance of Listeria spp. recovered from processed bison. Lett. Appl. Microbiol. 2007, 44, 86–91. [Google Scholar] [CrossRef]
- Poyart-Salmeron, C.; Carlier, C.; Trieu-Cuot, P.; Courtieu, A.L.; Courvalin, P. Transferable plasmid-mediated antibiotic resistance in Listeria monocytogenes. Lancet 1990, 335, 1422–1426. [Google Scholar] [CrossRef]
- Zhao, X.; Lin, C.-W.; Wang, J.; Oh, D.H. Advances in rapid detection methods for foodborne pathogens. J. Microbiol. Biotechnol. 2014, 24, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Ohk, S.-H.; Bhunia, A.K. Multiplex fiber optic biosensor for detection of Listeria monocytogenes, Escherichia coli O157:H7 and Salmonella enterica from ready-to-eat meat samples. Food Microbiol. 2013, 33, 166–171. [Google Scholar] [CrossRef]
- Pochop, J.; Kačániová, M.; Hleba, L.; Lopasovský, L.; Bobková, A.; Zeleňáková, L.; Stričík, M. Detection of Listeria monocytogenes in ready-to-eat food by Step One real-time polymerase chain reaction. J. Environ. Sci. Health B 2012, 47, 212–216. [Google Scholar] [CrossRef] [PubMed]
- Portanti, O.; Di Febo, T.; Luciani, M.; Pompilii, C.; Lelli, R.; Semprini, P. Development and validation of an antigen capture ELISA based on monoclonal antibodies specific for Listeria monocytogenes in food. Vet. Ital. 2011, 47, 281–290. [Google Scholar]
- Hof, H.; Hefner, P. Pathogenicity of Listeria monocytogenes in comparison to other Listeria species. Infection 1988, 16, S141–S1444. [Google Scholar] [CrossRef] [PubMed]
- Paoli, G.C.; Kleina, L.G.; Brewster, J.D. Development of Listeria monocytogenes-specific immunomagnetic beads using a single-chain antibody fragment. Foodborne Pathog. Dis. 2007, 4, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Bhunia, A.K.; Ball, P.H.; Fuad, A.T.; Kurz, B.W.; Emerson, J.W.; Johnson, M.G. Development and characterization of a monoclonal antibody specific for Listeria monocytogenes and Listeria innocua. Infect. Immun. 1991, 59, 3176–3184. [Google Scholar] [CrossRef]
- Charlier, C.; Perrodeau, É.; Leclercq, A.; Cazenave, B.; Pilmis, B.; Henry, B.; Lopes, A.; Maury, M.M.; Moura, A.; Goffinet, F.; et al. Clinical features and prognostic factors of listeriosis: The MONALISA national prospective cohort study. Lancet Infect. Dis. 2017, 17, 510–519. [Google Scholar] [CrossRef]
- Lathrop, A.A.; Jaradat, Z.W.; Haley, T.; Bhunia, A.K. Characterization and application of a Listeria monocytogenes reactive monoclonal antibody C11E9 in a resonant mirror biosensor. J. Immunol. Methods 2003, 281, 119–128. [Google Scholar] [CrossRef]
- Ojima-Kato, T.; Hashimura, D.; Kojima, T.; Minabe, S.; Nakano, H. In vitro generation of rabbit anti-Listeria monocytogenes monoclonal antibody using single cell based RT-PCR linked cell-free expression systems. J. Immunol. Methods 2015, 427, 58–65. [Google Scholar] [CrossRef]
- Sølve, M.; Boel, J.; Nørrung, B. Evaluation of a monoclonal antibody able to detect live Listeria monocytogenes and Listeria innocua. Int. J. Food Microbiol. 2000, 57, 219–224. [Google Scholar] [CrossRef]
- Brigati, J.R.; Samoylova, T.I.; Jayanna, P.K.; Petrenko, V.A. Phage display for generating peptide reagents. Curr. Protoc. Protein Sci. 2008, 51, 18.9.1–18.9.27. [Google Scholar] [CrossRef] [PubMed]
- Romanov, V.I.; Durand, D.B.; Petrenko, V.A. Phage display selection of peptides that affect prostate carcinoma cells attachment and invasion. Prostate 2001, 47, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P.; Petrenko, V.A. Phage Display. Chem. Rev. 1997, 97, 391–410. [Google Scholar] [CrossRef]
- Funke, S.A.; Willbold, D. Mirror image phage display—A method to generate D-peptide ligands for use in diagnostic or therapeutical applications. Mol. Biosyst. 2009, 5, 783–786. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.X.; Bishop-Hurley, S.L.; Cooper, M.A. Development of anti-infectives using phage display: Biological agents against bacteria, viruses, and parasites. Antimicrob. Agents Chemother. 2012, 56, 4569–4582. [Google Scholar] [CrossRef]
- Morton, J.; Karoonuthaisiri, N.; Charlermroj, R.; Stewart, L.D.; Elliott, C.T.; Grant, I.R. Phage display-derived binders able to distinguish Listeria monocytogenes from other Listeria species. PLoS ONE 2013, 8, e74312. [Google Scholar] [CrossRef]
- Yarbrough, D.K.; Hagerman, E.; Eckert, R.; He, J.; Choi, H.; Cao, N.; Le, K.; Hedger, J.; Qi, F.; Anderson, M.; et al. Specific binding and mineralization of calcified surfaces by small peptides. Calcif. Tissue Int. 2010, 86, 58–66. [Google Scholar] [CrossRef]
- Yarbrough, D.K.; Eckert, R.; He, J.; Hagerman, E.; Qi, F.; Lux, R.; Wu, B.; Anderson, M.H.; Shi, W. Rapid probing of biological surfaces with a sparse-matrix peptide library. PLoS ONE 2011, 6, e23551. [Google Scholar] [CrossRef]
- Gouin, E.; Mengaud, J.; Cossart, P. The virulence gene cluster of Listeria monocytogenes is also present in Listeria ivanovii, an animal pathogen, and Listeria seeligeri, a nonpathogenic species. Infect. Immun. 1994, 62, 3550–3553. [Google Scholar] [CrossRef]
- Flachbartova, Z.; Pulzova, L.; Bencurova, E.; Potocnakova, L.; Comor, L.; Bednarikova, Z.; Bhide, M. Inhibition of multidrug resistant Listeria monocytogenes by peptides isolated from combinatorial phage display libraries. Microbiol. Res. 2016, 188–189, 34–41. [Google Scholar] [CrossRef] [PubMed]



| Peptide | Sequence |
|---|---|
| JB1 | H-GLHTSATNLYLH-K(FAM)-NH2 |
| JB2 | H-DSQFNKYSIATV-K(FAM)-NH2 |
| JB3 | H-SGVYKVAYDWQH-K(FAM)-NH2 |
| JB4 | H-QFDYMRPANDTH-K(FAM)-NH2 |
| JB5 | H-SNSIDKVNRPIN-K(FAM)-NH2 |
| Clone | Sequence | Frequency | Net Charge | Hydrophobicity | MW (Da) |
|---|---|---|---|---|---|
| JB1 | GLHTSATNLYLH | 3/11 | 0 | 33% | 1326.47 |
| JB2 | DSQFNKYSIATV | 1/11 | 0 | 33% | 1372.49 |
| JB3 | SGVYKVAYDWQH | 1/11 | 0 | 33% | 1452.58 |
| JB4 | QFDYMRPANDTH | 1/11 | −1 | 25% | 1488.61 |
| JB5 | SNSIDKVNRPIN | 1/11 | +1 | 25% | 1350.50 |
| VVSPDMNLLLTN | 2/11 | −1 | 50% | 1309.557 | |
| SLDGAGAALRTS | 1/11 | 0 | 41% | 1118.214 | |
| GHYTNSEWGFQE | 1/11 | −2 | 16% | 1454.476 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kenzel, J.; Brüggemann, D.A.; Funke, S.A. Selection of Listeria monocytogenes InlA-Binding Peptides Using Phage Display—Novel Compounds for Diagnostic Applications? Appl. Microbiol. 2022, 2, 921-933. https://doi.org/10.3390/applmicrobiol2040070
Kenzel J, Brüggemann DA, Funke SA. Selection of Listeria monocytogenes InlA-Binding Peptides Using Phage Display—Novel Compounds for Diagnostic Applications? Applied Microbiology. 2022; 2(4):921-933. https://doi.org/10.3390/applmicrobiol2040070
Chicago/Turabian StyleKenzel, Julia, Dagmar Adeline Brüggemann, and Susanne Aileen Funke. 2022. "Selection of Listeria monocytogenes InlA-Binding Peptides Using Phage Display—Novel Compounds for Diagnostic Applications?" Applied Microbiology 2, no. 4: 921-933. https://doi.org/10.3390/applmicrobiol2040070
APA StyleKenzel, J., Brüggemann, D. A., & Funke, S. A. (2022). Selection of Listeria monocytogenes InlA-Binding Peptides Using Phage Display—Novel Compounds for Diagnostic Applications? Applied Microbiology, 2(4), 921-933. https://doi.org/10.3390/applmicrobiol2040070







