Genomic Features of Pseudomonas putida PCL1760: A Biocontrol Agent Acting via Competition for Nutrient and Niche
Abstract
1. Introduction
2. Materials and Methods
| Microbial Strains | NCBI GenBank | Source of Isolation | Reference |
|---|---|---|---|
| P. putida PCL1760 | CP099727.1 | From the rhizosphere of the avocado plant | In this study |
| P. simiae PCL1751 | NZ_CP010896.1 | From the rhizosphere potato | Kamilova et al. [17] |
| P. fluorescens Pt14 | CP017296.1 | From the rhizosphere of rice plants in acidic soil | Rani et al. [18] |
| P. simiae WCS417 | NZ_CP007637.1 | From lesions of wheat roots growing in a take all disease-suppressive soil | Pieterse et al. [19] |
| P. putida S12 | CP009974.1 | From soil using styrene as a sole carbon source | Hartmans et al. [20] |
| P. fluorescens W-6 | CP058533.1 | From the Napahai plateau wetland | Xiang et al. [21] |
2.1. Genomic DNA Preparation
2.2. Library Preparation, Genome Sequencing, and Annotation
2.3. Genome Annotation and Comparison
2.4. Cell Suspension and Cell-Free Suspension Preparation
2.5. Antagonistic Activities
| Pathogenic Bacterial Strains | References |
|---|---|
| Pseudomonas syringae pv. tomato DC3000 | Cuppels and Ainsworth [35] |
| Staphylococcus aureus RN6390 | Khusainov et al. [36] |
| Pseudomonas aeruginosa IR1.5 | Egamberdieva et al. [37] |
3. Results
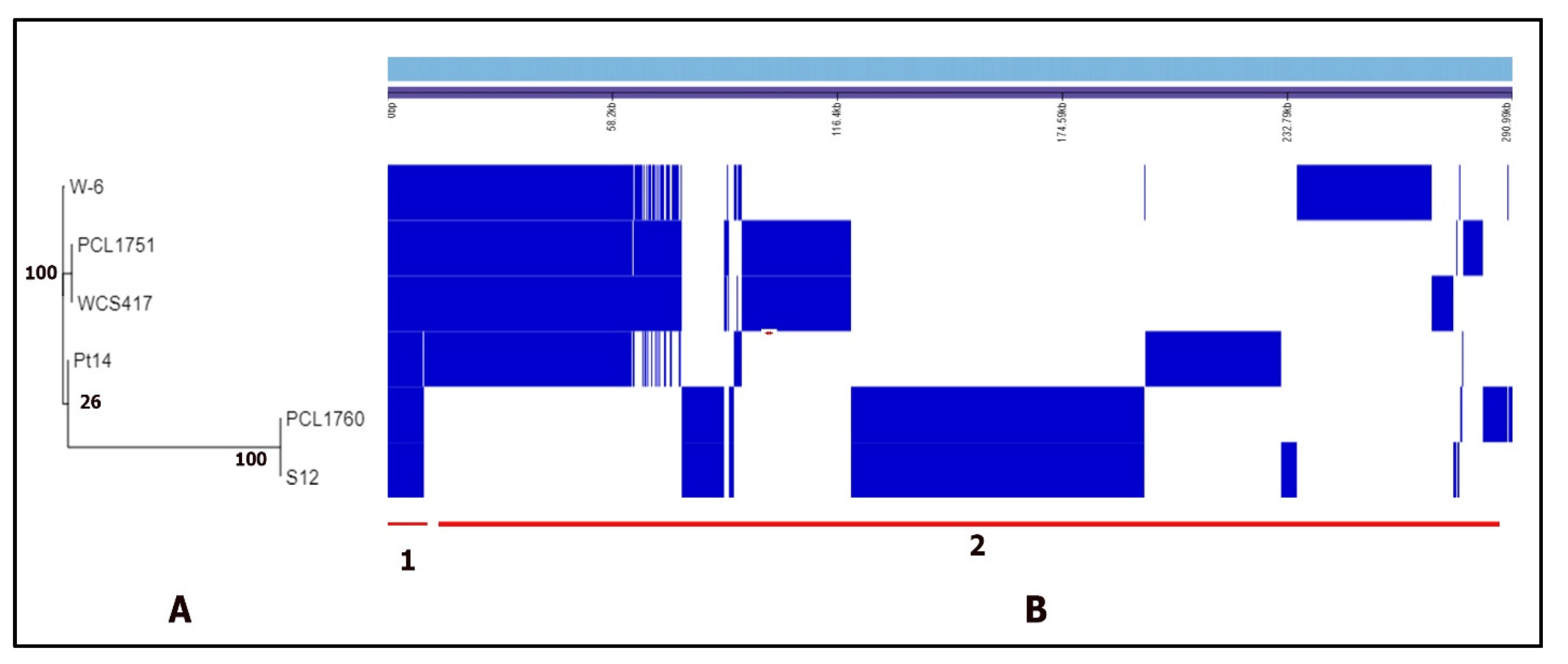
3.1. The Genome Assembly, Annotation, and Comparison
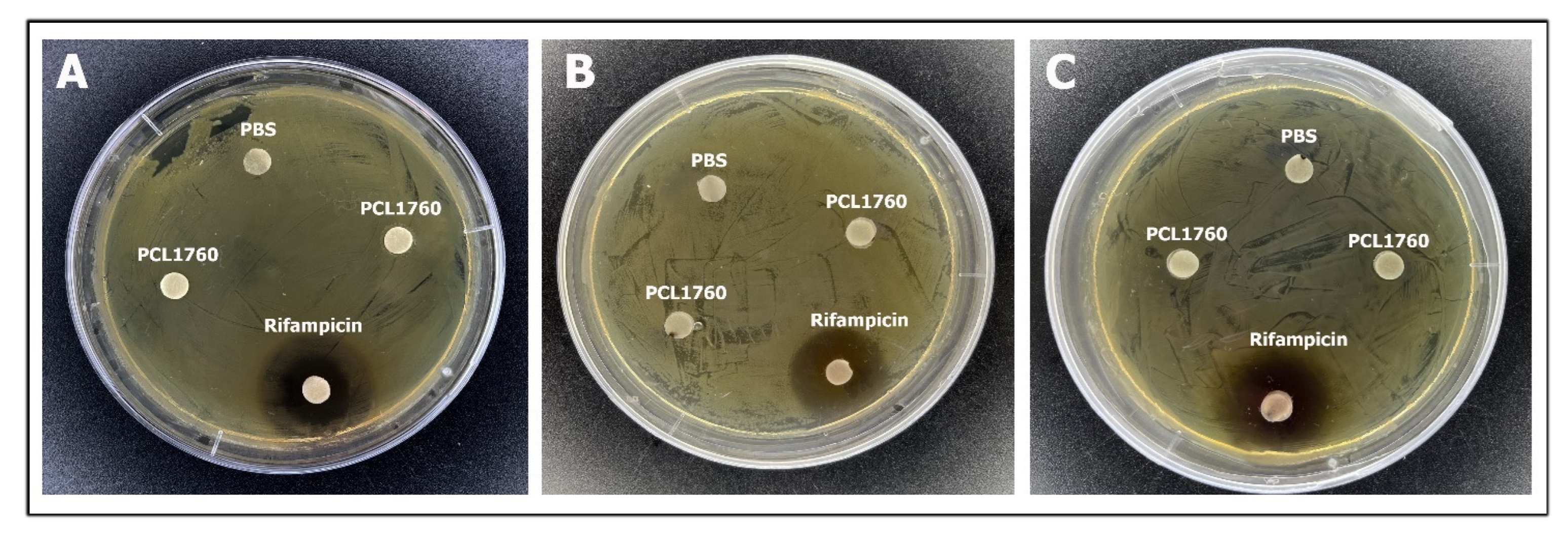
3.2. Antagonistic Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- O’Brien, P.A. Biological control of plant diseases. Australas. Plant Pathol. 2017, 46, 293–304. [Google Scholar] [CrossRef]
- Punja, Z.K.; Utkhede, R.S. Using fungi and yeasts to manage vegetable crop diseases. Trends Biotechnol. 2003, 21, 400–407. [Google Scholar] [CrossRef]
- Khabbaz, S.E.; Zhang, L.; Cáceres, L.A.; Sumarah, M.; Wang, A.; Abbasi, P.A. Characterisation of antagonistic Bacillus and Pseudomonas strains for biocontrol potential and suppression of damping-off and root rot diseases. Ann. Appl. Biol. 2015, 166, 456–471. [Google Scholar] [CrossRef]
- Validov, S.Z.; Kamilova, F.; Lugtenberg, B.J. Pseudomonas putida strain PCL1760 controls tomato foot and root rot in stonewool under industrial conditions in a certified greenhouse. Biol. Control. 2009, 48, 6–11. [Google Scholar] [CrossRef]
- Bernal, P.; Allsopp, L.P.; Filloux, A.; Llamas, M.A. The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 2017, 11, 97–2987. [Google Scholar] [CrossRef]
- Agisha, V.N.; Kumar, A.; Eapen, S.J.; Sheoran, N.; Suseelabhai, R. Broad-spectrum antimicrobial activity of volatile organic compounds from endophytic Pseudomonas putida BP25 against diverse plant pathogens. Biocontrol. Sci. Technol. 2019, 29, 1069–1089. [Google Scholar] [CrossRef]
- Patten, C.L.; Glick, B.R. Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 2002, 68, 3795–3801. [Google Scholar] [CrossRef]
- Rawat, P.; Das, S.; Shankhdhar, D.; Shankhdhar, S.C. Phosphate-solubilizing microorganisms: Mechanism and their role in phosphate solubilization and uptake. J. Plant Nutr. Soil Sci. 2021, 21, 49–68. [Google Scholar] [CrossRef]
- Shabayev, V.P. Effect of the introduction of the nitrogen-fixing bacteria Pseudomonas putida 23 on the nitrogen balance in soil. Eurasian Soil Sci. 2010, 43, 436–441. [Google Scholar] [CrossRef]
- Poblete-Castro, I.; Becker, J.; Dohnt, K.; Dos Santos, V.M.; Wittmann, C. Industrial biotechnology of Pseudomonas putida and related species. Appl. Microbiol. Biotechnol. 2012, 93, 2279–2290. [Google Scholar] [CrossRef]
- Schmid, A.; Dordick, J.S.; Hauer, B.; Kiener, A.; Wubbolts, M.; Witholt, B. Industrial biocatalysis today and tomorrow. Nature 2001, 409, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; Mendes, S.; Brissos, V.; Martins, L.O. New dye-decolorizing peroxidases from Bacillus subtilis and Pseudomonas putida MET94: Towards biotechnological applications. Appl. Microbiol. Biotechnol. 2014, 98, 2053–2065. [Google Scholar] [CrossRef] [PubMed]
- Mardani, G.; Ahankoub, M.; Alikhani Faradonbeh, M.; Raeisi Shahraki, H.; Fadaei, A. Biodegradation of ceftriaxone in soil using dioxygenase-producing genetically engineered Pseudomonas putida. Bioremediat. J. 2022, 26, 1–12. [Google Scholar] [CrossRef]
- Amiri, N.A.; Amiri, F.A.; Faravardeh, L.; Eslami, A.; Ghasemi, A.; Rafiee, M. Enhancement of MBBR reactor efficiency using effective microorganism for treatment of wastewater containing diazinon by engineered Pseudomonas putida KT2440 with manganese peroxidase 2 gene. J. Environ. Manag. 2022, 316, 115293. [Google Scholar] [CrossRef]
- Validov, S.Z.; Kamilova, F.; Qi, S.; Stephan, D.; Wang, J.J.; Makarova, N.; Lugtenberg, B.J. Selection of bacteria able to control Fusarium oxysporum f. sp. radicis-lycopersici in stonewool substrate. J. Appl. Microbiol. 2007, 102, 461–471. [Google Scholar] [CrossRef]
- Ye, L.; Hildebrand, F.; Dingemans, J.; Ballet, S.; Laus, G.; Matthijs, S.; Berendsen, R.; Cornelis, P. Draft genome sequence analysis of a Pseudomonas putida W15Oct28 strain with antagonistic activity to Gram-positive and Pseudomonas sp. pathogens. PLoS ONE 2014, 9, e110038. [Google Scholar] [CrossRef]
- Kamilova, F.; Validov, S.; Azarova, T.; Mulders, I.; Lugtenberg, B.j. Enrichment for enhanced competitive plant root tip colonizers selects for a new class of biocontrol bacteria. Appl. Environ. Microbiol. 2005, 7, 1809–1817. [Google Scholar] [CrossRef]
- Rani, P.; Mahato, N.K.; Sharma, A.; Rao, D.L.N.; Kamra, K.; Lal, R. Genome mining and predictive functional profiling of acidophilic rhizobacterium Pseudomonas fluorescens Pt14. Indian J. Microbiol. 2017, 57, 155–161. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Berendsen, R.L.; de Jonge, R.; Stringlis, I.A.; Van Dijken, A.J.; Van Pelt, J.A.; Van Wees, S.C.M.; Yu, K.; Zamioudis, C.; Bakker, H.M. Pseudomonas simiae WCS417: Star track of a model beneficial rhizobacterium. Plant Soil 2021, 461, 245–263. [Google Scholar] [CrossRef]
- Hartmans, S.; Van der Werf, M.J.; De Bont, J.A. Bacterial degradation of styrene involving a novel flavin adenine dinucleotide-dependent styrene monooxygenase. Appl. Environ. Microbiol. 1990, 56, 1347–1351. [Google Scholar] [CrossRef]
- Xiang, Y.; Wang, S.; Li, J.; Wei, Y.; Zhang, Q.; Lin, L.; Ji, X. Isolation and characterization of two lytic cold-active bacteriophages infecting Pseudomonas fluorescens from the Napahai plateau wetland. Can. J. Microbiol. 2018, 64, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e100559. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Li, X.; Wu, Y. GAPPadder: A sensitive approach for closing gaps on draft genomes with short sequence reads. BMC Genom. 2019, 20, 426. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Parkhill, J. Roary: Rapid large-scale prokaryote pan-genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Q.Y.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A Web Server for Whole-Genome Comparison and Annotation of Orthologous Clusters Across Multiple Species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef] [PubMed]
- Grissa, I.; Vergnaud, G.; Pourcel, C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinform. 2007, 8, 172. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Medema, M.H. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef] [PubMed]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Wright, G.D. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- Cuppels, D.A.; Ainsworth, T. Molecular and physiological characterization of Pseudomonas syringae pv. tomato and Pseudomonas syringae pv. maculicola strains that produce the phytotoxin coronatine. Appl. Environ. Microbiol. 1995, 61, 3530–3536. [Google Scholar] [CrossRef]
- Khusainov, I.; Fatkhullin, B.; Pellegrino, S.; Bikmullin, A.; Liu, W.; Gabdulkhakov, A.; Al Shebel, A.; Golubev, A.; Zeyer, D.; Trachtmann, N.; et al. Mechanism of ribosome shutdown by RsfS in Staphylococcus aureus revealed by integrative structural biology approach. Nat. Commun. 2020, 11, 1656. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Kamilova, F.; Validov, S.; Gafurova, L.; Kucharova, Z.; Lugtenberg, B. High incidence of plant growth-stimulating bacteria associated with the rhizosphere of wheat grown on salinated soil in Uzbekistan. Environ. Microbiol. 2008, 10, 1–9. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standard Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 7th ed.; Sixteenth Informational Supplement; CLSI: Wayne, PA, USA, 2006; pp. 15–130. [Google Scholar]
- Hadfield, J.; Croucher, N.J.; Goater, R.J.; Abudahab, K.; Aanensen, D.M.; Harris, S.R. Phandango: An Interactive Viewer for Bacterial Population Genomics. Bioinformatics 2018, 34, 292–293. [Google Scholar] [CrossRef]
- Diabankana, R.G.C.; Shulga, E.U.; Validov, S.Z.; Afordoanyi, D.M. Genetic Characteristics and Enzymatic Activities of Bacillus velezensis KS04AU as a Stable Biocontrol Agent against Phytopathogens. Int. J. Plant Biol. 2022, 13, 201–222. [Google Scholar] [CrossRef]
- Hussein, K.A.; Joo, J.H. Stimulation, purification, and chemical characterization of siderophores produced by the rhizospheric bacterial strain Pseudomonas putida. Rhizosphere 2017, 4, 16–21. [Google Scholar] [CrossRef]
- Weimer, A.; Kohlstedt, M.; Volke, D.C.; Nikel, P.I.; Wittmann, C. Industrial biotechnology of Pseudomonas putida: Advances and prospects. Appl. Microbiol. Biotechnol. 2020, 104, 7745–7766. [Google Scholar] [CrossRef] [PubMed]
- Loeschcke, A.; Thies, S. Pseudomonas putida—a versatile host for the production of natural products. Appl. Microbiol. Biotechnol. 2015, 99, 6197–6214. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ma, C.; Li, Q.; Li, Q.; He, Y.C. Efficient chemoenzymatic valorization of biobased D-fructose into 2, 5-bis (hydroxymethyl) furan with deep eutectic solvent Lactic acid: Betaine and Pseudomonas putida S12 whole cells. Bioresour. Technol. 2022, 344, 126299. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Campos, V.; Covarrubias-García, I.; Arriaga, S. Styrene bioconversion by Pseudomonas putida utilizing a non-aqueous phase for polyhydroxyalkanoate production. J. Chem. Technol. Biotechnol. 2022, 97, 1424–1435. [Google Scholar] [CrossRef]
- Kuatsjah, E.; Johnson, C.W.; Salvachúa, D.; Werner, A.Z.; Zahn, M.; Szostkiewicz, C.J.; Singer, C.A.; Dominick, G.; Okekeogbu, I.; Haugen, S.J.; et al. Debottlenecking 4-hydroxybenzoate hydroxylation in Pseudomonas putida KT2440 improves muconate productivity from p-coumarate. Metab. Eng. 2022, 70, 31–42. [Google Scholar] [CrossRef]
- Fernández, M.; Duque, E.; Pizarro-Tobías, P.; Van Dillewijn, P.; Wittich, R.M.; Ramos, J.L. Microbial responses to xenobiotic compounds. Identification of genes that allow Pseudomonas putida KT2440 to cope with 2, 4, 6-trinitrotoluene. Microb. Biotechnol. 2009, 2, 287–294. [Google Scholar] [CrossRef]
- Thomassen, G.M.B.; Reiche, T.; Tennfjord, C.E.; Mehli, L. Antibiotic Resistance Properties among Pseudomonas spp. Associated with Salmon Processing Environments. Microorganisms 2022, 10, 1420. [Google Scholar] [CrossRef]
- Igbinosa, I.H.; Nwodo, U.U.; Sosa, A.; Tom, M.; Okoh, A.I. Commensal Pseudomonas species isolated from wastewater and freshwater milieus in the Eastern Cape Province, South Africa, as reservoir of antibiotic resistant determinants. Int. J. Environ. Health Res. 2012, 9, 2537–2549. [Google Scholar] [CrossRef]
- Rojas Murcia, N.; Lee, X.; Waridel, P.; Maspoli, A.; Imker, H.J.; Chai, T.; Walsh, C.T.; Reimmann, C. The Pseudomonas aeruginosa antimetabolite L-2-amino-4-methoxy-trans-3-butenoic acid (AMB) is made from glutamate and two alanine residues via a thiotemplate-linked tripeptide precursor. Front. Microbiol. 2015, 6, 170. [Google Scholar] [CrossRef]
- McLeish, M.J.; Kneen, M.M.; Gopalakrishna, K.N.; Koo, C.W.; Babbitt, P.C.; Gerlt, J.A.; Kenyon, G.L. Identification and characterization of a mandelamide hydrolase and an NAD (P)+-dependent benzaldehyde dehydrogenase from Pseudomonas putida ATCC 12633. J. Bacteriol. 2003, 185, 2451–2456. [Google Scholar] [CrossRef]
- Tiso, T.; Wierckx, N.; Blank, L.M. Non-pathogenic Pseudomonas as platform for industrial biocatalysis. Ind. Biocatal. 2014, 1, 323–372. [Google Scholar]
- Jiang, W.; Marraffini, L.A. CRISPR-Cas: New tools for genetic manipulations from bacterial immunity systems. Annu. Rev. Microbiol. 2015, 69, 209–228. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, K.L. Phages fight back: Inactivation of the CRISPR-Cas bacterial immune system by anti-CRISPR proteins. PLoS Pathogens 2016, 12, e1005282. [Google Scholar] [CrossRef] [PubMed]
- Cumby, N.; Davidson, A.R.; Maxwell, K.L. The moron comes of age. Bacteriophage 2012, 2, 5012–5019. [Google Scholar] [CrossRef]
- Schroven, K.; Aertsen, A.; Lavigne, R. Bacteriophages as drivers of bacterial virulence and their potential for biotechnological exploitation. FEMS Microbiol. Rev. 2021, 45, fuaa041. [Google Scholar] [CrossRef]
- Wang, X.; Kim, Y.; Ma, Q.; Hong, S.H.; Pokusaeva, K.; Sturino, J.M.; Wood, T.K. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010, 1, 147. [Google Scholar] [CrossRef]
- Nobrega, F.L.; Walinga, H.; Dutilh, B.E.; Brouns, S.J. Prophages are associated with extensive CRISPR–Cas auto-immunity. Nucleic Acids Res. 2020, 48, 12074–12084. [Google Scholar] [CrossRef]
- Pleška, M.; Lang, M.; Refardt, D.; Levin, B.R.; Guet, C.C. Phage–host population dynamics promotes prophage acquisition in bacteria with innate immunity. Nat. Ecol. Evol. 2018, 2, 359–366. [Google Scholar] [CrossRef]


| Features | PCL1760 | Pt14 | W-6 | S12 | PCL1751 | WCS417 |
|---|---|---|---|---|---|---|
| Genome size (bp) | 6,002,785 | 5,841,722 | 6,190,190 | 6,382,434 | 6,143,950 | 6,169,071 |
| G + C (%) | 61.80 | 60.30 | 62.70 | 61.44 | 60.40 | 60.30 |
| Genes (total) | 5402 | 5301 | 5674 | 5888 | 5668 | 5692 |
| CDSs (total) | 5303 | 5210 | 5571 | 5805 | 5575 | 5615 |
| Genes (coding) | 5215 | 5140 | 5484 | 5708 | 5507 | 5534 |
| Genes (RNA) | 99 | 91 | 103 | 83 | 93 | 77 |
| tRNAs | 72 | 68 | 76 | 61 | 70 | 64 |
| ncRNAs | 4 | 4 | 5 | 4 | 4 | 4 |
| Pseudo genes (total) | 88 | 70 | 87 | 97 | 68 | 81 |
| Plasmid | 0 | 0 | 0 | 0 | 0 | 0 |
| CRISPR-elements | 5 | 11 | 1 | 3 | 2 | 1 |
| Cas3 elements type I | 10 | 16 | 16 | 12 | 14 | 14 |
| PCL1760 | S12 | Pt14 | W-6 | PCL1751 | WCS417 | |
| PCL1760 | – | 99.30 (94.29) | 76.06 (51.55) | 75.82 (52.16) | 76.26 (52.29) | 76.26 (52.27) |
| S12 | 99.75 (94.84) | – | 76.32 (51.84) | 76.03 (52.22) | 76.38 (52.58) | 76.35 (52.68) |
| Pt14 | 76.40 (50.52) | 76.45 (50.53) | – | 85.65 (75.31) | 86.08 (77.14) | 86.04 (77.26) |
| W-6 | 75.78 (50.40) | 75.77 (50.22) | 85.44 (72.36) | – | 86.57 (77.32) | 86.59 (77.25) |
| PCL1751 | 76.30 (50.29) | 76.32 (50.11) | 85.82 (74.17) | 86.73 (76.41) | – | 99.50 (95.70) |
| WCS417 | 76.15 (49.73) | 76.12 (49.60) | 85.70 (73.71) | 86.57 (76.28) | 99.41 (95.25) | – |
| Predicted Prophages | The Presence (+) or Absence (−) in Related Bacterial Strains | |||||
|---|---|---|---|---|---|---|
| PCL1760 | S12 | Pt14 | W-6 | PCL1751 | WCS417 | |
| PHAGE Escher 500465 1 NC 049342 | + | + | + | + | − | |
| PHAGE Pseudo YMC11/02/R656 NC 028657 | + | − | + | + | + | + |
| PHAGE Pseudo JBD44 NC 030929 | + | + | − | − | − | − |
| PHAGE Bacill vB BtS BMBtp14 NC 048640 | − | + | − | − | − | − |
| PHAGE Vibrio VP882 NC 009016 | − | − | + | − | + | + |
| PHAGE Pseudo F10 NC 007805 | − | − | + | − | − | − |
| PHAGE Vibrio vB VpaM MAR NC 019722 | − | − | − | − | + | − |
| PHAGE Salmon SJ46 NC 031129 | − | − | − | − | + | − |
| ARO Term | AMR Gene Family | Drug Class | Resistance Mechanism | The Presence (+) or Absence (−) in Related Bacterial Strains | |||||
|---|---|---|---|---|---|---|---|---|---|
| PCL1760 | S12 | Pt14 | W-6 | PCL1751 | WCS417 | ||||
| adeF | Resistance-nodulation-cell division (RND) antibiotic efflux pump | fluoroquinolone antibiotic, tetracycline antibiotic | antibiotic efflux | + | + | + | + | + | + |
| Pseudomo-nas aeruginosa soxR | ATP-binding cassette (ABC) antibiotic efflux pump, major facilitator superfamily (MFS) antibiotic efflux pump, resistance-nodulation-cell division (RND) antibiotic efflux pump | fluoroquinolone antibiotic, cephalosporin, glycylcycline, penam, tetracycline antibiotic, rifamycin antibiotic, phenicol antibiotic, disinfecting agents and antiseptics | antibiotic target alteration, antibiotic efflux | − | − | + | + | + | + |
| Acinetobacter baumannii AbaQ | major facilitator superfamily (MFS) antibiotic efflux pump | fluoroquinolone antibiotic | antibiotic efflux | − | − | + | + | + | + |
| Secondary Metabolites Related to the Most Known Biosynthesis Cluster Genes | The Presence (+) or Absence (−) in Related Bacterial Strains | |||||
|---|---|---|---|---|---|---|
| PCL1760 | S12 | Pt14 | W-6 | PCL1751 | WCS417 | |
| O-antigen | + | + | + | + | + | + |
| lankacidin C | + | + | + | + | + | + |
| pyoverdin | + | + | + | + | + | + |
| fengycin | − | − | + | + | + | + |
| APE-vf | − | − | + | + | + | + |
| Coelibactin | − | − | − | + | + | + |
| L-2-amin0-4-methoxy-trans-3-butenioc acid | − | − | − | + | − | + |
| Pseudomonine | − | − | + | − | − | − |
| Ambactin | − | − | + | − | − | − |
| Species | Cluster | Singletons |
|---|---|---|
| P. putida PCL1760 | 4920 | 278 |
| P. simiae PCL1751 | 4702 | 469 |
| P. fluorescens Pt14 | 4817 | 613 |
| P. simiae WCS417 | 4890 | 161 |
| P. putida S12 | 5345 | 172 |
| P. fluorescens W-6 | 5375 | 192 |
| Antibiotics | Masses of Antibiotics and Susceptibility of P. putida PCL1760 | |||||
|---|---|---|---|---|---|---|
| 0.75 µg | 1.5 µg | 3 µg | 6 µg | 12 µg | 24 µg | |
| Erythromycin | R | R | R | R | R | R |
| Ciprofloxacin | R | I | I | S | S | S |
| Moxifloxacin | S | S | S | S | S | S |
| Ampicillin | R | R | R | R | R | R |
| Chloramphenicol | R | R | R | R | R | R |
| Kanamycin | I | I | I | S | S | S |
| Ceftriaxone | I | I | S | S | S | S |
| Spectinomycin | R | R | R | R | R | R |
| Tetracycline | R | R | R | R | R | I |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Afordoanyi, D.M.; Diabankana, R.G.C.; Miftakhov, A.K.; Kuchaev, E.S.; Validov, S.Z. Genomic Features of Pseudomonas putida PCL1760: A Biocontrol Agent Acting via Competition for Nutrient and Niche. Appl. Microbiol. 2022, 2, 749-765. https://doi.org/10.3390/applmicrobiol2040057
Afordoanyi DM, Diabankana RGC, Miftakhov AK, Kuchaev ES, Validov SZ. Genomic Features of Pseudomonas putida PCL1760: A Biocontrol Agent Acting via Competition for Nutrient and Niche. Applied Microbiology. 2022; 2(4):749-765. https://doi.org/10.3390/applmicrobiol2040057
Chicago/Turabian StyleAfordoanyi, Daniel Mawuena, Roderic Gilles Claret Diabankana, Aynur Kamilevich Miftakhov, Evgenii Sergeyevich Kuchaev, and Shamil Zavdatovich Validov. 2022. "Genomic Features of Pseudomonas putida PCL1760: A Biocontrol Agent Acting via Competition for Nutrient and Niche" Applied Microbiology 2, no. 4: 749-765. https://doi.org/10.3390/applmicrobiol2040057
APA StyleAfordoanyi, D. M., Diabankana, R. G. C., Miftakhov, A. K., Kuchaev, E. S., & Validov, S. Z. (2022). Genomic Features of Pseudomonas putida PCL1760: A Biocontrol Agent Acting via Competition for Nutrient and Niche. Applied Microbiology, 2(4), 749-765. https://doi.org/10.3390/applmicrobiol2040057






