Differences at Species Level and in Repertoires of Secondary Metabolite Biosynthetic Gene Clusters among Streptomyces coelicolor A3(2) and Type Strains of S. coelicolor and Its Taxonomic Neighbors
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Taxnomic Positions of S. coelicolor and Related Strains
3.2. PKS and NRPS Gene Clusters in Genomes
3.3. Prediction of Products Synthesized by Orphan PKS and NRPS Gene Clusters
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hatano, K.; Tamura, T.; Nishii, T. Taxonomic status of Streptomyces coelicolor A3(2) and Streptomyces lividans 66. Actinomycetologica 1994, 8, 47–50. [Google Scholar] [CrossRef]
- Müller, R. Eine Diphtheridee und eine Streptothrix mit gleichen blauen Farbstoff sowie Untersuchungen über Streptothrixarten in allgemeinen. Zent. Für Bakteriol. Parasitenkd. Infekt. Und Hyg. Abt. I 1908, 46, 195–212. [Google Scholar]
- Waksman, S.A.; Henrici, A.T. Family III. Streptomycetaceae Waksman and Henrici. In Bergey’s Manual of Determinative Bacteriology, 6th ed.; Breed, R.S., Murray, E.G.D., Hitchens, A.P., Eds.; The Williams & Wilkins Co: Baltimore, MD, USA, 1948; pp. 929–980. [Google Scholar]
- Skerman, V.B.D.; McGowan, V.; Sneath, P.H.A. Approved lists of bacterial names. Int. J. Syst. Bacteriol. 1980, 30, 225–420. [Google Scholar] [CrossRef]
- Erikson, D. Loss of aerial mycelium and other changes in streptomycete development due to physical variations of cultural conditions. J. Gen. Microbiol. 1955, 13, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Hopwood, D.A. Linkage and the mechanism of recombination in Streptomyces coelicolor. Ann. N. Y. Acad. Sci. 1959, 81, 887–898. [Google Scholar] [CrossRef]
- Hopwood, D.A.; Chater, K.F.; Dowding, J.E.; Vivian, A. Advances in Streptomyces coelicolor genetics. Bacteriol. Rev. 1973, 37, 371–405. [Google Scholar] [CrossRef] [PubMed]
- Bentley, S.D.; Chater, K.F.; Cerdeno-Tarraga, A.M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef]
- Lee, N.; Hwang, S.; Kim, J.; Cho, S.; Palsson, B.; Cho, B.K. Mini review: Genome mining approaches for the identification of secondary metabolite biosynthetic gene clusters in Streptomyces. Comput. Struct. Biotechnol. J. 2020, 18, 1548–1556. [Google Scholar] [CrossRef]
- Nett, M.; Ikeda, H.; Moore, B.S. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 2009, 26, 1362–1384. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Ma, J. The Global Catalogue of Microorganisms (GCM) 10K type strain sequencing project: Providing services to taxonomists for standard genome sequencing and annotation. Int. J. Syst. Evol. Microbiol. 2019, 69, 895–898. [Google Scholar] [CrossRef] [PubMed]
- Komaki, H.; Ichikawa, N.; Oguchi, A.; Hamada, M.; Tamura, T.; Fujita, N. Genome-based analysis of non-ribosomal peptide synthetase and type-I polyketide synthase gene clusters in all type strains of the genus Herbidospora. BMC Res. Notes 2015, 8, 548. [Google Scholar] [CrossRef][Green Version]
- Komaki, H.; Ichikawa, N.; Tamura, T.; Oguchi, A.; Hamada, M.; Fujita, N. Genome-based survey of nonribosomal peptide synthetase and polyketide synthase gene clusters in type strains of the genus Microtetraspora. J. Antibiot. 2016, 69, 712–718. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Komaki, H.; Oguchi, A.; Tamura, T.; Hamada, M.; Ichikawa, N. Diversity of nonribosomal peptide synthetase and polyketide synthase gene clusters in the genus Acrocarpospora. J. Gen. Appl. Microbiol. 2021, 66, 315–322. [Google Scholar] [CrossRef]
- Komaki, H.; Sakurai, K.; Hosoyama, A.; Kimura, A.; Igarashi, Y.; Tamura, T. Diversity of nonribosomal peptide synthetase and polyketide synthase gene clusters among taxonomically close Streptomyces strains. Sci. Rep. 2018, 8, 6888. [Google Scholar] [CrossRef]
- Komaki, H.; Tamura, T. Polyketide synthase and nonribosomal peptide synthetase gene clusters in type strains of the genus Phytohabitans. Life 2020, 10, 257. [Google Scholar] [CrossRef]
- Komaki, H.; Tamura, T.; Ichikawa, N.; Oguchi, A.; Hamada, M.; Suzuki, K.; Fujita, N. Genome-based analysis of type-I polyketide synthase and nonribosomal peptide synthetase gene clusters in a novel strain taxonomically close to the genus Salinispora. J. Antibiot. 2015, 68, 767–770. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Rong, X.; Huang, Y. Taxonomic evaluation of the Streptomyces hygroscopicus clade using multilocus sequence analysis and DNA-DNA hybridization, validating the MLSA scheme for systematics of the whole genus. Syst. Appl. Microbiol. 2012, 35, 7–18. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Saito, H.; Miura, K.I. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta 1963, 72, 619–629. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed]
- Wayne, L.; Brenner, D.; Colwell, R.R.; Grimont, P.A.D.; Kandler, O.; Krichevsky, M.I.; Moore, M.H.; Moore, W.E.C.; Murray, R.G.E.; Stackebrandt, E.; et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 1987, 37, 463–464. [Google Scholar] [CrossRef]
- Rong, X.; Guo, Y.; Huang, Y. Proposal to reclassify the Streptomyces albidoflavus clade on the basis of multilocus sequence analysis and DNA-DNA hybridization, and taxonomic elucidation of Streptomyces griseus subsp. Solvifaciens Syst. Appl. Microbiol. 2009, 32, 314–322. [Google Scholar] [CrossRef]
- Wendt-Pienkowski, E.; Huang, Y.; Zhang, J.; Li, B.; Jiang, H.; Kwon, H.; Hutchinson, C.R.; Shen, B. Cloning, sequencing, analysis, and heterologous expression of the fredericamycin biosynthetic gene cluster from Streptomyces griseus. J. Am. Chem. Soc. 2005, 127, 16442–16452. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Ishikawa, J.; Hanamoto, A.; Shinose, M.; Kikuchi, H.; Shiba, T.; Sakaki, Y.; Hattori, M.; Omura, S. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 2003, 21, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Xiong, Z.; Chu, J.; Wang, Y. Enhanced production of avermectin by deletion of type III polyketide synthases biosynthetic cluster rpp in Streptomyces avermitilis. Lett. Appl. Microbiol. 2016, 63, 384–390. [Google Scholar] [CrossRef]
- Becerril, A.; Alvarez, S.; Brana, A.F.; Rico, S.; Diaz, M.; Santamaria, R.I.; Salas, J.A.; Mendez, C. Uncovering production of specialized metabolites by Streptomyces argillaceus: Activation of cryptic biosynthesis gene clusters using nutritional and genetic approaches. PLoS ONE 2018, 13, e0198145. [Google Scholar] [CrossRef]
- Lee, M.Y.; Ames, B.D.; Tsai, S.C. Insight into the molecular basis of aromatic polyketide cyclization: Crystal structure and in vitro characterization of WhiE-ORFVI. Biochemistry 2012, 51, 3079–3091. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, D.; Marahiel, M.A. Multimodular biocatalysts for natural product assembly. Naturwissenschaften 2001, 88, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Wildermuth, H. Development and organization of the aerial mycelium in Streptomyces coelicolor. J. Gen. Microbiol. 1970, 60, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Faddetta, T.; Renzone, G.; Vassallo, A.; Rimini, E.; Nasillo, G.; Buscarino, G.; Agnello, S.; Licciardi, M.; Botta, L.; Scaloni, A.; et al. Streptomyces coelicolor vesicles: Many molecules to be delivered. Appl. Environ. Microbiol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Kim, T.J.; Kwon, H.J.; Suh, J.W. Effects of extracellular ATP on the physiology of Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 2008, 286, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Jankevics, A.; Merlo, M.E.; de Vries, M.; Vonk, R.J.; Takano, E.; Breitling, R. Metabolomic analysis of a synthetic metabolic switch in Streptomyces coelicolor A3(2). Proteomics 2011, 11, 4622–4631. [Google Scholar] [CrossRef]
- Sulheim, S.; Kumelj, T.; van Dissel, D.; Salehzadeh-Yazdi, A.; Du, C.; van Wezel, G.P.; Nieselt, K.; Almaas, E.; Wentzel, A.; Kerkhoven, E.J. Enzyme-constrained models and omics analysis of Streptomyces coelicolor reveal metabolic changes that enhance heterologous production. iScience 2020, 23, 101525. [Google Scholar] [CrossRef] [PubMed]
- Elliot, M.; Damji, F.; Passantino, R.; Chater, K.; Leskiw, B. The bldD gene of Streptomyces coelicolor A3(2): A regulatory gene involved in morphogenesis and antibiotic production. J. Bacteriol. 1998, 180, 1549–1555. [Google Scholar] [CrossRef]
- Martin, J.F.; Liras, P. Molecular mechanisms of phosphate sensing, transport and signalling in Streptomyces and related actinobacteria. Int. J. Mol. Sci. 2021, 22, 1129. [Google Scholar] [CrossRef]
- Shiffman, D.; Cohen, S.N. Role of the imp operon of the Streptomyces coelicolor genetic element SLP1: Two imp-encoded proteins interact to autoregulate imp expression and control plasmid maintenance. J. Bacteriol. 1993, 175, 6767–6774. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Baltz, R.H. Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J. Ind. Microbiol. Biotechnol. 2010, 37, 759–772. [Google Scholar] [CrossRef] [PubMed]
- Eustaquio, A.S.; Gust, B.; Li, S.M.; Pelzer, S.; Wohlleben, W.; Chater, K.F.; Heide, L. Production of 8’-halogenated and 8’-unsubstituted novobiocin derivatives in genetically engineered Streptomyces coelicolor strains. Chem. Biol. 2004, 11, 1561–1572. [Google Scholar] [CrossRef][Green Version]
- Kumar, K.; Bruheim, P. A comparative study at bioprocess and metabolite levels of superhost strain Streptomyces coelicolor M1152 and its derivative M1581 heterologously expressing chloramphenicol biosynthetic gene cluster. Biotechnol. Bioeng. 2021. [Google Scholar] [CrossRef] [PubMed]
- Mitousis, L.; Thoma, Y.; Musiol-Kroll, E.M. An update on molecular tools for genetic engineering of actinomycetes-the source of important antibiotics and other valuable compounds. Antibiotics 2020, 9, 494. [Google Scholar] [CrossRef]
- Pfeifer, B.A.; Khosla, C. Biosynthesis of polyketides in heterologous hosts. Microbiol. Mol. Biol. Rev. 2001, 65, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Bednarz, B.; Millan-Oropeza, A.; Kotowska, M.; Swiat, M.; Quispe Haro, J.J.; Henry, C.; Pawlik, K. Coelimycin synthesis activatory proteins are key regulators of specialized metabolism and precursor flux in Streptomyces coelicolor A3(2). Front. Microbiol. 2021, 12, 616050. [Google Scholar] [CrossRef]
- Falke, D.; Fischer, M.; Ihling, C.; Hammerschmidt, C.; Sinz, A.; Sawers, G. Co-purification of nitrate reductase 1 with components of the cytochrome bcc-aa3 oxidase supercomplex from spores of Streptomyces coelicolor A3(2). FEBS Open Bio. 2021, 11, 652–669. [Google Scholar] [CrossRef] [PubMed]
- Honma, S.; Ito, S.; Yajima, S.; Sasaki, Y. Nitric oxide signaling for actinorhodin production in Streptomyces coelicolor A3(2) via the DevS/R two-component system. Appl. Environ. Microbiol. 2021, 87, e0048021. [Google Scholar] [CrossRef]
- Tsevelkhoroloo, M.; Shim, S.H.; Lee, C.R.; Hong, S.K.; Hong, Y.S. LacI-family transcriptional regulator DagR acts as a repressor of the agarolytic pathway genes in Streptomyces coelicolor A3(2). Front. Microbiol. 2021, 12, 658657. [Google Scholar] [CrossRef] [PubMed]
- Streptomyces coelicolor A3(2), Taxonomy, NCBI. Available online: https://www.ncbi.nlm.nih.gov/taxonomy/100226 (accessed on 18 July 2021).
- Streptomyces Coelicolor A3(2) Complete Genome, Nucleotide, NCBI. Available online: https://www.ncbi.nlm.nih.gov/nuccore/AL645882.2 (accessed on 18 July 2021).
- Komaki, H.; Hosoyama, A.; Igarashi, Y.; Tamura, T. Streptomyces lydicamycinicus sp. nov. and its secondary metabolite biosynthetic gene clusters for polyketide and nonribosomal peptide compounds. Microorganisms 2020, 8, 370. [Google Scholar] [CrossRef] [PubMed]
- Komaki, H.; Sakurai, K.; Hosoyama, A.; Kimura, A.; Trujilo, M.E.; Igarashi, Y.; Tamura, T. Diversity of PKS and NRPS gene clusters between Streptomyces abyssomicinicus sp. nov. and its taxonomic neighbor. J. Antibiot. 2020, 73, 141–151. [Google Scholar] [CrossRef]
- Komaki, H.; Tamura, T. Reclassification of Streptomyces diastaticus subsp. ardesiacus, Streptomyces gougerotii and Streptomyces rutgersensis. Int. J. Syst. Evol. Microbiol. 2020, 70, 4291–4297. [Google Scholar] [CrossRef]
- Komaki, H.; Tamura, T. Reclassification of Streptomyces fulvissimus as a later heterotypic synonym of Streptomyces microflavus. Int. J. Syst. Evol. Microbiol. 2020, 70, 5156–5162. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Komaki, H. Reclassification of 15 Streptomyces species as synonyms of Streptomyces albogriseolus, Streptomyces althioticus, Streptomyces anthocyanicus, Streptomyces calvus, Streptomyces griseoincarnatus, Streptomyces mutabilis, Streptomyces pilosus or Streptomyces rochei. Int. J. Syst. Evol. Microbiol. 2019, 71, 004718. [Google Scholar] [CrossRef]
- Smanski, M.J.; Schlatter, D.C.; Kinkel, L.L. Leveraging ecological theory to guide natural product discovery. J. Ind. Microbiol. Biotechnol. 2016, 43, 115–128. [Google Scholar] [CrossRef] [PubMed]
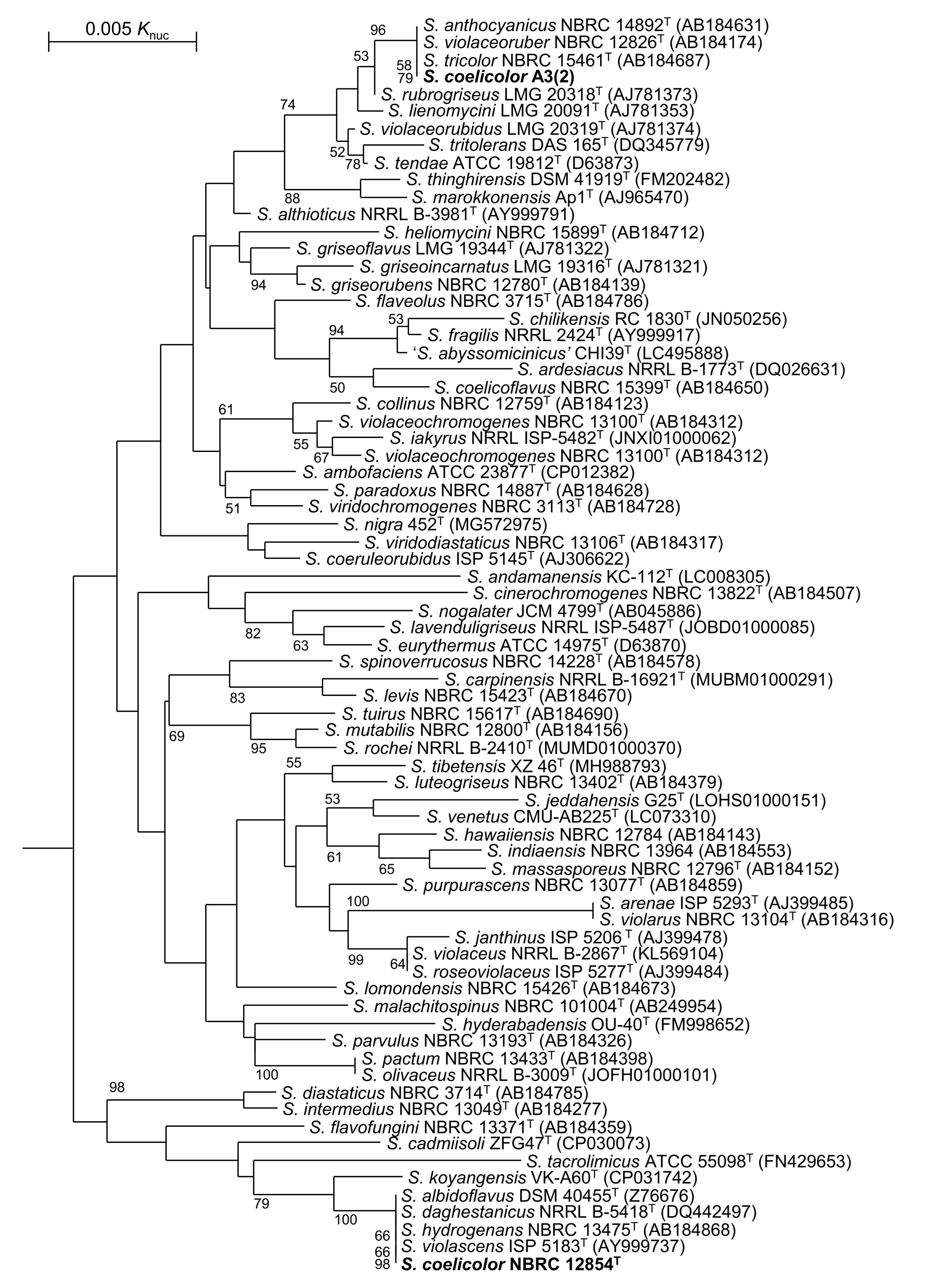
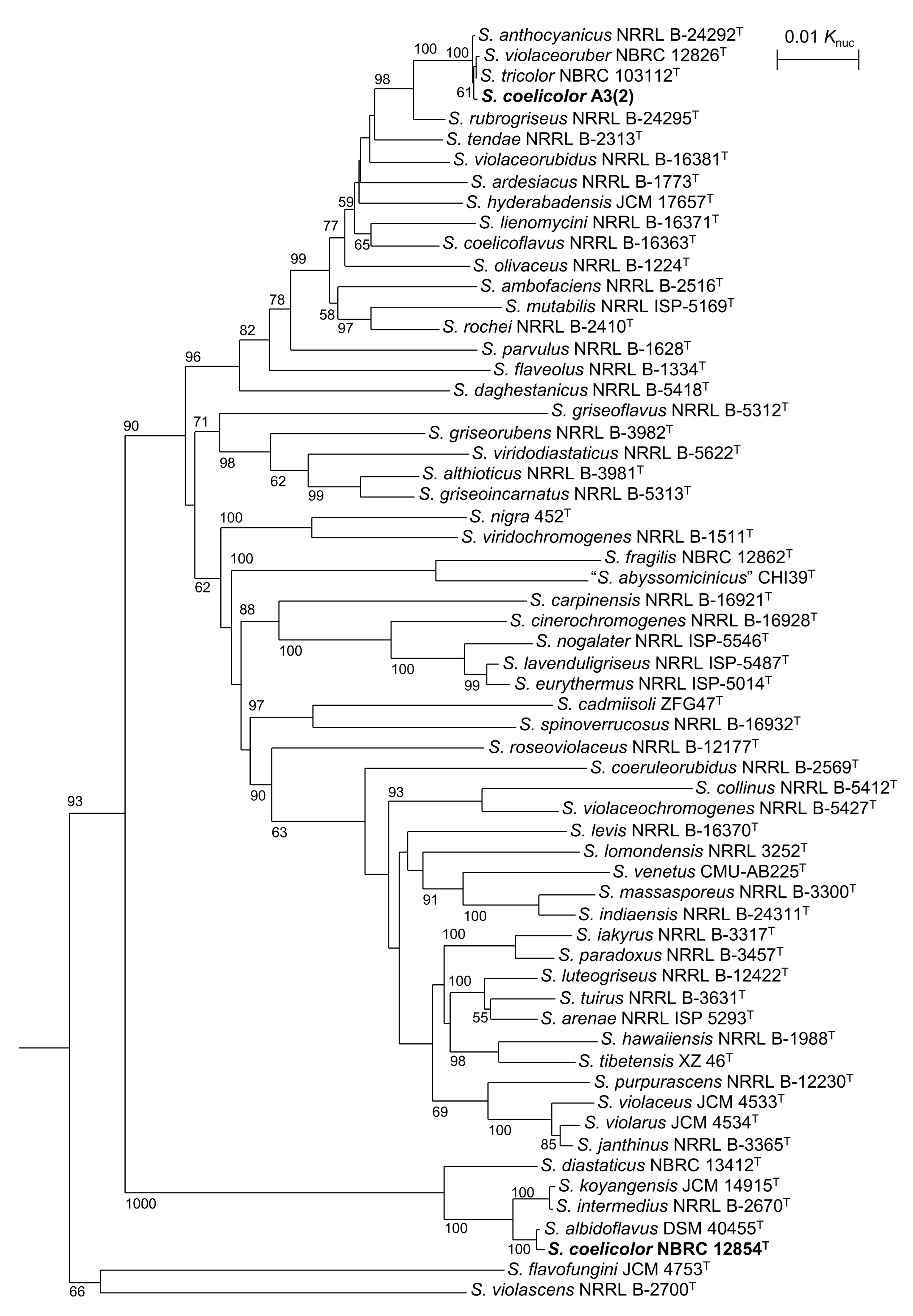
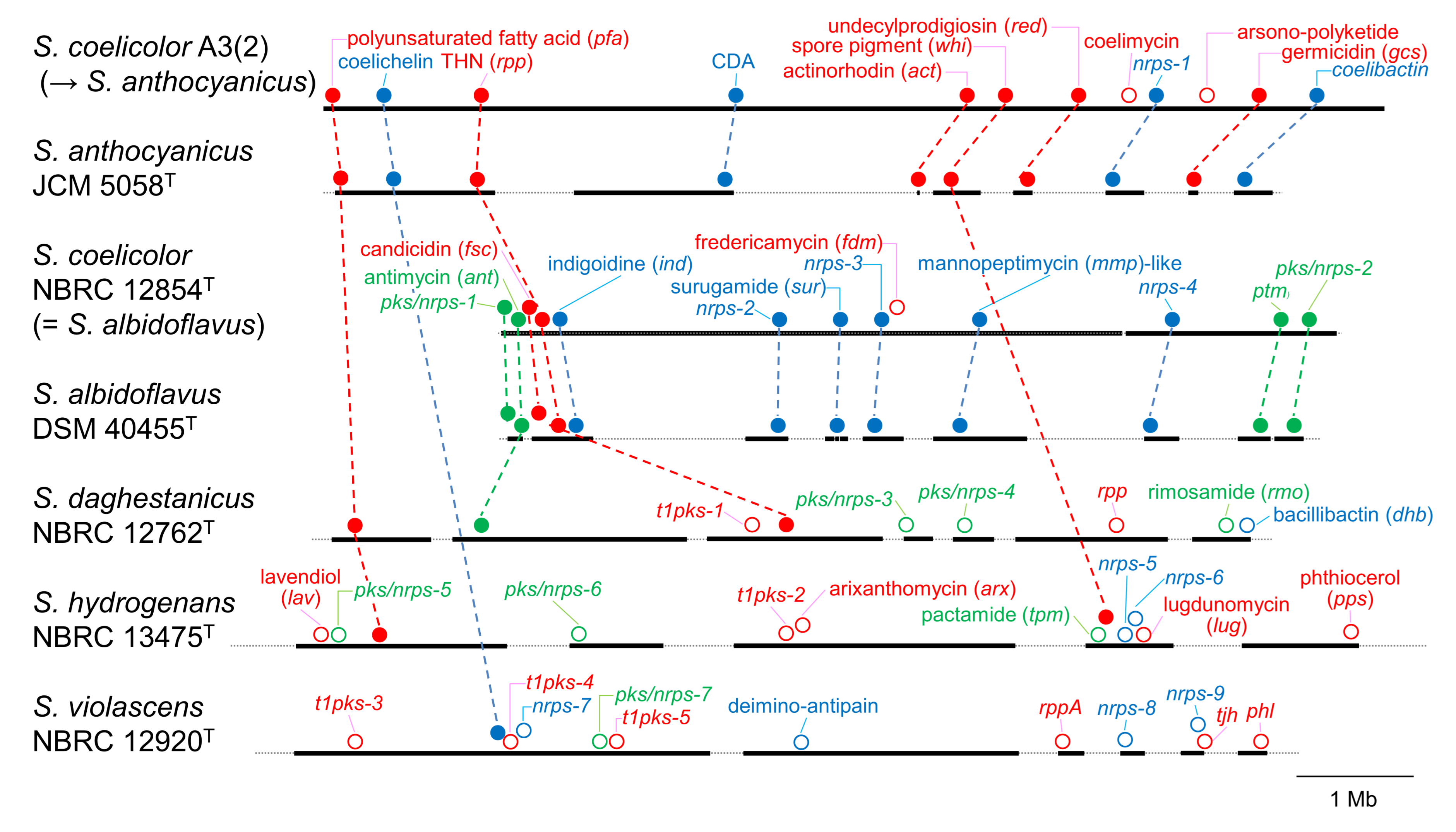
| Strain | atpD | gyrB | recA | rpoB | trpB |
|---|---|---|---|---|---|
| S. albus NBRC 13014T | BBQG01000033 | BBQG01000007 | BBQG01000035 | BBQG01000012 | BBQG01000017 |
| “S. abyssomicinicus” CHI39T | BBZI01000006 | BBZI01000009 | BBZI01000014 | BBZI01000006 | BBZI01000008 |
| S. albidoflavus DSM 40455T | FJ406416 | FJ406427 | FJ406438 | FJ406449 | FJ406460 |
| S. althioticus NRRL B-3981T | KT384460 | KT384809 | KT385157 | KT388779 | KT389129 |
| S. ambofaciens NRRL B-2516T | KT384462 | KT384811 | KT385159 | KT388781 | KT389131 |
| S. anthocyanicus NRRL B-24292T | KT384465 | KT384814 | KT385162 | KT388784 | KT389134 |
| S. ardesiacus NRRL B-1773T | KT384534 | KT384883 | KT385231 | KT388853 | KT389203 |
| S. arenae NRRL ISP 5293T | KT384470 | KT384819 | KT385167 | KT388789 | KT389139 |
| S. cadmiisoli ZFG47T | CP030073 | CP030073 | CP030073 | CP030073 | CP030073 |
| S. carpinensis NRRL B-16921T | KT384503 | KT384852 | KT385200 | KT388822 | KT389172 |
| S. cinerochromogenes NRRL B-16928T | KT384514 | KT384863 | KT385211 | KT388833 | KT389183 |
| S. coelicoflavus NRRL B-16363T | KT384524 | KT384873 | KT385221 | KT388843 | KT389193 |
| S. coelicolor NBRC 12854T | BNDZ01000005 | BNDZ01000005 | BNDZ01000003 | BNDZ01000005 | BNDZ01000005 |
| S. coelicolor A3(2) | AL939123 | AL939118 | AL939125 | AL939121 | AL939121 |
| S. coeruleorubidus NRRL B-2569T | KT384528 | KT384877 | KT385225 | KT388847 | KT389197 |
| S. collinus NRRL B-5412T | KT384529 | KT384878 | KT385226 | KT388848 | KT389198 |
| S. daghestanicus NRRL B-5418T | KJ137021 | KJ137038 | KJ137055 | KJ996779 | KJ137089 |
| S. diastaticus NBRC 13412T | BLLN01000005 | BLLN01000002_ | BLLN01000005 | BLLN01000002 | BLLN01000003 |
| S. eurythermus NRRL ISP-5014T | KT384544 | KT384893 | KT385242 | KT388863 | KT389213 |
| S. flaveolus NRRL B-1334T | KT384550 | KT384899 | KT385248 | KT388869 | KT389219 |
| S. flavofungini JCM 4753T | JAEKOZ01000002 | JAEKOZ01000001 | JAEKOZ010000015 | JAEKOZ010000008 | JAEKOZ010000005 |
| S. fragilis NBRC 12862T | BEVZ01000004 | BEVZ01000008 | BEVZ01000002 | BEVZ01000004 | BEVZ01000003 |
| S. griseoflavus NRRL B-5312T | KT384578 | KT384927 | KT385276 | KT388897 | KT389247 |
| S. griseoincarnatus NRRL B-5313T | KT384580 | KT384929 | KT385278 | KT388899 | KT389249 |
| S. griseorubens NRRL B-3982T | KT384583 | KT384932 | KT385281 | KT388903 | KT389252 |
| S. hawaiiensis NRRL B-1988T | KT384592 | KT384941 | KT385290 | KT388912 | KT389261 |
| S. hyderabadensis JCM 17657T | JAIOJZ010000004 | JAIOJZ010000004 | JAIOJZ010000060 | JAIOJZ010000079 | JAIOJZ010000001 |
| S. hydrogenans NBRC 13475T | BNDW01000004 | BNDW01000040 | BNDW01000004 | BNDW01000004 | BNDW01000019 |
| S. iakyrus NRRL B-3317T | KT384600 | KT384949 | KT385298 | KT388920 | KT389269 |
| S. indiaensis NRRL B-24311T | KT384601 | KT384950 | KT385300 | KT388921 | KT389270 |
| S. intermedius NRRL B-2670T | KT384602 | KT384951 | KT385301 | KT388922 | KT389271 |
| S. janthinus NRRL B-3365T | KT384604 | KT384953 | KT385303 | KT388924 | KT389273 |
| S. koyangensis JCM 14915T | LC381971 | LC381972 | LC381973 | LC381974 | LC413709 |
| S. lavenduligriseus NRRL ISP-5487T | JOBD01000030 | JOBD01000019 | JOBD01000017 | JOBD01000005 | JOBD01000013 |
| S. levis NRRL B-16370T | KT384621 | KT384970 | KT385320 | KT388941 | KT389290 |
| S. lienomycini NRRL B-16371T | KT384622 | KT384971 | KT385321 | KT388942 | KT389291 |
| S. lomondensis NRRL 3252T | KT384626 | KT384975 | KT385326 | KT388946 | KT389295 |
| S. luteogriseus NRRL B-12422T | KT384632 | KT384981 | KT385332 | KT388952 | KT389301 |
| S. massasporeus NRRL B-3300T | KT384636 | KT384985 | KT385336 | KT388956 | KT389305 |
| S. mutabilis NRRL ISP-5169T | KT384652 | KT385002 | KT385353 | KT388972 | KT389321 |
| S. nigra 452T | CP029043 | CP029043 | CP029043 | CP029043 | CP029043 |
| S. nogalater NRRL ISP-5546T | KT384664 | KT385014 | KT385365 | KT388984 | KT389333 |
| S. olivaceus NRRL B-1224T | KT384667 | KT385017 | KT385368 | KT388987 | KT389336 |
| S. pactum ATCC 27456T | JACYXC010000001 | JACYXC010000001 | JACYXC010000001 | JACYXC010000001 | JACYXC010000001 |
| S. paradoxus NRRL B-3457T | KT384674 | KT385024 | KT385375 | KT388994 | KT389343 |
| S. parvulus NRRL B-1628T | KJ196367 | KJ196369 | KJ196371 | KJ196373 | KJ196375 |
| S. purpurascens NRRL B-12230T | KT384696 | KT385046 | KT385397 | KT389017 | KT389365 |
| S. rochei NRRL B-2410T | KT384704 | KT385054 | KT385405 | KT389025 | KT389373 |
| S. roseoviolaceus NRRL B-12177T | KT384710 | KT385060 | KT385411 | KT389031 | KT389379 |
| S. rubrogriseus NRRL B-24295T | KT384715 | KT385065 | KT385416 | KT389036 | KT389384 |
| S. spinoverrucosus NRRL B-16932T | KT384725 | KT385074 | KT385426 | KT844525 * | KT389394 |
| S. tendae NRRL B-2313T | KT384733 | KT385082 | KT385434 | KT389053 | KT389402 |
| S. tibetensis XZ 46T | SZVR01000021 | SZVR01000021 | SZVR01000011 | SZVR01000023 | SZVR01000005 |
| S. tricolor NBRC 103112T | LC634004 | LC634005 | LC634006 | LC634007 | LC634008 |
| S. tuirus NRRL B-3631T | KT384742 | KT385090 | KT385444 | KT389062 | KT389411 |
| S. venetus CMU-AB225T | LC381976 | LC381977 | LC381978 | LC381979 | LC381980 |
| S. violaceochromogenes NRRL B-5427T | KT384748 | KT385096 | KT385450 | KT389068 | KT389417 |
| S. violaceoruber NBRC 12826T | LC634009 | LC634010 | LC634011 | LC634012 | LC634013 |
| S. violaceorubidus NRRL B-16381T | JODM01000021 | JODM01000004 | JODM01000011 | JODM01000001 | JODM01000015 |
| S. violaceus JCM 4533T | LC381981 | LC381982 | LC381983 | LC381984 | LC381985 |
| S. violarus JCM 4534T | BMUP01000003 | BMUP01000013 | BMUP01000001 | BMUP01000012 | BMUP01000009 |
| S. violascens NRRL B-2700T | KT384752 | KT385100 | KT385454 | KT389072 | KT389421 |
| S. viridochromogenes NRRL B-1511T | KT384756 | KT385104 | KT385458 | KT389076 | KT389425 |
| S. viridodiastaticus NRRL B-5622T | KT384757 | KT385105 | KT385459 | KT389077 | KT389426 |
| Strain | DDH Estimate (%) | ||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| - | 91.8 | 23.2 | 23.2 | 22.7 | 22.7 | 23.1 |
| - | 23.0 | 23.1 | 22.7 | 22.7 | 22.7 | |
| - | 23.0 | 22.7 | 28.0 | 28.1 | ||
| - | 23.4 | 22.8 | 22.9 | |||
| - | 22.6 | 22.8 | ||||
| - | 94.2 | |||||
| - | ||||||
| Strain | Gene Cluster | ORF (Locus Tag) | Domain Organization | Predicted Product |
|---|---|---|---|---|
| S. coelicolor A3(2) | nrps-1 | SCO6431 SCO6432 | A/T-C/T C/Acys/T-Te | Tripeptide (x-y-cys) |
| S. coelicolor NBRC 12854T | nrps-2 | ScoT_42860 | A/T/E-TD | Unknown |
| nrps-3 | ScoT_34810 | C/A/T-C/A/T-C/A/T-Te | Tripeptide | |
| nrps-4 | ScoT_12950 ScoT_12970 | Aphe/T-C/A/T/E-C/Aval/T C/A/T/E-C/Athr/T-C/Athr/T/E | Hexapeptide (phe-x-val-x-thr-thr) | |
| pks/nrps-1 | ScoT_62740 ScoT_62750 ScoT_62760 ScoT_62780 | Agly/T-KS/DH/ACP-KS/ATm DH/KR/ACP-KS/ACP-KS/KR/ACP KS/DH/ACP-KS/ATm DH/KR/ACP-AmT | Hexaketide including gly | |
| pks/nrps-2 | ScoT_02700 ScoT_02710 ScoT_02720 | A/T KS/ACP-C/Aala/T/C-Te C/Athr/T-C/T/E-C/Aval/T-C | Pentapeptide with polyketide moiety (thr-y-val-x-pk-ala) | |
| S. daghestanicus NBRC 12762T | t1pks-1 | Sdagh_09690 Sdagh_09680 Sdagh_09670 | CoL/KR/ACP-KS/ATmm/KR/ACP-KS/ATmm/DH/KR/ACP KS/ATm/DH/KR/ACP KS/ATm/ACP | Tetraketide with a starter |
| pks/nrps-3 | Sdagh_50890 Sdagh_50900 Sdagh_50940 Sdagh_50980 | C A/KR/ACP A T | Unknown | |
| pks/nrps-4 | Sdagh_52260 Sdagh_52060 Sdagh_51920 Sdagh_51910 Sdagh_51900 | T-C/Acys/MT/T KS (type-III PKS) A/T/E-C/A/T/E-C/A/T/E-C/A/T/E-C/Athr/T/E C/A/T/E C/A/T-C/A/T-C/A/T-X/A-Te | Undecapeptide including cys, thr, and a polyketide moiety | |
| S. hydrogenans NBRC 13475T | t1pks-2 | Shyd_35460 | KS/ATmm/ACP- KS/ATmm/DH/ER/KR/ACP | Unknown |
| nrps-5 | Shyd_62130 Shyd_62150 Shyd_62160 | Aphe/MT/T C/Aval/T-C/Agly/T/E C/A/T-Te | Tetrapeptide (methyl phe-val-gly-x) | |
| nrps-6 | Shyd_62460 Shyd_62450 | A/MT/T-C/A/T-C/Aval/T C/A/T-C/Athr/T-Te | Pentapeptide (methyl x-x-val-x-thr) | |
| pks/nrps-5 | Shyd_13440 Shyd_13450 Shyd_13460 Shyd_13470 Shyd_13490 Shyd_13520 Shyd_13530 | Aval/T C/T-C T KS/ACP-TD T FkbH A | Dipeptide with a polyketide moiety | |
| pks/nrps-6 | Shyd_48540 Shyd_48670 Shyd_48680 | Aval/T-C Atyr/T KS/ATm/ACP | Dipeptide with a polyketide moiety (val-tyr-pk) | |
| S. violascens NBRC 12920T | t1pks-3 | Sviol_48010 Sviol_48000 Sviol_47990 | KS/ATm/ACP-KR KS/ATm ACP | Unknown |
| t1pls-4 | Sviol_60680 | KS/AT/DH/MT/ER/KR/ACP | Unknown | |
| t1pks-5 | Sviol_68600 | KS/AT/DH/KR/ACP | Unknown | |
| tjh (t2pks) | Sviol_35480 Sviol_35470 Sviol_35460 Sviol_35440 | KSα KSβ (CLF) ACP KR | Unidentified aromatic polyketide | |
| phlD | Sviol_40150 | KS (type-III PKS) | Unidentified polyketide | |
| nrps-7 | Sviol_61030 | C/Athr/T-Te | Unknown including thr | |
| nrps-8 | Sviol_25110 | C/Aphe/T/E-C/Aphe/T-C/A/T/T-Te | Tripeptide (phe-phe-x) | |
| nrps-9 | Sviol_35280 Sviol_35230 Sviol_35220 | A/T-C T A | Dipeptide | |
| nrps-10 | Sviol_76120 Sviol_76140 Sviol_76160 | A/KR/ACP Acys/T-C/A/T C/Aala/T/E-C/A/T-C/A/T/E-C/A/T-Te | Heptapeptide including cys and ala | |
| pks/nrps-7 | Sviol_68100 Sviol_68090 Sviol_68080 Sviol_68070 Sviol_68030 Sviol_68020 Sviol_67990 Sviol_67980 | A/T C/A/T-C/T KS/DH/ACP-TD A ATm TE ACP KS | Tripeptide with a polyketide moiety |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komaki, H.; Tamura, T. Differences at Species Level and in Repertoires of Secondary Metabolite Biosynthetic Gene Clusters among Streptomyces coelicolor A3(2) and Type Strains of S. coelicolor and Its Taxonomic Neighbors. Appl. Microbiol. 2021, 1, 573-585. https://doi.org/10.3390/applmicrobiol1030037
Komaki H, Tamura T. Differences at Species Level and in Repertoires of Secondary Metabolite Biosynthetic Gene Clusters among Streptomyces coelicolor A3(2) and Type Strains of S. coelicolor and Its Taxonomic Neighbors. Applied Microbiology. 2021; 1(3):573-585. https://doi.org/10.3390/applmicrobiol1030037
Chicago/Turabian StyleKomaki, Hisayuki, and Tomohiko Tamura. 2021. "Differences at Species Level and in Repertoires of Secondary Metabolite Biosynthetic Gene Clusters among Streptomyces coelicolor A3(2) and Type Strains of S. coelicolor and Its Taxonomic Neighbors" Applied Microbiology 1, no. 3: 573-585. https://doi.org/10.3390/applmicrobiol1030037
APA StyleKomaki, H., & Tamura, T. (2021). Differences at Species Level and in Repertoires of Secondary Metabolite Biosynthetic Gene Clusters among Streptomyces coelicolor A3(2) and Type Strains of S. coelicolor and Its Taxonomic Neighbors. Applied Microbiology, 1(3), 573-585. https://doi.org/10.3390/applmicrobiol1030037





