Prevalence of invA Gene of Salmonella spp. in Fish and Fishery Resources from Manila Bay Aquaculture Farms Using Real-Time PCR
Abstract
1. Introduction
2. Materials and Methods
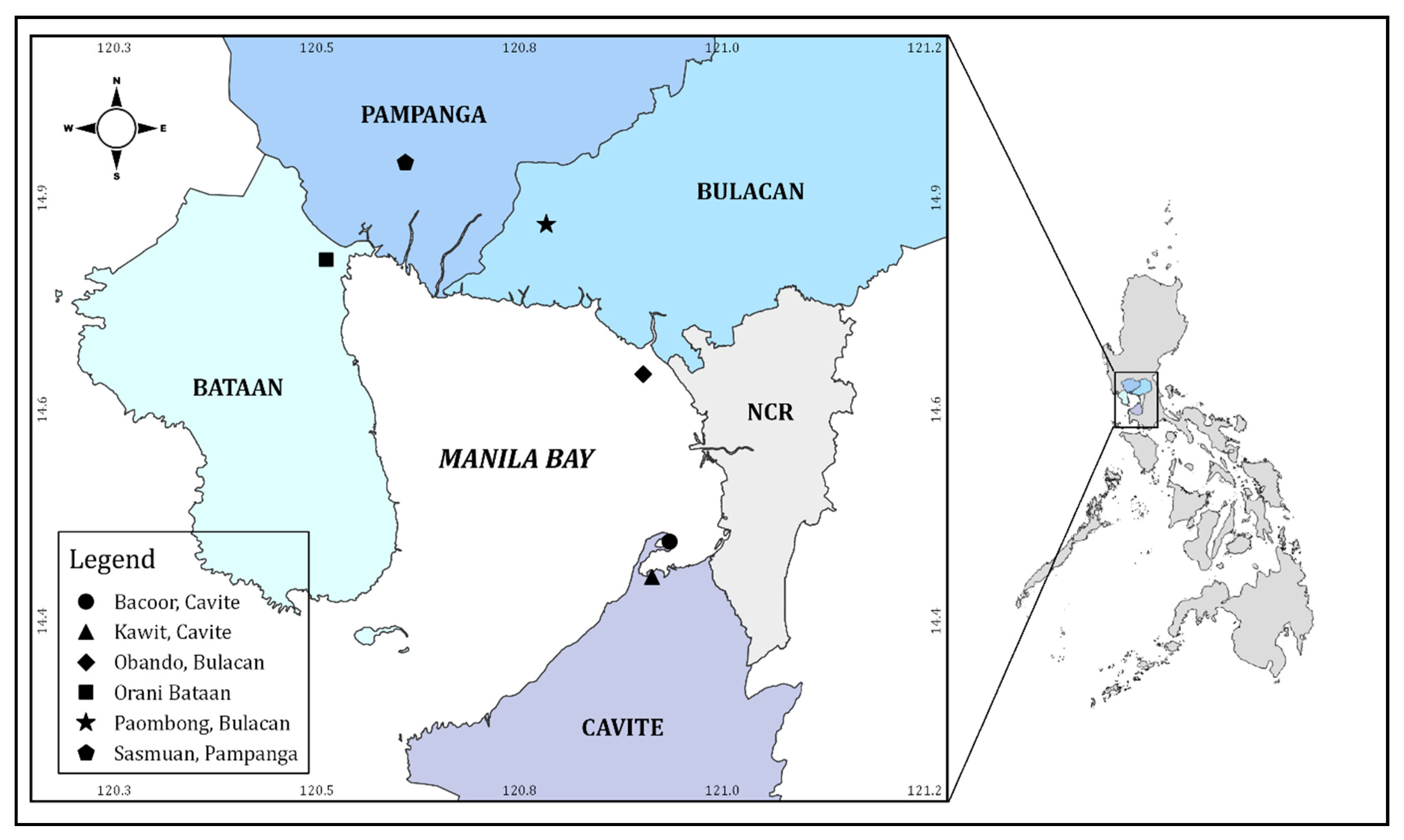
2.1. Sampling Area
2.2. Sample Collection
2.3. Detection of Salmonella
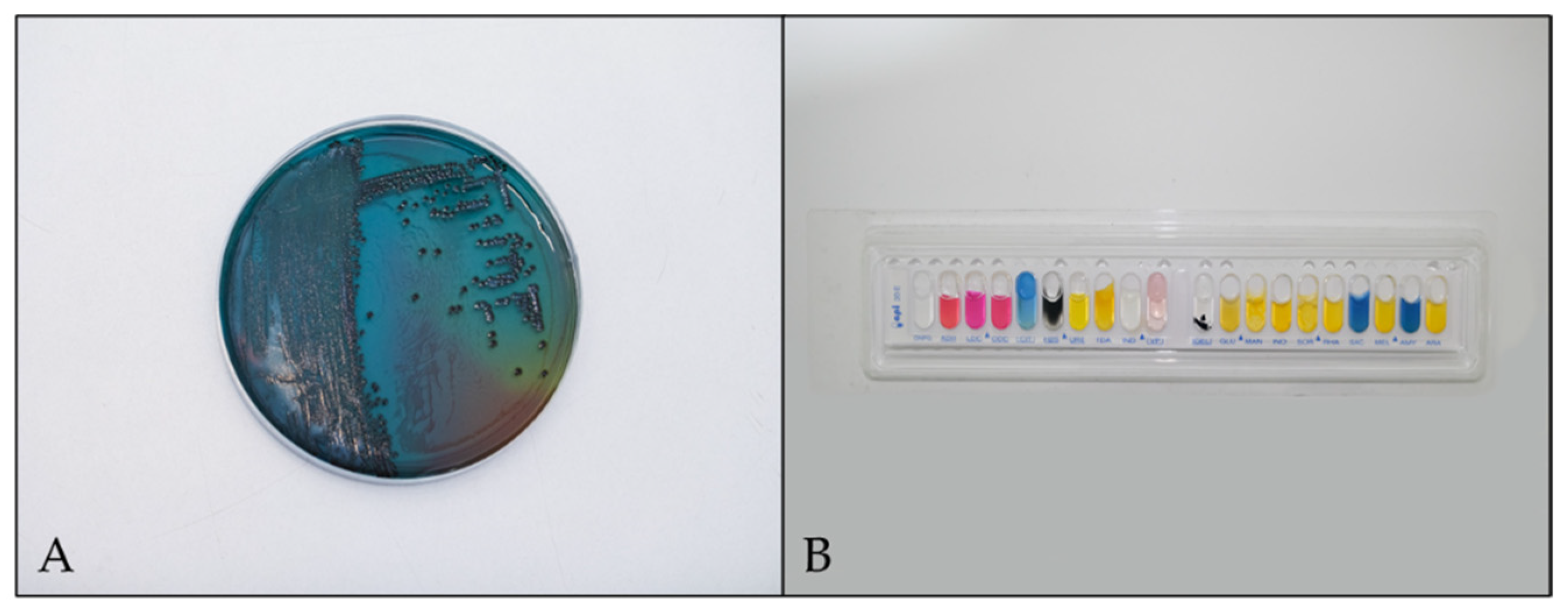
2.3.1. Sample Preparation and Isolation of Salmonella
2.3.2. Blanks and Controls
2.3.3. DNA Extraction
2.3.4. Identification and Confirmation of Salmonella spp.
Culture-Based Method
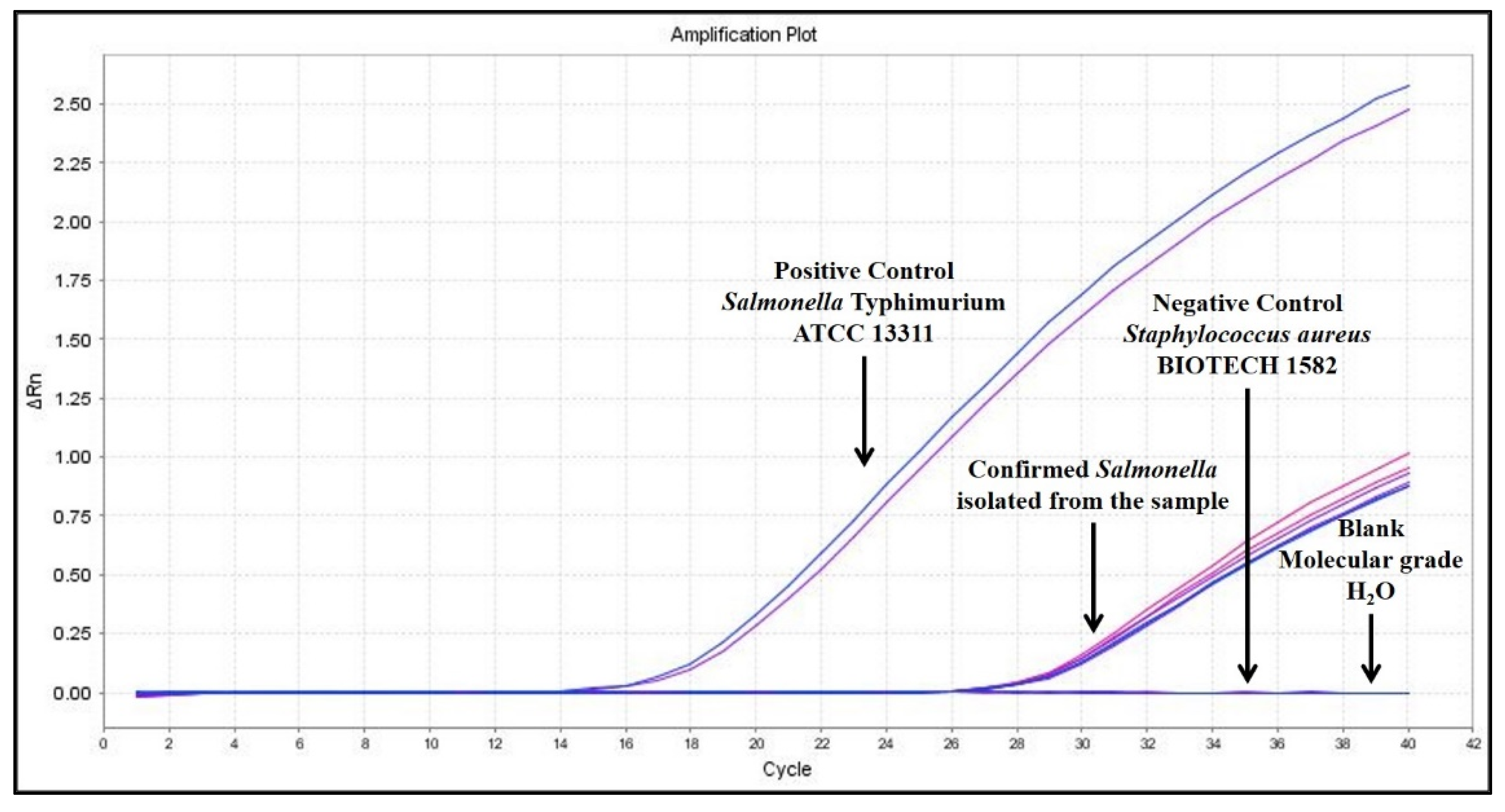
Real-Time PCR Taqman Assay
2.4. Statistical Analysis
3. Results and Discussions
3.1. Detection of Salmonella by Culture-Based and Real-Time PCR Methods
3.2. Prevalence of Salmonella in Fish and Fishery Resources and Environmental Waters
3.3. Prevalence of Salmonella between Sampling Locations
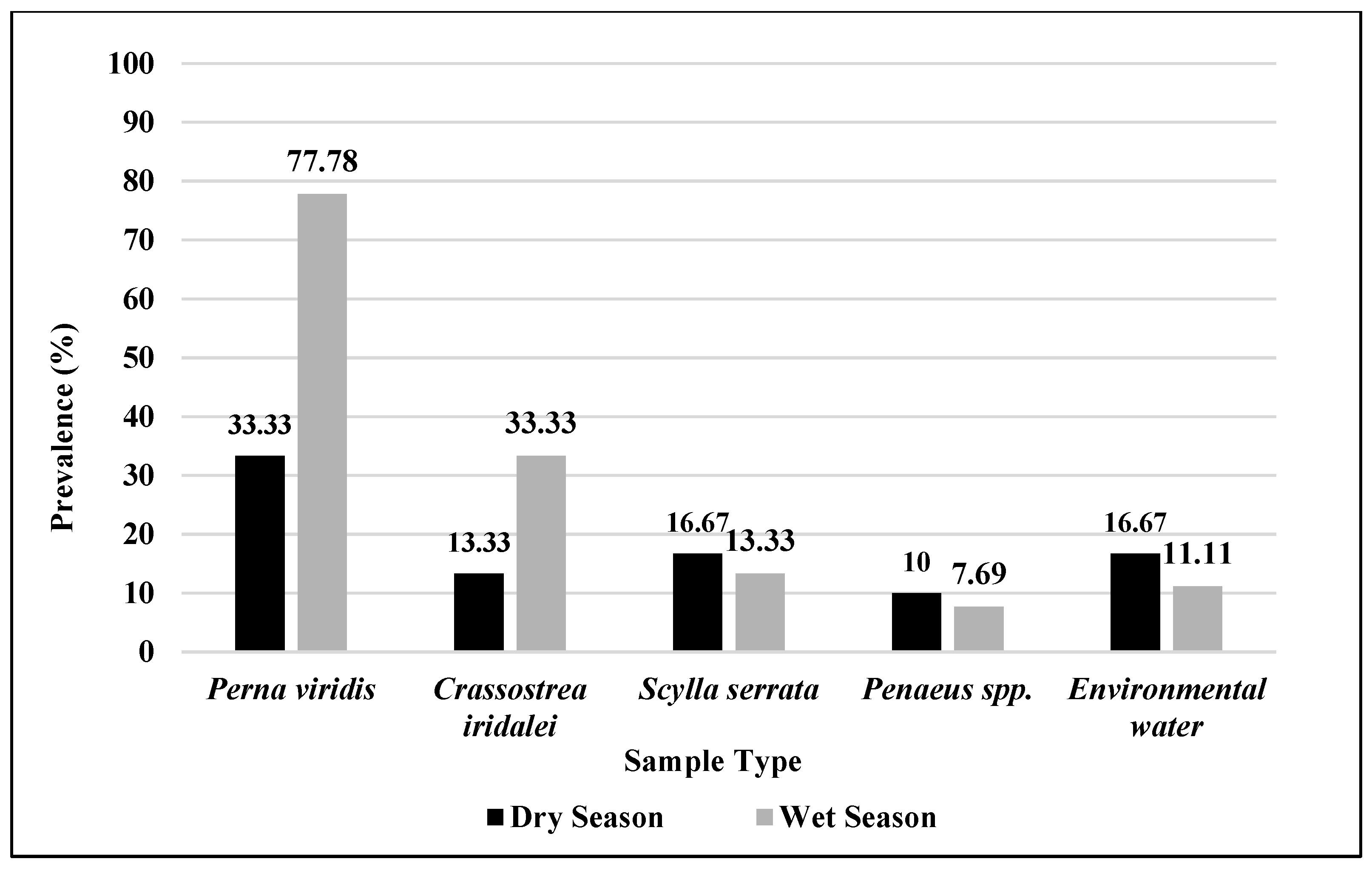
3.4. Seasonal Distribution of Salmonella in Fish and Fishery Resources and Environmental Waters
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization. FAO Aquaculture Newsletter No. 61. FAO Aquac. Newletter. 2020. Available online: https://www.fao.org/3/ca8302en/CA8302EN.pdf (accessed on 30 May 2021).
- Food and Agriculture Organization. The State of World Fisheries and Aquaculture; FAO: Rome, Italy, 2014. [Google Scholar]
- Philippine Statistics Authority. Fisheries Statistics of the Philippines 2017–2019; PSA: Manila, Philippines, 2020. [Google Scholar]
- PEMSEA; DENR. Initial Valuation of Selected Uses and Habitats and Damage Assessment of Manila Bay; Global Environment Facility/United Nations Development Programme/International Maritime Organization Regional Programme on Building Partnerships in Environmental Management for the Seas of East Asia (GEF/UNDP/PEM SEA): Quezon City, Philippines, 2005; p. 165. [Google Scholar]
- Raña, J.; Domingo, J.; Opinion, A.G.; Cambia, F. Contamination of Coliform Bacteria in Water and Fishery Resources in Manila Bay Aquaculture Farms. Philipp. J. Fish. 2017, 24, 98–126. [Google Scholar] [CrossRef]
- Brooks, G.F.; Carroll, K.C.; Butel, J.S.; Morse, S.A.; Mietzner, T.A. (Eds.) Jawetz, Melnick & Adelberg’s Medical Microbiology, 26th ed.; The McGraw-Hill Companies, Inc.: New York, NY, USA, 2010. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Typhoid Fever and Paratyphoid Fever. Available online: https://www.cdc.gov/typhoid-fever/ (accessed on 21 June 2021).
- World Health Organization. Typhoid. Available online: https://www.who.int/news-room/fact-sheets/detail/typhoid (accessed on 30 May 2021).
- Amagliani, G.; Brandi, G.; Schiavano, G. Incidence and role of Salmonella in seafood safety. Food Res. Int. 2012, 45, 780–788. [Google Scholar] [CrossRef]
- Azanza, M.P.V. Philippine Foodborne-Disease Outbreaks (1995–2004). J. Food Saf. 2006, 26, 92–102. [Google Scholar] [CrossRef]
- Azanza, M.P.V.; Membrebe, B.N.Q.; Sanchez, R.G.R.; Estilo, E.E.C.; Dollete, U.G.M.; Feliciano, R.J.; Garcia, N.K.A. Foodborne Disease Outbreaks in the Philippines (2005–2018). Philipp. J. Sci. 2019, 148, 20. [Google Scholar]
- Baird, R.B.; Eaton, A.D.; Rice, E.W.; Bridgewater, L.L. (Eds.) Standard Methods for the Examination of Water and Wastewater, 23rd ed.; American Public Health Association: Washington, DC, USA, 2017. [Google Scholar]
- Food and Drug Administration. BAM Chapter 1: Food Sampling/Preparation of Sample Homogenate. Available online: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-1-food-samplingpreparation-sample-homogenate (accessed on 21 June 2021).
- Sharma, I. Detection of invA Gene in Isolated Salmonella from Marketed Poultry Meat by PCR Assay. J. Food Process. Technol. 2016, 7, 2. [Google Scholar] [CrossRef]
- Yanestria, S.M.; Rahmaniar, R.P.; Wibisono, F.J.; Effendi, M.H. Detection of invA gene of Salmonella from milkfish (Chanos chanos) at Sidoarjo wet fish market, Indonesia, using polymerase chain reaction technique. Veter-World 2019, 12, 170–175. [Google Scholar] [CrossRef]
- El-Sebay, N.A.; Abu Shady, H.M.; El-Zeedy, S.A.E.-R.; Samy, A. InvA Gene Sequencing of Salmonella typhimurium Isolated from Egyptian Poultry. Asian J. Sci. Res. 2017, 10, 194–202. [Google Scholar] [CrossRef][Green Version]
- Eng, S.-K.; Pusparajah, P.; Ab Mutalib, N.-S.; Ser, H.-L.; Chan, K.-G.; Lee, L.-H. Salmonella: A review on pathogenesis, epidemiology and antibiotic resistance. Front. Life Sci. 2015, 8, 284–293. [Google Scholar] [CrossRef]
- Kumar, R.; Surendran, P.; Thampuran, N. Distribution and genotypic characterization of Salmonellaserovars isolated from tropical seafood of Cochin, India. J. Appl. Microbiol. 2009, 106, 515–524. [Google Scholar] [CrossRef]
- Ben Hassena, A.; Siala, M.; Guermazi, S.; Zormati, S.; Gdoura, R.; Sellami, H. Occurrence and Phenotypic and Molecular Characterization of Antimicrobial Resistance of Salmonella Isolates from Food in Tunisia. J. Food Prot. 2019, 82, 1166–1175. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Urtaza, J.; Saco, M.; de Novoa, J.; Perez-Piñeiro, P.; Peiteado, J.; Lozano-Leon, A.; Garcia-Martin, O. Influence of Environmental Factors and Human Activity on the Presence of Salmonella Serovars in a Marine Environment. Appl. Environ. Microbiol. 2004, 70, 2089–2097. [Google Scholar] [CrossRef]
- Rubini, S.; Galletti, G.; D’Incau, M.; Govoni, G.; Boschetti, L.; Berardelli, C.; Barbieri, S.; Merialdi, G.; Formaglio, A.; Guidi, E.; et al. Occurrence of Salmonella enterica subsp. enterica in bivalve molluscs and associations with Escherichia coli in molluscs and faecal coliforms in seawater. Food Control. 2018, 84, 429–435. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, X.; Kuang, D.; Shi, X.; Xiao, W.; Zhang, J.; Gu, Z.; Xu, X.; Meng, J. Prevalence of antimicrobial resistance of non-typhoidal Salmonella serovars in retail aquaculture products. Int. J. Food Microbiol. 2015, 210, 47–52. [Google Scholar] [CrossRef]
- Ferrari, R.; Rosario, D.K.A.; Neto, A.C.; Mano, S.B.; Figueiredo, E.E.S.; Conte-Junior, C.A. Worldwide Epidemiology of Salmonella Serovars in Animal-Based Foods: A Meta-analysis. Appl. Environ. Microbiol. 2019, 85, e00591-19. [Google Scholar] [CrossRef] [PubMed]
- Sheng, L.; Wang, L. The microbial safety of fish and fish products: Recent advances in understanding its significance, contamination sources, and control strategies. Compr. Rev. Food Sci. Food Saf. 2020, 20, 738–786. [Google Scholar] [CrossRef] [PubMed]
- Hatem, A.; Alameer, A.; Faidullah Atshan, O.; Mahmood, M.M.; Al-Jewari, M.; Mohammed, A.A. Detection of Salmonella Species in Viscera of Carp Fish. Plant Arch. 2020, 20, 2683–2686. [Google Scholar]
- Bureau of Fisheries and Aquatic Resources. Fisheries Administrative Order No. 210. Available online: https://www.bfar.da.gov.ph/LAW?fi=355 (accessed on 5 July 2021).
- European Union. Commission Regulation (EC) No 2073/2005; European Union: Maastricht, The Netherlands, 2005. [Google Scholar]
- Philippine Statistics Authority. Philippine Statistics Authority|Republic of the Philippines; Philippine Statistics Authority: Manila, Philippines, 2021. [Google Scholar]
- Levantesi, C.; Bonadonna, L.; Briancesco, R.; Grohmann, E.; Toze, S.; Tandoi, V. Salmonella in surface and drinking water: Occurrence and water-mediated transmission. Food Res. Int. 2012, 45, 587–602. [Google Scholar] [CrossRef]
- Smith, B.A.; Meadows, S.; Meyers, R.; Parmley, E.J.; Fazil, A. Seasonality and zoonotic foodborne pathogens in Canada: Relationships between climate and Campylobacter, E. coli and Salmonella in meat products. Epidemiol. Infect. 2019, 147, e190. [Google Scholar] [CrossRef] [PubMed]
- Kovats, R.S.; Edwards, S.J.; Hajat, S.; Armstrong, B.G.; Ebi, K.L.; Menne, B.; Cowden, J.; Gerner-Smidt, P.; Hernández Pezzi, G.; Kristufkova, Z.; et al. The effect of temperature on food poisoning: A time-series analysis of salmonellosis in ten European countries. Epidemiol. Infect. 1999, 132, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Ravel, A.; Smolina, E.; Sargeant, J.M.; Cook, A.; Marshall, B.; Fleury, M.D.; Pollari, F. Seasonality in Human Salmonellosis: Assessment of Human Activities and Chicken Contamination as Driving Factors. Foodborne Pathog. Dis. 2010, 7, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, H.A.; Akil, L. Effects of Environment and Socioeconomics on Salmonella Infections. In Current Topics in Salmonella and Salmonellosis; InTech: London, UK, 2017. [Google Scholar] [CrossRef]
- Liu, H.; Whitehouse, C.A.; Li, B. Presence and Persistence of Salmonella in Water: The Impact on Microbial Quality of Water and Food Safety. Front. Public Health 2018, 6, 159. [Google Scholar] [CrossRef] [PubMed]
- Simental, L.; Martinez-Urtaza, J. Climate Patterns Governing the Presence and Permanence of Salmonellae in Coastal Areas of Bahia de Todos Santos, Mexico. Appl. Environ. Microbiol. 2008, 74, 5918–5924. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Gu, G.; Ginn, A.; Giurcanu, M.C.; Adams, P.; Vellidis, G.; van Bruggen, A.H.C.; Danyluk, M.D.; Wright, A.C. Distribution and Characterization of Salmonella enterica Isolates from Irrigation Ponds in the Southeastern United States. Appl. Environ. Microbiol. 2015, 81, 4376–4387. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Jackson, C.R.; Frye, J.G. The Prevalence and Antimicrobial Resistance Phenotypes of Salmonella, Escherichia Coli and Enterococcus Sp. in Surface Water. In Letters in Applied Microbiology; Blackwell Publishing Ltd.: Hoboken, NJ, USA, 2020; pp. 3–25. [Google Scholar] [CrossRef]
| invA Gene | Bases | Sequence | |
|---|---|---|---|
| Primers | Forward | 20 | 5′-AAC GTG TTT CCG TGC GTA AT-3′ |
| Reverse | 20 | 5′-TCC ATC AAA TTA GCG GAG GC-3′ | |
| Probe | 20 | 5′/FAM/TGG AAG CGC TCG CAT TGT GG/BHQ/-3′ |
| Sample Type | No. of Samples Examined | Positive (%) |
|---|---|---|
| Bivalves | ||
| Perna viridis * | 18 | 10 (55.55) |
| Crassostrea iridalei | 21 | 4 (19.05) |
| Crustaceans | ||
| Scylla serrata | 27 | 4 (14.81) |
| Penaeus spp. | 23 | 2 (8.70) |
| Finfishes | ||
| Oreochromis niloticus | 16 | 0 (0) |
| Chanos chanos | 18 | 0 (0) |
| Environmental Water | 81 | 11 (13.58) |
| Total | 204 | 31 (15.20) |
| Sample Type | No. of Samples Examined | Positive (%) |
|---|---|---|
| Bulacan | 48 | 11 (22.92) |
| Bataan | 48 | 10 (20.83) |
| Cavite | 50 | 5 (10.00) |
| Pampanga | 58 | 5 (8.62) |
| Total | 204 | 31 (15.20) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanyag, B.; Quiambao, J.J.; Ko, A.A.; Singh, A.; Cambia, F.; Montojo, U. Prevalence of invA Gene of Salmonella spp. in Fish and Fishery Resources from Manila Bay Aquaculture Farms Using Real-Time PCR. Appl. Microbiol. 2021, 1, 510-519. https://doi.org/10.3390/applmicrobiol1030033
Tanyag B, Quiambao JJ, Ko AA, Singh A, Cambia F, Montojo U. Prevalence of invA Gene of Salmonella spp. in Fish and Fishery Resources from Manila Bay Aquaculture Farms Using Real-Time PCR. Applied Microbiology. 2021; 1(3):510-519. https://doi.org/10.3390/applmicrobiol1030033
Chicago/Turabian StyleTanyag, Bryan, Jerick Jann Quiambao, Aira Angeline Ko, Amarjet Singh, Flordeliza Cambia, and Ulysses Montojo. 2021. "Prevalence of invA Gene of Salmonella spp. in Fish and Fishery Resources from Manila Bay Aquaculture Farms Using Real-Time PCR" Applied Microbiology 1, no. 3: 510-519. https://doi.org/10.3390/applmicrobiol1030033
APA StyleTanyag, B., Quiambao, J. J., Ko, A. A., Singh, A., Cambia, F., & Montojo, U. (2021). Prevalence of invA Gene of Salmonella spp. in Fish and Fishery Resources from Manila Bay Aquaculture Farms Using Real-Time PCR. Applied Microbiology, 1(3), 510-519. https://doi.org/10.3390/applmicrobiol1030033





