Abstract
With fast-growing polymerase chain reaction (PCR) technologies and various application methods, the technique has benefited science and medical fields. While having strengths and limitations on each technology, there are not many studies comparing the efficiency and specificity of PCR technologies. The objective of this review is to summarize a large amount of scattered information on PCR technologies focused on the two majorly used technologies: qPCR (quantitative polymerase chain reaction) and ddPCR (droplet-digital polymerase chain reaction). Here we analyze and compare the two methods for (1) efficiency, (2) range of detection and limitations under different disciplines and gene targets, (3) optimization, and (4) status on antibiotic resistance genes (ARGs) analysis. It has been identified that the range of detection and quantification limit varies depending on the PCR method and the type of sample. Careful optimization of target gene analysis is essential for building robust analysis for both qPCR and ddPCR. In our era where mutation of genes may lead to a pandemic of viral infectious disease or antibiotic resistance-induced health threats, this study hopes to set guidelines for meticulous detection, quantification, and analysis to help future prevention and protection of global health, the economy, and ecosystems.
1. Introduction
Many studies and global communities noted that the prevalence and ubiquity of antibiotic resistance genes (ARGs) are increasing over time due to human activities of producing and overdosing antibiotics [1,2,3,4,5,6,7,8,9] (Figure 1). Antibiotics are widely used in our daily lives. From agriculture to industrial level manufacturing, we often use them to prevent disease spreading among livestock, promote growth in farming, consume them to treat illness, or combine them with over-the-counter medicine and hygiene products at home. At the industrial level, especially, maximization of profits by promoting production and selling of antibiotics is intensifying antibiotic resistance (AR), which is contradictory to policies of governments and health services that incentivize conservation of the common good [10]. The wastewater and soil surrounding humans, agricultural land, and industrial manufacturing are contaminated with antibiotics, and AR is accumulating in the environment and in organisms over time (Figure 1). According to the World Health Organization (WHO), we are lacking on global surveillance of AR, which poses a threat not only to human health but also can lead to overloading and systemic failure of environmental systems [11,12]. The WHO urges public awareness, national action plans, optimizing the use of antimicrobials, innovation on research and development, and access, surveillance, and global governance with sustainable development goals (SDGs) [13]. The WHO’s guidance is consistent with recent studies showing that anthropogenic antibiotic usage influences the accumulation of antibiotic-resistant bacteria (ARB) and ARGs in sediments of estuary [14,15], marine and river [5,16,17,18] environments, as well as throughout water reuse cycles [19]. ARGs have been hypothesized to spread through horizontal gene transfer (HGT) with the use of mobile genetic elements (MGEs) [20] and are often linked to other gene elements associated with heavy metal resistance such as class 1 integron-integrase gene (intl1) [21]. MGEs have been linked to ARGs in studies using high-throughput qPCR, ddPCR, and 16S rRNA sequencing [22,23]. To uncover and assess more characteristics of AR and its relation to human activity, identification and quantification of ARB and ARGs are important, especially in different environments and at different genetic targets.
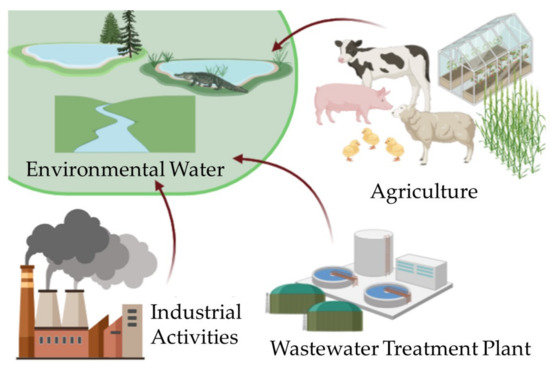
Figure 1.
Sources of anthropogenic influence of AR in the water system.
2. Evolution/Expansion of Nucleic Acid Detection Methods for Molecular Targets
2.1. The Initial State of Polymerase Chain Reaction (PCR)
The ability to identify, detect, and quantify nucleic acids has significantly improved since the mid-1980s. The groundbreaking first-generation polymerase chain reaction (PCR) device used thermal cycling to amplify specific regions of DNA in the presence of Taq polymerase [24], deoxynucleotide triphosphates (dNTPs), and primers of the desired target. The thermal cycling process denatures DNA, anneals the primers and the desired target, and amplifies DNA using polymerase and dNTPs over multiple cycles [25].
This rapid amplification process used in PCR reduced the time and cost of nucleic acid detection in medicine and research, and it boosted most major fields of science. However, PCR has been reported with its downside that it is hard to separate incomplete and damaged genes amplified in the detection of signals [26]. In other cases, incomplete DNA template fragments (for example if DNA is extremely degraded) may not be amplified by PCR if the primer site is missing. Other examples of limitation in PCR include presence of primer-dimers [27] and different levels of inhibition observed depending on target-primer binding structures [28]. A study in the late 1980s by Clewley summarized the approaches to minimize contamination and erroneous issues of PCR by suggesting practical measures such as using well-characterized controls, improving lab techniques and practices, optimizing primer selection, and testing of plasmids [26]. Throughout the years, researchers also evaluated PCR for forensic applications [29], variant identification [30], and primer and temperature optimization [31,32], and PCR has been continually optimized for various applications [33,34,35]. What researchers expected as potential applications of the PCR technique have become more general nowadays. The following paragraphs describe new generations of PCR techniques and their uniqueness.
2.2. The Second-Generation PCR—qPCR
The emergence of real-time PCR allowed for the fluorescent detection of nucleotides during amplification. The real-time amplification fluorescence detection enabled the relative quantification of target genes with the use of known standard genes. As the fluorescence signal is detected throughout the multiple cycles of gene amplification, when the intensity is over the threshold, a cycle threshold (Ct) value is recorded [36]. Based on the Ct values of samples, the relative nucleic concentration can be estimated by comparing against a known standard. Real-time PCR is more commonly known as quantitative polymerase chain reaction, qPCR.
Currently, qPCR is the most widely used technique for identification and quantification of nucleic acids. This technique has had broad use in scientific analyses in food, agricultural, medical, and environmental fields. Multiplex qPCR, which involves the tracking of multiple genotypes within a sample, is also possible, as shown by Alia et al. (2020) who studied four major foodborne pathogen genotypes from meat processing plants using multiplex qPCR [37].
With various applications for qPCR, direct comparison of qPCR performance remains difficult due to variations in the initial template amount, partitioning of sample, and sample preparation methods. Still, the downfall of the qPCR method is that it is susceptible to inhibitors from environmental contaminants that can arise from the extraction of nucleic acids and reaction mix compositions [38]. Such inhibitors can lead to inaccurate or overestimated results depending on the type of inhibitor, number of amplicons, or biological sample type [38,39,40]. In addition, all PCR application techniques are subject to poor primer/probe conditions requiring multiple rounds of probe and primer optimization.
2.3. The Next Generation—ddPCR
Droplet digital PCR (ddPCR) is the third-generation PCR technique that sequesters probes and nucleic acids within droplets in an oil emulsion [41]. ddPCR can generate up to 20,000 droplets in a single-tube reaction, with a singular PCR reaction occurring in each droplet. The number of droplets with (and without) amplification can be counted allowing for absolute quantification at a molecular level. ddPCR detects fluorescence at the endpoint of amplification, identifying the number of positive droplets and determining total concentration by Poisson distribution. Examining the number of negative and positive droplets together helps obtain the absolute copies per reaction [42].
While the concept of reading the fluorescent signals from amplified genes is the same compared to qPCR, ddPCR is better at preserving the initial reaction mix conditions and clearly representing amplification progress, minimizing the occurrence of primer-dimers, and false amplification reading. ddPCR widely applies to many fields due to its high specificity and lower limit of detection across many applications. It has been used for better detection of copy number variants (CNV) in samples compared to conventional PCR [43,44], gene expression of RNAs from viral targets and pathogens [45,46], and food research [47,48]. Some clinical study examples are on cancer and tuberculosis research [41], and other examples include product quality determination in the food industry such as olive oil [49], fish [50], and meat [51] products. Other studies compared the use of qPCR and ddPCR methods for detecting different targets such as meat products or Cryptosporidium and reported that ddPCR had a lower limit of quantification (LoQ) for determining meat product identification and better relative standard deviation (RSD%) to detect Cryptosporidium [52,53]. Moreover, ddPCR has been used to detect and quantify amounts of H7N9 influenza virus cDNA and was able to do so at a resolution of 3 to 5 copies/uL [54,55]. On the other hand, qPCR will detect fluorescent signals at the exponential phase of amplification [56], which does not easily allow for absolute quantification and such low limit of detection (LoD). For this reason, it is the most important benefit of ddPCR that it is free from factors such as requiring a standard for calibration or PCR efficiency, and the result is only dependent on the molecular interactions within the droplets [57].
Though the results of analysis for both qPCR and ddPCR are represented as copies per microliter (copies/volume), the output of ddPCR is different from the result produced from qPCR. An example is shown in Figure 2. The qPCR produces a real-time amplification curve in a 1D plot for each polymerase amplification reaction (Figure 2A, adapted from another study) showing relative fluorescence, which also can be represented as the Ct value, of different samples. On the other hand, ddPCR measures fluorescence signals at post amplification from each droplet, which captures individual target genes and can be presented in 1D or 2D plots. This shows groups of similar amplification, which makes it easier to detect negative and positive populations of droplets. Figure 2B shows a 1D plot of ddPCR with the automated threshold line shown in pink color by the ddPCR Quantasoft software (adapted from Ibekwe M.A. 2020 [58]). The clear numeration of negative and positive, as mentioned earlier, is key factor for absolute quantification by Poisson distribution.

Figure 2.
Example of qPCR (A) and ddPCR (B) output showing differences in amplification detection analysis.
2.3.1. The ddPCR Specificity
Furthermore, for rare targets, a single ddPCR sample can be subdivided across a 96-well plate, allowing for the detection of a single target among 2 million [59]. One application of ddPCR is single-cell ddPCR (sc-ddPCR), where it is possible to detect a single nucleotide polymorphism (SNP) in an enclosed single cell [60]. The sc-ddPCR saves time and costs of the assay for detection of SNPs in mitochondrial DNA because researchers can skip cloning procedures, which takes 2–3 days to complete. The procedure of sc-ddPCR is similar to performing conventional PCR reactions, which takes a few hours. The high-throughput ddPCR has been used in parallel comparisons with PCR for testing efficiencies of systematic evolution of ligands by exponential enrichment (SELEX) [61]. In this study, PCR showed a bias for C/T-rich nucleotide regions when forming initial RNA libraries. More detailed comparison studies are needed because both PCR and ddPCR showed amplification bias in forming initial RNA libraries. In addition, researchers should pay close attention to the details for such verification because the sequence and structure of a target gene can influence PCR amplification efficiency.
2.3.2. Multiplex ddPCR
Multiplexing (targeting multiple loci within a single reaction) is another consideration for accurate and multi-target detection. Multiplex qPCR was verified to provide 100% efficiency and an R2 value of 0.99 for parasite detection, such as Giardia and Cryptosporidium [62], and between 92.5% to 105.8% with an R2 value of >0.98 in detecting Listeria monocytogenes serotype in ready-to-eat meat products [37]. Multiplex qPCR has been mainly used to identify different species within sample, with a benefit of enabling detection as low as one organism/40 mL of soil in the case of sand flies [63]. On the other hand, use of multiplex ddPCR benefits assays of atypical transcripts such as detecting the presence of the fusion transcript BCR-ABL1, a marker for chronic myeloid leukemia [64]. The utilization of ddPCR in multiplexing is important because such mutation and variance is hard to detect using multiplex qPCR, which is capable of targeting up to four different targets within a single reaction [46]. Another study tracked epigenetic subtype markers using methylation-specific multiplex ddPCR [65]. Application of ddPCR to multiplexing has enabled detecting methylation site variations of the target gene. The results described above suggest greater potential of multiplex ddPCR over qPCR for application use.
This paper explores and compares the two PCR application methods, qPCR and ddPCR, to investigate the findings in the suitability of methods to target ARGs, including specificity, benefits, and downfalls of each method from the literature. The purpose of this study is to discover suitable conditions to best utilize both qPCR and ddPCR, in an effort to reduce erroneous research results and to suggest optimized procedures for different sample types. Our primary literature search criteria involve the keywords such as ddPCR, qPCR, resistance/antimicrobial, comparison, etc., but careful review criteria used in comparison of qPCR and ddPCR techniques are as follows: (1) applications, (2) limit of detection and sensitivity, (3) reproducibility, (4) measurement variance, (5) cost, (6) biases, and (7) ARG detection.
3. Comparison of qPCR and ddPCR Methods and Their Applications
With qPCR as a conventional method and ddPCR as the emerging technique, findings in comparison of both methods in different types of studies are collected in Table 1. Most studies were done in human diseases such as HIV and viral infections as well as areas related to bacteria, plants, and food. More of the studies reported that the LoD and sensitivity are better performed in ddPCR compared to qPCR, and accuracy was up to 20 times higher at a lower limit of detection. qPCR was performed at wider ranges of gene concentrations, especially at higher concentration ranges. Some ddPCR detections were more precise and better at LoD but could not cover the broad LoQ.
There are some controversies in the use of ddPCR for genetic analysis and quantification since it is shown that ddPCR sensitivity can vary depending on the sequence of a target material, and the quantification range does not surpass the performance level of qPCR. In HIV-1 and human DNA detection studies, both ddPCR and qPCR performed similarly for the lower limit of detection on target genes 50% of the time [66]. A study done with norovirus reported that there is no advantage of choosing either qPCR or ddPCR methods if research focuses on a lower detection limit [67]. Some research showed that ddPCR may have better LoD but has a limitation in detecting a high number of target genes, and both qPCR and ddPCR showed a similar coefficient of determination of standards in the gyrB gene [68]. Other research with bacterial genetic markers stated that while ddPCR shows better reproducibility for marker detection in fecal composites, qPCR shows a higher sensitivity for markers with environmental and composite samples with less than 10% sensitivity difference [69]. Nevertheless, both methods rely on target amplification, so certain genetic targets may work better in one or the other technology, and marker optimization is critical to both technologies.
Under the presence of inhibitors, the performance of reverse transcriptase ddPCR in detection of pepper mild mottle virus (PMMoV) was better than that of qPCR under complex matrices of seeds, plants, soil, and wastewater, as reported by Racki N. et al. (2014) [70]. Another study also supported this finding, that ddPCR is less sensitive to inhibitors compared to qPCR in viral detection [71]. In addition, it was suggested that the multiplexing capability of ddPCR technology is helpful for library preparation and next-generation sequencing applications to accommodate a large number of samples, while showing robustness on inhibitors [72].
Research cases mentioned above imply qPCR and ddPCR performances change depending on the type, structure, and initial concentration of samples. One study presented a challenge in ddPCR analysis; it is hard to find validation of the trueness of the sample reading since it is difficult to find representative samples with reference values [73]. Depending on the ddPCR setup, the reporting value may change drastically, and it is hard to compare between studies. Therefore, careful comparison studies focusing on performances of the two qPCR and ddPCR methods need to be explored in varying areas of study.

Table 1.
Findings in ddPCR and qPCR analysis in LoD, reproducibility, and range of detection.
Table 1.
Findings in ddPCR and qPCR analysis in LoD, reproducibility, and range of detection.
| Author (Year) | Gene | Type | LOD | LOQ & Range | Reproducibility | |||
|---|---|---|---|---|---|---|---|---|
| qPCR | ddPCR | qPCR | ddPCR | qPCR | ddPCR | |||
| Laura Cavé et al. (2016) [74] # | sul1, qnrB | ARG | +, 10-fold | + | - | |||
| Cesare, Andrea Di et al. (2018) [75] # | sul2, Intl1 | ARG | - | + | ||||
| Ginn O. et al. (2021) [76] | tetA, qnrB, blaTEM, intl1 | ARG | N/A | + | * | |||
| Kimbell L. et al. (2021) [77] # | blaTEM, blaSHV, sul1, czcD, copA, intl1 | ARG | - | + | * | * | ||
| Sun Y. e al. (2021) [78] | quinolones, tetracyclines, sulfonamides, macrolides | ARG | N/A | * | ||||
| Srisutham S. et al. (2021) [79] | pfmdr1, pfplasmepsin2, pfgch1 | ARG | N/A | * | * | |||
| Xu J. et al. (2021) [80] | mcr-1, blaCTX-M-14, bla CTX-M-55 | ARG | - | + | ||||
| Yang et al. (2014) [53] | Cryptosporidium Oocysts, 18S rRNA | Parasite | N/A | N/A | + | + | ||
| Weerakoon K.G. et al. (2016) [81] # | S. japoricum, SjR2 and nad1 | Parasite | - | 0.05fg, + | - | |||
| Overall | ARG & Parasite | - | + | + | - | * | * | |
| Henrich T.J. et al. (2012) [66] # | HIV-1, human CCR5 DNA | Human Diseases | + | +, HIV-1 | ||||
| Heredia N.J. et al. (2013) [82] | HER2 (= erbB2), CEP17 | Human Diseases | N/A | + | + | |||
| Strain M.C. et al. (2013) [83] # | HIV, episomal 2-LTR | Human Diseases | + | + | - | + | ||
| Bharuthram A. (2014) [84] # | CCL4L, CCL4L1 and CCL4L2 encodes HIV-1 | Human Diseases | - | + | - | + | ||
| Jones M. et al. (2014) [85] | HIV-1 from 8E5/LAV cells | Human Diseases | - | + | + | -, lower target | + | |
| Coudray-Meunier et al. (2015) [86] # | Hepatitis A, Norovirus | Human Diseases | * | * | - | + | - | + |
| Taylor S.C. et al. (2015) [46] # | H275-WT and H275Y-MUT of H1N1 | Human Diseases | +, mutant | + | - | + | ||
| Yan Y. et al. (2016) [87] | H7N9 | Human Diseases | N/A | - | ||||
| Yang Q (2017) [88] # | PRRSV | Human Diseases | - | + | +, false positive | - | ||
| Link-Lenczowska D. et al. (2018) [89]# | JAK2 mutation on V617F | Human Diseases | 0.12% | 0.01%, + | * | * | + | |
| Persson S. et al. (2018) [67] # | norovirus GI (GI.4) and GII (GII.4) | Human Diseases | * | * | + | |||
| Pinheiro T.F. et al. (2018) [45] | foot-and-mouth disease virus RNA | Human Diseases | - | + | ||||
| Baume M. et al. (2019) [90] # | Legionella DNA reference material | Human Disease | + | * | * | |||
| Zhang Y. et al. (2019) [91] # | PCV3 | Human Diseases | - | + | - | + | ||
| Dong L. et al. (2020) [92] | Tumor DNA reference material, BRAF V600E | Human Diseases | N/A | 0.02% | 0.10% | + | ||
| Lin Q. et al. (2020) [50] # | ISKNV | Human Diseases | - | + | - | +, low | ||
| Petiti J. et al. (2020) [64] | BCR-ABL1 disease marker leukemia | Human Diseases | N/A | 0.001% | ||||
| Thwin KKM et al. (2020) [93] # | NB-mRNAs (CRMP1, DBH, DDC, GAP43, ISL1, PHOX2B, and TH mRNA) | Human Diseases | - | + | - | + | ||
| Overall | Human Disease | - | + | * | * | - | + | |
| Milbury C.A. et al. (2014) [94] | EGFR T790M, L858R | Mutation | + | |||||
| Zhao Y. (2019) [95] | MTRNR1-WT | Mutation | N/A | + | + | - | + | |
| Liu Q. et al. (2020) [44] # | CNVs causing somatic mosaicism | Mutation | + | - | * | * | N/A | N/A |
| Overall | Mutation | * | * | + | - | + | ||
| Burns et al. (2010) [96] # | ERM-AD413 carries Mon810 | Plant, Food | - | + | -, lower range | |||
| Coudray-Meunier et al. (2015) [86] # | Hepatitis A, Norovirus | Plant, Food | - | + | + | -, bias | ||
| Porcellato D. et al. (2016) [68] # | gyrB of B. cereus group | Plant, Food | - | + | + | - | * | * |
| Scollo F. et al. (2016) [49] # | 11C Chloroplast locus | Plant, Food | - | + | ||||
| Wang X. et al. (2019) [47] # | transgenic rice line TT51-1 | Plant, Food | - | + | ||||
| Demeke et al. (2020) [97] # | Canola and soybean | Plant, Food | * | * | * | * | + | |
| Overall | Plant & Food | - | + | * | * | + | * | |
| Pinheiro L.B. et al. (2012) [98] | Lambda DNA | Bacteria, Phage | + | + | ||||
| Xi Z. (2018) [48] # | 16S rRNA of Las | Bacteria, Phage | - | + | - | + | ||
| Sivagnesan et al. (2018) [99] | Std1_Xhol insert with M13 E coli plasmid DNA | Bacteria, Phage | - | + | * | * | ||
| Furuta-Hanawa B. et al. (2019) [100] # | rAAV2RSM, rAAV8RSM | Bacteria, Phage | * | * | - | + | - | + |
| Nshimyimana J.P. et al. (2019) [69] # | Bacteroidales, BacHum and B. theta | Bacteria, Phage | +, Environmental | +, sensitivity | +, fecal | |||
| Raurich et al. (2019) [101] | Bifidobacterium animalis (BAN) | Bacteria, Phage | + | * | + | - | ||
| Ahn Y. et al. (2020) [102] # | Burkholderia epacian | Bacteria, Phage | - | + | -, recovery | +, recovery | - | + |
| Ibekwe M.A. et al. (2020) [58] # | Shiga toxin-producing E. coli O157:H7 | Bacteria, Phage | + | * | * | - | + | |
| Voegel T.M. et al. (2020) [103] # | amoA, nirS, nirK, nosZI, nosZII | Bacteria, Phage | - | + | + | - | ||
| Overall | Bacteria & Phage | - | + | * | * | - | + | |
(+) represents better performance compared to the other method from the paper. (-) represents worse performance compared to the other method from the paper. The * represents similar efficiency and performance regarding criteria. The # represents literature that made direct comparison analyses of qPCR and ddPCR. Detailed comparison results are summarized and provided in Supplementary Information (SI) Excel spreadsheet. (Tables S1–S5 include Supplementary Tables S1–S5 for each type of target gene, and Table S6 includes a heatmap of all literature compared in this table).
4. ddPCR as the Future
ddPCR is reported to show a lower detection limit with 1 log unit better in LoD than qPCR [48,49]; a greater number of samples tested positive [91], and it has better sensitivity compared to qPCR when it is optimized [46,48,74,91,101,104]. For this reason, ddPCR has the potential to be applied to a broader and more generalized field, which is supported by a review paper [56]. Therefore, depending on its use, ddPCR can be applied to many studies improving the sensitivity of the measurement, determining fine variances between sample sequences, or analyzing samples with low initial DNA concentration. The unknown outcomes and variances of ddPCR efficiency depending on sample types, DNA/RNA concentrations, gene structures, and assay methods can be defined as the “case specificity” of ddPCR. While having the merit of broad applicability for PCR, ddPCR needs more fine-tuning than qPCR to find the true performances matched to each target analysis. Collective knowledge from more research is needed to discover the advantages and unknown characteristics of ddPCR technology.
ddPCR is a promising method with better performance in detecting transgenic components due to its stability, accuracy, and resistance to PCR inhibitors, and there is no need for reference materials [47]. If studied in careful experimental design and comparison analyses, there will be numerous potentials to be used in the future. The current global situation of the pandemic on viral pathogens and the increasing amount of ARGs promote the need for a faster, more efficient, and more precise analysis method for genetic materials. The occurrence of pathogens in the environment needs to be tested and studied in timely manner with continuous mutation/evolution and adaptation of microbes to the ever-changing environment. Careful implementation of new technologies such as ddPCR may bring benefits with its properties of absolute quantification and sensitivity.
5. Current Findings on ddPCR Analysis on Genetic Targets Compared with Other Methods
5.1. Sensitivity
To our knowledge, general reporting on sensitivity in studies using qPCR and ddPCR methods is that the lower limit of detection of ddPCR is 10 times more effective compared to qPCR [42,46,48,50,74,91,101,104]. The target genes of such studies vary from bacterial 16S RNA gene and ARGs to HIV and viral genes. Due to the diversity of target genes, the sensitivity performance resulted in a broad spectrum in quantification and detection limits. Therefore, it is asserted that optimization of the target gene will enhance detection at a lower level of genetic materials, such as viral genes that cause disease [66]. This means the optimization process of primer/probes and concentration of reagents are required for increased sensitivity, especially for ddPCR. Depending on the target of interest for study, one must optimize testing conditions to fairly compare and assess quantification capabilities of qPCR and ddPCR.
Particularly, higher accessibility and amplifiability govern ddPCR results. The use of restriction enzymes can increase accessibility for PCR amplification and detection, while fragmentation of DNA can decrease positive readings in ddPCR, although such fragmentation can reduce rain effects, where the droplet readings are located in between positives and negatives. The smaller the fragments, the greater the reduction in positive readings [105]. This implies that assay characteristics, target gene size, and GC contents are important determinants for reproducibility in quantitative analyses.
During their case study using ddPCR and comparing its performance to qPCR, Guitierrez et al. mentioned that sensitivity can vary depending on the assay characteristics, presence of inhibitors, SNPs in the probe annealing region, etc. [72]. It is plausible that these factors will affect the testing lower limit range of genes, as increased sensitivity needs improvement in marginal errors. Additionally, Quan et al. (2018) mentioned that sensitivity or lower LoD is governed by partitioned volume, the standard deviation of the volume, and partitions of the reaction mix [56]. This is because a single nucleic acid in a partition will be detected in the lowest amount, which can be quantified as the lower limit. For this reason, the study mentioned that ddPCR can reach higher sensitivity if the reaction volume can be adjusted [56].
It was also stated that ddPCR detection may be troublesome when genes have high GC contents [44]. This is due to the nature of ddPCR analysis methods of encapsulating genetic materials within oil droplets; genes with higher GC content tend to have a hydrophobic site, staying intact within hydrophobic oil droplets can be unstable because the oil and higher GC region repels each other and carries the risk of incomplete encapsulation throughout amplification cycles. One study on the detection of Leishmania infection using ddPCR found that ddPCR may not be suitable for the assay, as the target sequence has more than 50% higher GC content [106]. A review study on the diagnosis of leishmaniasis reported that qPCR reached a lower detection limit of 10 pg, equivalent to ~120 parasites, in insect vectors, which verifies the method is precise in quantification and identification of species [107]. Noticing the differences in methods, curated planning for more effective evaluation and verification is recommended in studies handled with ddPCR and/or qPCR analysis. Each method has its differences in strength and weakness, and proper application of these methods will bring significant developments in the sensitivity of targeting genetic matter and lessen the noise of erroneous results. Importantly, it brings to our attention that assay-specific performances can differ depending on reaction volume, reaction mixes, and target nucleotide content.
5.2. Dynamic Range of Detection and Measurement Variance
In most cases, qPCR has a broader dynamic range of quantification, especially higher than 4 or 5 log units of concentration, compared to ddPCR [83,88]. The degree of dynamic range seems to change depending on the types of the gene and analysis settings [68,88,98]. This may be coming from the number of partitions that ddPCR can make, which is up to 20,000 droplets in the case of ddPCR. When the DNA concentration increases more than the number of partitions it can be in, there is no possible number of negative droplets to conduct the Poisson distribution analysis. Then, the analysis may be no longer valid. Theoretically, based on 95% confidence intervals, the partitioning error significantly increases when the average number of targets per partition, λ, is outside of range of 0.001 to 65.38, where λ equals the sample concentration (C) multiplied by partition volume (n) [108]. To prevent this overpopulation of positive over negative droplets, researchers need to dilute the concentrated samples before processing by measuring the DNA/RNA contents to make sure the experimental plan is suitable for ddPCR application.
While the concentration of the target gene is important for absolute quantification based on Poisson distribution calculations, % coefficient of variance (%CV), a calculated parameter of standard deviation/average*100, may vary depending on the dilution factor. From ddPCR and qPCR assessments, the %CV of the same dilution series ranges from 0 to 8.26, and 1.45 to 12.37 for qPCR and ddPCR, respectively [104]. Such variances may be coming from inaccurate dilution of residual protein and chemical contaminants [104]. Digital PCR (dPCR) is regarded as a technique that will have less variance compared to qPCR if it is free of any upstream errors derived from sampling and extraction processes [57]. With the development of technology, having a higher number of partitions of ddPCR will minimize partitioning errors, and much improvement in precision of detection can be expected.
Multiple studies proved that qPCR can be applied to various types of genes with a broader range of quantification. The qPCR performance is quite competitive compared to ddPCR. For example, the range of quantification was reported to be similar or better for qPCR depending on samples, where Cave et al. (2016) reported a broader quantification range compared to ddPCR [74]. Zhang Y. et al. (2019) reported that ddPCR and qPCR readings were highly correlated, by 95% [91].
In the case of the lower limit of detection, ddPCR seems to perform either similar or better with detection. Both ddPCR and qPCR showed 95% similarity in finding the LoD on a norovirus gene [67] and showed similar detection capabilities on genetically modified canola and soybean genes [97]. Depending on the gene types, ddPCR can detect down to 1 log unit better compared to qPCR [48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,73,74]. To our best knowledge, studies with qPCR showed LoD and LoQ as low as 2 log units and 3 log units, respectively [48,101]. For overall ranges of concentration where both methods can be used, ddPCR shows a positive correlation to qPCR results at an R-value around 0.85 [109] and shows high linearity [91].
5.3. Reproducibility
Reproducibility of an analysis method is important, especially when assaying samples that have low concentrations or are highly unstable, such as RNA. It is particularly important to reduce human error that could result from pipetting, cross-contamination, RNA degradation, or other factors. Establishing an internationally recognized reference system for achieving consistent results between laboratories is critical to ensure that analyses are comparable and informative [105]. To develop a good practice, consistent DNA or RNA extraction methods are critical to obtaining trustworthy and reproducible data [49]. In addition, the importance of quality and purity control of the target genetic material is stressed enough for testing reproducibility [97].
The ddPCR platform provides automated robotic droplet generation and sample mix preparation that can filter out human biases. In a study using QX200 automated ddPCR for developing miRNA markers, authors were able to determine the miRNA copy number repeatedly (p < 0.05) [32]. If there are issues observed with the expected reproducible rate, one may check if the target genetic concentration is at a proper range, as described in the previous section of measurement variance, because a too high or low partitioning may increase errors in measurements. Additionally, proper optimization steps for each method are essential because an approach to detect the same target sample showed varying results without optimization [99].
Overall, both ddPCR and qPCR measurements have been shown to be highly correlated in many different studies, consistent with high experimental reproducibility and repeatability for both [110]. In one study, ddPCR measurements showed better reproducibility for quantification in fecal composites, while qPCR showed a higher reproducibility for environmental and composite samples [69]. In another study, ddPCR achieved a higher reproducibility for specific species such as P. falciparum, while it showed similar sensitivity to qPCR in P. vivax [111]. Although ddPCR appears to be slightly more reproducible [104], and the automated platform can reduce human error, the improvement in reproducibility does not appear to be significant.
5.4. Cost
The cost-effectiveness of ddPCR is one of its main concerns [41,69]. It is reported that ddPCR costs two times as much as qPCR [53], and its lesser availability has prevented distribution of ddPCR technology in developing countries since its introduction in 2011 [41]. In addition, despite Sanger sequencing and ddPCR having their own benefits and irreplaceability in microbiological analyses, when compared, ddPCR is capable of detecting low-frequency mutations better than Sanger sequencing, while the cost to detect mutations was higher in ddPCR [95]. For microbiological applications that require direct knowledge of the DNA sequence of the microbe that extends beyond the locus, assaying unknown mutants, or developing primers and probes for detection, Sanger or high-throughput sequencing technologies cannot be replaced. However, if the mutant is known, ddPCR or qPCR can be a cost-effective method for microbiological identification.
In the case of multiplex ddPCR, the cost differential becomes less pronounced. Multiplex ddPCR allows for multiple assays within a single assay (at least four) and does not need standards, while qPCR will require multiple reactions with standards [89]. The ability to target multiple samples concurrently can level out the higher cost of the ddPCR reaction. High-throughput ddPCR with automated analysis settings also reduces the processing, labor time, and cost for analysis [60]. Development of single-use, low-cost injection molds for ddPCR have begun to be put into use in field applications [65]. Over time, there is a possibility that cost differences between qPCR and ddPCR methods may be decreased with improvements in technology in reagents and consumables production, increased availability of primers and probes, multiplexing, and price balancing by competition within the markets.
5.5. Risk of Bias in qPCR and ddPCR
Although the high correlation and linearity between qPCR and ddPCR methods are true [64,91], there is detection bias in the qPCR method, which stems from its analysis method. The qPCR technique requires standards developed before sample analysis, which makes the assay very dependent on the reading of the standards. One study on bacterial plasmids in feces reported that quantification errors in the concentration of original standards may lead to reading biases in qPCR [99]. In addition, qPCR tends to be more affected by contaminants such as SDS and heparin compared to ddPCR [45]. It is plausible because ddPCR separates nucleic acids into single droplets, which prevents the reaction from being interrupted by contaminants that may be captured in a different droplet or in the oil emulsion. The absolute quantification of genes makes ddPCR more independent of systematic errors of standard curves, unlike qPCR [69]. Therefore, quantification methods using qPCR will need well-developed standard and analysis methods put in place for justifiable analysis and comparison between samples. When the standard and/or sample purity is compromised, test results can be biased even when other types of optimization factors such as primer design, temperatures, and reaction volume are considered.
The ddPCR is not immune to the risks of bias. As discussed in the sensitivity section above, one should consider finding the right types of sample genes with GC contents less than 50%, understanding the physico-chemical nature of ddPCR reactions, and adapting the known information of reaction specificity of primers to genes, temperature cycles, and the ratio of positive/negative counts for fine outcomes. If possible, testing with known positive standards with ddPCR may be necessary to develop a robust analysis on specific target genes. While knowing different types of genes may have different ranges of concentration to suit ddPCR analysis, researchers can pinpoint experimental conditions and record optimized settings for the prosperity of future research.
5.6. Applicability
While ddPCR is used in more and more gene expression analysis, still, qPCR has been the main method used in the field. Transitioning to different technology may require standardization and verification efforts, and this would probably be the cause for qPCR technology being mainstream in analysis, despite ddPCR offering higher sensitivity and better view of gene amplification environments. Application of ddPCR in gene expression analysis was explored in cases of clinical study and detection of targets of low abundance [82,104]. It would be best used in situations where there are no well-developed positive standards available, or the target gene is present in low concentrations, or when detection attempts using qPCR are not successful. Since ddPCR does not require standards for each run, it is suitable for analysis of many samples of low abundance with fewer preparation steps, which facilitate efficiency and productivity [63].
6. Current Status in ARG Detection Methods with ddPCR
To our best knowledge, as reviewed in this paper, there are only a few studies that have analyzed ARG using ddPCR and qPCR methods in comparison. More comparisons of detection methods on ARGs are needed for precise quantification, better understanding of ARG, and optimized conditions for target genes. The conventional methods used for quantitative measurement of ARG in the environment are the following: (1) PCR, (2) real-time PCR (rtPCR) or quantitative PCR (qPCR) for genetic target materials, (3) ddPCR, (4) flow cytometry methods, and (5) cell counting for in vitro cultures of bacteria samples [53]. Out of these methods, ddPCR is the only method that does not need positive controls for quantification, and it is free of bias originating from the quality of the positive control.
By far, most ARG analyses are done with qPCR, and are coupled with sequencing methods such as Illumina, to comprehensively investigate the relationship of the microbiome of targeted environments to the spread of ARG. Some research has been done on the impact of nanoparticles on ARGs and surrounding microbial communities in the estuary [112], analysis of metagenomic correlation of AR in anaerobic digestors [113], and ARGs in bioaerosols of municipal sewage [114]. One study applied ddPCR and 16S rRNA sequencing to track ARGs, MGEs, and bacterial compositions in the air of composting plants. The reported detection of ARGs ranged from 1 to 7 log units [23]; within this range, a lower LoD is hard to find if qPCR is used.
Many researchers have been searching for a better method for precise detection and efficiency in such analyses, but each detection method has different limitations. There is a need for more studies to be done on absolute quantification of ARG using droplet digital PCR (ddPCR) and comparison of techniques on differences in specificity for optimization, the suitability of measurement, and the possibility of usage in broader fields. If optimized appropriately, ddPCR has the potential to be applied in many different fields with greater efficiency and sensitivity.
7. Conclusions and Perspectives
The consensus observed from the literature on the use of qPCR and ddPCR technologies is that both methods have great potential for multiple applications. qPCR’s strength lies in its broader detection range of genetic materials, lower upfront costs, and shows specificity to some target genes over ddPCR. The ddPCR method, on the other hand, shows an enhanced lower limit of detection for many different sample types, and it is a powerful tool for other biological assays such as mutation tracking. While both methods require optimization steps, ddPCR requires more optimization to develop a robust analysis.
Transferring the exact qPCR primers to ddPCR may be possible without losing amplification efficiency if the general requirements of primer design criteria are met, such as length of target gene, GC contents of binding sites, and avoidance of secondary structures. Additional considerations recommended for ddPCR reaction by the industry include avoiding repeats of G/C longer than three bases, addition of GC repeats at the 3’ end of primers, and design of 50–60% of GC content in the target region [42]. Experimental conditions such as concentrations of target gene, primers, and probes (if necessary) may need to be changed depending on optimization process of detection. If the result of ddPCR is not as expected, the reading may show the potential presence of impurities in the reaction mix, or that the detection setting is not optimized.
The detection and quantification of ARGs require more studies on different types of ARGs to find the suitability of each method on specific types and to determine the optimal settings for a reaction (e.g., DNA concentration, GC contents, reaction volumes, partitioned number of reactions, and suitability of primers/probes to the analysis). Although there are only a few studies that have employed ddPCR to detect ARGs, ddPCR shows similar or better LoD and sensitivity predictions when compared to qPCR (Table 1). ddPCR excelled in detection for human diseases and viral genes, and given time, we expect ddPCR could be a more effective solution for ARG tracing and quantification in the environment.
Recent studies have shown increasing accumulation of ARGs in municipal wastewater treatment plant effluent [115] (3–5 unit copies/mL) as well as in the air [23]; thus, better surveillance and treatment methods are needed. To track ARGs’ spread, peak, and attenuation, the technical power to detect lower gene concentrations is important. Clinical and environmental samples are expected to be found at lower concentrations when compared with cultured samples. Therefore, the ddPCR gene quantification method can be more suitable in the following situations: (1) when more precise detection is required, such as genetic mutation detection; (2) for new types of strains or genes that do not have reliable positive controls; and (3) limited amounts of genetic material can be extracted from collected sample.
However, researchers who choose to use the ddPCR technique must not ignore that ddPCR can have varying results, which can be influenced by the experimental setup and process, such as annealing temperature and threshold [116]. Therefore, the optimization step is necessary, and optimization settings should be adjusted to a level that is most suitable for the characteristic of the target gene. Without this optimization, ddPCR will not outperform qPCR.
Despite its merits, the use of ddPCR still holds some disadvantages, such as higher operation cost, reagent costs, and availability, when compared to conventional methods [44,95]. Despite these limitations, the high accuracy and resolution of ddPCR has led it to be widely used in food sciences [31,45,47,48,49,51,52,67,68,91,116,117] and disease and evolution studies [23,46,53,66,69,73,85,90,92,93,94,99,101,118,119,120,121,122,123]. These studies are continually developing the technology and creating an extensive collection and recording of the optimization process for different genes. In the future, with more and more usage, one can organize a database of ddPCR genes and their optimum conditions depending on primer sets, temperature, and probes. This information will help future researchers to reach the best output in a shorter timeframe and at a lower cost.
The global community has experienced a pandemic where a novel pathogen has threatened global health and the economy. Detecting, tracking, and forecasting these microbes and their genes in nature is very hard due to the different characteristics of species. Development of biotechnologies that enhance genetic detection methods with more precision and sensitivity is required. Studies show that with proper optimization steps and verification with positive and negative controls, ddPCR analysis will allow researchers to capture biological information that could have been missed in conventional methods. There is a strong case for more ddPCR application studies to be done with many other gene targets because these data will contribute to better usage of ddPCR technology and boost future work in this field.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/applmicrobiol1030028/s1, Table S1: List of literature on ARG and Parasite, Table S2: List of literature on Human Disease, Table S3: List of literature on Mutation, Table S4: List of literature on Plant and Food, Table S5: List of literature on Bacteria and Bacteriophage, Table S6: Heatmap-Option1.
Author Contributions
Conceptualization, S.P., W.S. and M.M.; methodology, S.P.; validation, S.P., A.R., W.S. and M.M.; formal analysis, S.P.; data curation and structure formatting, S.P., W.S. and M.M.; writing—original draft preparation, S.P.; writing—review and editing, S.P., A.R., W.S. and M.M.; visualization, S.P., A.R., W.S. and M.M.; supervision, W.S. and M.M.; funding acquisition, W.S. and M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Any further data related to this manuscript and Supplementary information are available to share upon request to the authors.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Knapp, C.; Dolfing, J.; Ehlert, P.A.I.; Graham, D.W. Evidence of Increasing Antibiotic Resistance Gene Abundances in Archived Soils since 1940. Environ. Sci. Technol. 2009, 44, 580–587. [Google Scholar] [CrossRef]
- Bergeron, S.; Brown, R.; Homer, J.; Rehage, S.; Boopathy, R. Presence of antibiotic resistance genes in different salinity gradients of freshwater to saltwater marshes in southeast Louisiana, USA. Int. Biodeterior. Biodegrad. 2016, 113, 80–87. [Google Scholar] [CrossRef]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Genet. 2010, 8, 251–259. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Huang, Y.-H.; Liu, Y.; Du, P.-P.; Zeng, L.-J.; Mo, C.-H.; Li, Y.-W.; Lü, H.; Cai, Q.-Y. Occurrence and distribution of antibiotics and antibiotic resistant genes in water and sediments of urban rivers with black-odor water in Guangzhou, South China. Sci. Total Environ. 2019, 670, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Di Cesare, A.; Eckert, E.; Teruggi, A.; Fontaneto, D.; Bertoni, R.; Callieri, C.; Corno, G. Constitutive presence of antibiotic resistance genes within the bacterial community of a large subalpine lake. Mol. Ecol. 2015, 24, 3888–3900. [Google Scholar] [CrossRef]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pruden, A.; Pei, R.; Storteboom, H.; Carlson, K.H. Antibiotic Resistance Genes as Emerging Contaminants: Studies in Northern Colorado. Environ. Sci. Technol. 2006, 40, 7445–7450. [Google Scholar] [CrossRef] [PubMed]
- Baquero, F.; Martinez, J.L.; Cantón, R. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 2008, 19, 260–265. [Google Scholar] [CrossRef]
- Morel, C.M.; Lindahl, O.; Harbarth, S.; De Kraker, M.E.A.; Edwards, S.; Hollis, A. Industry incentives and antibiotic resistance: An introduction to the antibiotic susceptibility bonus. J. Antibiot. 2020, 73, 421–428. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Antimicrobial Resistance: Global Report on Surveillance; WHO: Geneva, Switzerland, 2014. [Google Scholar]
- World Health Organization (WHO). Sanitation Safety Planning, Greywater and Excreta; WHO: Geneva, Switzerland, 2016. [Google Scholar]
- World Health Organization (WHO). No Time to Wait: Securing the Future from Drug-Resistant Infections; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Bhattacharyya, A.; Haldar, A.; Bhattacharyya, M.; Ghosh, A. Anthropogenic influence shapes the distribution of antibiotic resistant bacteria (ARB) in the sediment of Sundarban estuary in India. Sci. Total Environ. 2018, 647, 1626–1639. [Google Scholar] [CrossRef]
- Guo, X.-P.; Yang, Y.; Lu, D.-P.; Niu, Z.-S.; Feng, J.-N.; Chen, Y.-R.; Tou, F.-Y.; Garner, E.; Xu, J.; Liu, M.; et al. Biofilms as a sink for antibiotic resistance genes (ARGs) in the Yangtze Estuary. Water Res. 2018, 129, 277–286. [Google Scholar] [CrossRef]
- Reddy, B.; Dubey, S.K.; Reddy, B.; Dubey, S.K. River Ganges water as reservoir of microbes with antibiotic and metal ion resistance genes: High throughput metagenomic approach. Environ. Pollut. 2018, 246, 443–451. [Google Scholar] [CrossRef]
- Fiorentino, A.; De Luca, G.; Rizzo, L.; Viccione, G.; Lofrano, G.; Carotenuto, M. Simulating the fate of indigenous antibiotic resistant bacteria in a mild slope wastewater polluted stream. J. Environ. Sci. 2018, 69, 95–104. [Google Scholar] [CrossRef]
- Pruden, A.; Arabi, M.; Storteboom, H.N. Correlation between Upstream Human Activities and Riverine Antibiotic Resistance Genes. Environ. Sci. Technol. 2012, 46, 11541–11549. [Google Scholar] [CrossRef] [PubMed]
- Christou, A.; Agüera, A.; Bayona, J.M.; Cytryn, E.; Fotopoulos, V.; Lambropoulou, D.; Manaia, C.M.; Michael, C.; Revitt, M.; Schröder, P.; et al. The potential implications of reclaimed wastewater reuse for irrigation on the agricultural environment: The knowns and unknowns of the fate of antibiotics and antibiotic resistant bacteria and resistance genes—A review. Water Res. 2017, 123, 448–467. [Google Scholar] [CrossRef] [Green Version]
- Van Hoek, A.H.A.M.; Mevius, D.; Guerra, B.; Mullany, P.; Roberts, A.P.; Aarts, H.J.M. Acquired Antibiotic Resistance Genes: An Overview. Front. Microbiol. 2011, 2, 203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillings, M.R.; Gaze, W.H.; Pruden, A.; Smalla, K.; Tiedje, J.M.; Zhu, Y.-G. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2014, 9, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Stedtfeld, R.D.; Guo, X.; Bhalsod, G.D.; Jeon, S.; Tiedje, J.M.; Li, H.; Zhang, W. Pharmaceutical exposure changed antibiotic resistance genes and bacterial communities in soil-surface- and overhead-irrigated greenhouse lettuce. Environ. Int. 2019, 131, 105031. [Google Scholar] [CrossRef]
- Gao, M.; Qiu, T.; Sun, Y.; Wang, X. The abundance and diversity of antibiotic resistance genes in the atmospheric environment of composting plants. Environ. Int. 2018, 116, 229–238. [Google Scholar] [CrossRef]
- De Carvalho, C.C.C.R.; Da Fonseca, M.M.R. Biotransformations. In Reference Module in Life Sciences; Elsevier: Amsterdam, The Netherlands, 2017; pp. 574–585. [Google Scholar]
- Garibyan, L.; Avashia, N. Polymerase Chain Reaction. J. Investig. Dermatol. 2013, 133, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Clewley, J.P. The polymerae chain reaction, a review of the practical limitations for human immunodeficiency virus diagnosis. J. Virol. Methods 1989, 25, 179–188. [Google Scholar] [CrossRef]
- Bustin, S.; Huggett, J. qPCR primer design revisited. Biomol. Detect. Quantif. 2017, 14, 19–28. [Google Scholar] [CrossRef]
- Chandler, D.P.; Wagnon, C.A.; Bolton, H. Reverse Transcriptase (RT) Inhibition of PCR at Low Concentrations of Template and Its Implications for Quantitative RT-PCR. Appl. Environ. Microbiol. 1998, 64, 669–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romsos, E.L.; Vallone, P. Estimation of extraction efficiency by droplet digital PCR. Forensic Sci. Int. Genet. Suppl. Ser. 2019, 7, 515–517. [Google Scholar] [CrossRef]
- Perry, D.J.; Harper, P.L.; Fairham, S.; Daly, M.; Carrell, R.W. Antithrombin Cambridge, 384 Ala to Pro: A new variant identified using the polymerase chain reaction. FEBS Lett. 1989, 254, 174–176. [Google Scholar] [CrossRef] [Green Version]
- Cao, W.; Li, Y.; Chen, X.; Chang, Y.; Li, L.; Shi, L.; Bai, W.; Ye, L. Species identification and quantification of silver pomfret using the droplet digital PCR assay. Food Chem. 2019, 302, 125331. [Google Scholar] [CrossRef] [PubMed]
- Vishnuraj, M.; Devatkal, S.; Vaithiyanathan, S.; Kumar, R.U.; Srinivas, C.; Mendiratta, S. Detection of giblets in chicken meat products using microRNA markers and droplet digital PCR assay. LWT 2020, 140, 110798. [Google Scholar] [CrossRef]
- Ekman, S. Pcr Optimization and Troubleshooting, with Special Reference to the Amplification of Ribosomal DNA in Lichenized Fungi. Lichenologist 1999, 31, 517–531. [Google Scholar] [CrossRef]
- Butler, J.M.; Ruitberg, C.M.; Vallone, P. Capillary electrophoresis as a tool for optimization of multiplex PCR reactions. Anal. Bioanal. Chem. 2001, 369, 200–205. [Google Scholar] [CrossRef]
- Wong, W.H.; Tay, Y.C.; Puniamoorthy, J.; Balke, M.; Cranston, P.S.; Meier, R. ‘Direct PCR’ optimization yields a rapid, cost-effective, nondestructive and efficient method for obtaining DNA barcodes without DNA extraction. Mol. Ecol. Resour. 2014, 14, 1271–1280. [Google Scholar] [CrossRef]
- Ponchel, F.; Toomes, C.; Bransfield, K.; Leong, F.T.; Douglas, S.H.; Field, S.L.; Bell, S.M.; Combaret, V.; Puisieux, A.; Mighell, A.J.; et al. Real-time PCR based on SYBR-Green I fluorescence: An alternative to the TaqMan assay for a relative quantification of gene rearrangements, gene amplifications and micro gene deletions. BMC Biotechnol. 2003, 3, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alía, A.; Andrade, M.J.; Córdoba, J.J.; Martín, I.; Rodríguez, A. Development of a multiplex real-time PCR to differentiate the four major Listeria monocytogenes serotypes in isolates from meat processing plants. Food Microbiol. 2019, 87, 103367. [Google Scholar] [CrossRef]
- Huggett, J.F.; Novak, T.; Garson, J.; Green, C.; Morris-Jones, S.D.; Miller, R.F.; Zumla, A. Differential susceptibility of PCR reactions to inhibitors: An important and unrecognised phenomenon. BMC Res. Notes 2008, 1, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suslov, O. PCR inhibition by reverse transcriptase leads to an overestimation of amplification efficiency. Nucleic Acids Res. 2005, 33, e181. [Google Scholar] [CrossRef] [PubMed]
- Karlen, Y.; McNair, A.; Perseguers, S.; Mazza, C.; Mermod, N. Statistical significance of quantitative PCR. BMC Bioinform. 2007, 8, 131. [Google Scholar] [CrossRef] [Green Version]
- Nyaruaba, R.; Mwaliko, C.; Kering, K.K.; Wei, H. Droplet digital PCR applications in the tuberculosis world. Tuberculosis 2019, 117, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Biorad. Droplet Digital TM PCR Droplet Digital TM PCR Applications Guide; Biorad: Hercules, CA, USA, 2018; p. 145. [Google Scholar]
- Tone, M.; Torunn, K. Presence and Levels of Antibiotic Resistance Genes in Saliva from Dental Students in Tromsø. Master’s Thesis, The Arctic University of Norway, Tromsø, Norway, May 2016. [Google Scholar]
- Liu, Q.; Karolak, J.; Grochowski, C.M.; Wilson, T.A.; Rosenfeld, J.A.; Bacino, C.A.; Lalani, S.R.; Patel, A.; Breman, A.; Smith, J.L.; et al. Parental somatic mosaicism for CNV deletions—A need for more sensitive and precise detection methods in clinical diagnostics settings. Genomics 2020, 112, 2937–2941. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, T.; Fonseca, A.A.; Camargos, M.F.; Laguardia-Nascimento, M.; de Oliveira, A.M.; Cottorello, A.C.; Goes-Neto, A.; Barbosa-Stancioli, E.F. Development of a droplet digital RT-PCR for the quantification of foot-and-mouth virus RNA. J. Virol. Methods 2018, 259, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.C.; Carbonneau, J.; Shelton, D.N.; Boivin, G. Optimization of Droplet Digital PCR from RNA and DNA extracts with direct comparison to RT-qPCR: Clinical implications for quantification of Oseltamivir-resistant subpopulations. J. Virol. Methods 2015, 224, 58–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Tang, T.; Miao, Q.; Xie, S.; Chen, X.; Tang, J.; Peng, C.; Xu, X.; Wei, W.; You, Z.; et al. Detection of transgenic rice line TT51-1 in processed foods using conventional PCR, real-time PCR, and droplet digital PCR. Food Control. 2018, 98, 380–388. [Google Scholar] [CrossRef]
- Zhong, X.; Liu, X.-L.; Lou, B.-H.; Zhou, C.-Y.; Wang, X.-F. Development of a sensitive and reliable droplet digital PCR assay for the detection of ‘Candidatus Liberibacter asiaticus’. J. Integr. Agric. 2018, 17, 483–487. [Google Scholar] [CrossRef] [Green Version]
- Scollo, F.; Egea, L.A.; Gentile, A.; La Malfa, S.; Dorado, G.; Hernandez, P. Absolute quantification of olive oil DNA by droplet digital-PCR (ddPCR): Comparison of isolation and amplification methodologies. Food Chem. 2016, 213, 388–394. [Google Scholar] [CrossRef]
- Lin, Q.; Fu, X.; Liu, L.; Liang, H.; Niu, Y.; Wen, Y.; Huang, Z.; Li, N. Development and application of a sensitive droplet digital PCR (ddPCR) for the detection of infectious spleen and kidney necrosis virus. Aquaculture 2020, 529, 735697. [Google Scholar] [CrossRef]
- Naaum, A.M.; Shehata, H.R.; Chen, S.; Li, J.; Tabujara, N.; Awmack, D.; Lutze-Wallace, C.; Hanner, R. Complementary molecular methods detect undeclared species in sausage products at retail markets in Canada. Food Control 2018, 84, 339–344. [Google Scholar] [CrossRef]
- Floren, C.; Wiedemann, I.; Brenig, B.; Schütz, E.; Beck, J. Species identification and quantification in meat and meat products using droplet digital PCR (ddPCR). Food Chem. 2015, 173, 1054–1058. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.; Paparini, A.; Monis, P.; Ryan, U. Comparison of next-generation droplet digital PCR (ddPCR) with quantitative PCR (qPCR) for enumeration of Cryptosporidium oocysts in faecal samples. Int. J. Parasitol. 2014, 44, 1105–1113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahrberg, C.D.; Lee, J.M.; Chung, B.G. Microwell Array-based Digital PCR for Influenza Virus Detection. BioChip J. 2019, 13, 269–276. [Google Scholar] [CrossRef]
- Heyries, K.; Tropini, C.; VanInsberghe, M.; Doolin, C.; Petriv, O.; Singhal, A.; Leung, K.; Hughesman, C.B.; Hansen, C.L. Megapixel digital PCR. Nat. Methods 2011, 8, 649–651. [Google Scholar] [CrossRef] [PubMed]
- Quan, P.-L.; Sauzade, M.; Brouzes, E. dPCR: A Technology Review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef] [Green Version]
- Huggett, J.F.; Cowen, S.; Foy, C.A. Considerations for Digital PCR as an Accurate Molecular Diagnostic Tool. Clin. Chem. 2015, 61, 79–88. [Google Scholar] [CrossRef] [Green Version]
- Ibekwe, A.M.; Murinda, S.E.; Park, S.; Obayiuwana, A.; Murry, M.A.; Schwartz, G.; Lundquist, T. Comparative Use of Quantitative PCR (qPCR), Droplet Digital PCR (ddPCR), and Recombinase Polymerase Amplification (RPA) in the Detection of Shiga Toxin-Producing E. coli (STEC) in Environmental Samples. Water 2020, 12, 3507. [Google Scholar] [CrossRef]
- Hindson, B.J.; Ness, K.D.; Masquelier, N.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-Throughput Droplet Digital PCR System for Absolute Quantitation of DNA Copy Number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Maeda, R.; Kami, D.; Maeda, H.; Shikuma, A.; Gojo, S. High throughput single cell analysis of mitochondrial heteroplasmy in mitochondrial diseases. Sci. Rep. 2020, 10, 10821. [Google Scholar] [CrossRef]
- Takahashi, M.; Wu, X.; Ho, M.; Chomchan, P.; Rossi, J.J.; Burnett, J.C.; Zhou, J. High throughput sequencing analysis of RNA libraries reveals the influences of initial library and PCR methods on SELEX efficiency. Sci. Rep. 2016, 6, 33697. [Google Scholar] [CrossRef] [Green Version]
- Marangi, M.; Giangaspero, A.; Lacasella, V.; Lonigro, A.; Gasser, R.B. Multiplex PCR for the detection and quantification of zoonotic taxa of Giardia, Cryptosporidium and Toxoplasma in wastewater and mussels. Mol. Cell. Probes 2015, 29, 122–125. [Google Scholar] [CrossRef] [PubMed]
- Giantsis, I.; Chaskopoulou, A. Broadening the tools for studying sand fly breeding habitats: A novel molecular approach for the detection of phlebotomine larval DNA in soil substrates. Acta Trop. 2018, 190, 123–128. [Google Scholar] [CrossRef]
- Petiti, J.; Iacono, M.L.; Dragani, M.; Pironi, L.; Fantino, C.; Rapanotti, M.C.; Quarantelli, F.; Izzo, B.; Divona, M.; Rege-Cambrin, G.; et al. Novel Multiplex Droplet Digital PCR Assays to Monitor Minimal Residual Disease in Chronic Myeloid Leukemia Patients Showing Atypical BCR-ABL1 Transcripts. J. Clin. Med. 2020, 9, 1457. [Google Scholar] [CrossRef]
- Malic, L.; Daoud, J.; Geissler, M.; Boutin, A.; Lukic, L.; Janta, M.; Elmanzalawy, A.; Veres, T. Epigenetic subtyping of white blood cells using a thermoplastic elastomer-based microfluidic emulsification device for multiplexed, methylation-specific digital droplet PCR. Analyst 2019, 144, 6541–6553. [Google Scholar] [CrossRef]
- Henrich, T.J.; Gallien, S.; Li, J.Z.; Pereyra, F.; Kuritzkes, D.R. Low-level detection and quantitation of cellular HIV-1 DNA and 2-LTR circles using droplet digital PCR. J. Virol. Methods 2012, 186, 68–72. [Google Scholar] [CrossRef] [Green Version]
- Persson, S.; Eriksson, R.; Lowther, J.; Ellström, P.; Simonsson, M. Comparison between RT droplet digital PCR and RT real-time PCR for quantification of noroviruses in oysters. Int. J. Food Microbiol. 2018, 284, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Porcellato, D.; Narvhus, J.; Skeie, S.B. Detection and quantification of Bacillus cereus group in milk by droplet digital PCR. J. Microbiol. Methods 2016, 127, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Nshimyimana, J.P.; Cruz, M.C.; Wuertz, S.; Thompson, J.R. Variably improved microbial source tracking with digital droplet PCR. Water Res. 2019, 159, 192–202. [Google Scholar] [CrossRef]
- NejcRački, N.; Dreo, T.; Gutierrez-Aguirre, I.; Blejec, A.; Ravnikar, M. Reverse transcriptase droplet digital PCR shows high resilience to PCR inhibitors from plant, soil and water samples. Plant Methods 2014, 10, 1–10. [Google Scholar] [CrossRef]
- Rački, N.; Morisset, D.; Gutierrez-Aguirre, I.; Ravnikar, M. One-step RT-droplet digital PCR: A breakthrough in the quantification of waterborne RNA viruses. Anal. Bioanal. Chem. 2013, 406, 661–667. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez-Aguirre, I.; Rački, N.; Dreo, T.; Ravnikar, M. Droplet Digital PCR for Absolute Quantification of Pathogens. Methods Mol. Biol. 2015, 1302, 331–347. [Google Scholar] [CrossRef]
- Deprez, L.; Corbisier, P.; Kortekaas, A.-M.; Mazoua, S.; Hidalgo, R.B.; Trapmann, S.; Emons, H. Validation of a digital PCR method for quantification of DNA copy number concentrations by using a certified reference material. Biomol. Detect. Quantif. 2016, 9, 29–39. [Google Scholar] [CrossRef]
- Cavé, L.; Brothier, E.; Abrouk, D.; Bouda, P.S.; Hien, E.; Nazaret, S. Efficiency and sensitivity of the digital droplet PCR for the quantification of antibiotic resistance genes in soils and organic residues. Appl. Microbiol. Biotechnol. 2016, 100, 10597–10608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Cesare, A.; Petrin, S.; Fontaneto, D.; LoSasso, C.; Eckert, E.M.; Tassistro, G.; Borello, A.; Ricci, A.; Wilson, W.H.; Pruzzo, C.; et al. ddPCR applied on archived Continuous Plankton Recorder samples reveals long-term occurrence of class 1 integrons and a sulphonamide resistance gene in marine plankton communities. Environ. Microbiol. Rep. 2018, 10, 458–464. [Google Scholar] [CrossRef]
- Ginn, O.; Nichols, D.; Rocha-Melogno, L.; Bivins, A.; Berendes, D.; Soria, F.; Andrade, M.; Deshusses, M.A.; Bergin, M.; Brown, J. Antimicrobial resistance genes are enriched in aerosols near impacted urban surface waters in La Paz, Bolivia. Environ. Res. 2021, 194, 110730. [Google Scholar] [CrossRef]
- Kimbell, L.K.; LaMartina, E.L.; Kappell, A.D.; Huo, J.; Wang, Y.; Newton, R.J.; McNamara, P.J. Cast iron drinking water pipe biofilms support diverse microbial communities containing antibiotic resistance genes, metal resistance genes, and class 1 integrons. Environ. Sci. Water Res. Technol. 2021, 7, 584–598. [Google Scholar] [CrossRef]
- Sun, Y.; Guo, Y.; Shi, M.; Qiu, T.; Gao, M.; Tian, S.; Wang, X. Effect of antibiotic type and vegetable species on antibiotic accumulation in soil-vegetable system, soil microbiota, and resistance genes. Chemosphere 2020, 263, 128099. [Google Scholar] [CrossRef] [PubMed]
- Srisutham, S.; Suwannasin, K.; Sugaram, R.; Dondorp, A.M.; Imwong, M. Measurement of gene amplifications related to drug resistance in Plasmodium falciparum using droplet digital PCR. Malar. J. 2021, 20, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, N.; Luo, M.; Wang, M.; Wang, L.; Li, J.; Li, Z.; Zhao, H.; Li, Z.; Kan, B.; et al. Rapid Identification of Plasmid Replicon Type and Coexisting Plasmid-Borne Antimicrobial Resistance Genes by S1-Pulsed-Field Gel Electrophoresis-Droplet Digital Polymerase Chain Reaction. Foodborne Pathog. Dis. 2021, 18, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Weerakoon, K.G.; Gordon, C.; Gobert, G.; Cai, P.; McManus, D.P. Optimisation of a droplet digital PCR assay for the diagnosis of Schistosoma japonicum infection: A duplex approach with DNA binding dye chemistry. J. Microbiol. Methods 2016, 125, 19–27. [Google Scholar] [CrossRef] [Green Version]
- Heredia, N.J.; Belgrader, P.; Wang, S.; Koehler, R.; Regan, J.; Cosman, A.M.; Saxonov, S.; Hindson, B.; Tanner, S.C.; Brown, A.S.; et al. Droplet Digital™ PCR quantitation of HER2 expression in FFPE breast cancer samples. Methods 2012, 59, S20–S23. [Google Scholar] [CrossRef]
- Strain, M.C.; Lada, S.M.; Luong, T.; Rought, S.E.; Gianella, S.; Terry, V.H.; Spina, C.A.; Woelk, C.H.; Richman, D.D. Highly Precise Measurement of HIV DNA by Droplet Digital PCR. PLoS ONE 2013, 8, e55943. [Google Scholar] [CrossRef]
- Bharuthram, A.; Paximadis, M.; Picton, A.C.; Tiemessen, C. Comparison of a quantitative Real-Time PCR assay and droplet digital PCR for copy number analysis of the CCL4L genes. Infect. Genet. Evol. 2014, 25, 28–35. [Google Scholar] [CrossRef]
- Jones, M.; Williams, J.; Gärtner, K.; Phillips, R.; Hurst, J.; Frater, J. Low copy target detection by Droplet Digital PCR through application of a novel open access bioinformatic pipeline, ‘definetherain’. J. Virol. Methods 2014, 202, 46–53. [Google Scholar] [CrossRef] [Green Version]
- Coudray-Meunier, C.; Fraisse, A.; Martin-Latil, S.; Guillier, L.; Delannoy, S.; Fach, P.; Perelle, S. A comparative study of digital RT-PCR and RT-qPCR for quantification of Hepatitis A virus and Norovirus in lettuce and water samples. Int. J. Food Microbiol. 2015, 201, 17–26. [Google Scholar] [CrossRef]
- Yan, Y.; Jia, X.-J.; Wang, H.-H.; Fu, X.-F.; Ji, J.-M.; He, P.-Y.; Chen, L.-X.; Luo, J.-Y.; Chen, Z.-W. Dynamic quantification of avian influenza H7N9(A) virus in a human infection during clinical treatment using droplet digital PCR. J. Virol. Methods 2016, 234, 22–27. [Google Scholar] [CrossRef]
- Yang, Q.; Xi, J.; Chen, X.; Hu, S.; Chen, N.; Qiao, S.; Wan, S.; Bao, D. The development of a sensitive droplet digital PCR for quantitative detection of porcine reproductive and respiratory syndrome virus. Int. J. Biol. Macromol. 2017, 104, 1223–1228. [Google Scholar] [CrossRef]
- Link-Lenczowska, D.; Pallisgaard, N.; Cordua, S.; Zawada, M.; Czekalska, S.; Krochmalczyk, D.; Kanduła, Z.; Sacha, T. A comparison of qPCR and ddPCR used for quantification of the JAK2 V617F allele burden in Ph negative MPNs. Ann. Hematol. 2018, 97, 2299–2308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baume, M.; Cariou, A.; Leveau, A.; Fessy, N.; Pastori, F.; Jarraud, S.; Pierre, S. Quantification of Legionella DNA certified reference material by digital droplet PCR. J. Microbiol. Methods 2018, 157, 50–53. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Z.; Wang, Z.; Wang, Z.; Wang, C.; Feng, C.; Yuan, W.; Lin, X.; Wu, S. Development of a droplet digital PCR assay for sensitive detection of porcine circovirus 3. Mol. Cell. Probes 2018, 43, 50–57. [Google Scholar] [CrossRef]
- Dong, L.; Wang, X.; Wang, S.; Du, M.; Niu, C.; Yang, J.; Li, L.; Zhang, G.; Fu, B.; Gao, Y.; et al. Interlaboratory assessment of droplet digital PCR for quantification of BRAF V600E mutation using a novel DNA reference material. Talanta 2019, 207, 120293. [Google Scholar] [CrossRef] [PubMed]
- Thwin, K.K.; Ishida, T.; Uemura, S.; Yamamoto, N.; Lin, K.S.; Tamura, A.; Kozaki, A.; Saito, A.; Kishimoto, K.; Mori, T.; et al. Level of Seven Neuroblastoma-Associated mRNAs Detected by Droplet Digital PCR Is Associated with Tumor Relapse/Regrowth of High-Risk Neuroblastoma Patients. J. Mol. Diagn. 2020, 22, 236–246. [Google Scholar] [CrossRef] [Green Version]
- Milbury, C.A.; Zhong, Q.; Lin, J.; Williams, M.; Olson, J.; Link, D.R.; Hutchison, B. Determining lower limits of detection of digital PCR assays for cancer-related gene mutations. Biomol. Detect. Quantif. 2014, 1, 8–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y. A comparative study of ddPCR and sanger sequencing for quantitative detection of low-frequency mutation rate. IOP Conf. Ser. Earth Environ. Sci. 2019, 332, 032023. [Google Scholar] [CrossRef]
- Burns, M.J.; Burrell, A.M.; Foy, C.A. The applicability of digital PCR for the assessment of detection limits in GMO analysis. Eur. Food Res. Technol. 2010, 231, 353–362. [Google Scholar] [CrossRef]
- Demeke, T.; Beecher, B.; Eng, M. Assessment of genetically engineered events in heat-treated and non-treated samples using droplet digital PCR and real-time quantitative PCR. Food Control 2020, 115, 107291. [Google Scholar] [CrossRef]
- Pinheiro, L.B.; Coleman, V.A.; Hindson, C.M.; Herrmann, J.; Hindson, B.J.; Bhat, S.; Emslie, K.R. Evaluation of a Droplet Digital Polymerase Chain Reaction Format for DNA Copy Number Quantification. Anal. Chem. 2011, 84, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Sivaganesan, M.; Varma, M.; Siefring, S.; Haugland, R. Quantification of plasmid DNA standards for U.S. EPA fecal indicator bacteria qPCR methods by droplet digital PCR analysis. J. Microbiol. Methods 2018, 152, 135–142. [Google Scholar] [CrossRef]
- Furuta-Hanawa, B.; Yamaguchi, T.; Uchida, E. Two-Dimensional Droplet Digital PCR as a Tool for Titration and Integrity Evaluation of Recombinant Adeno-Associated Viral Vectors. Hum. Gene Ther. Methods 2019, 30, 127–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raurich, S.; Weber, B.; Klose, V.; Mohnl, M.; Petri, D.; Fibi-Smetana, S. Optimisation of a droplet digital PCR for strain specific quantification of a probiotic Bifidobacterium animalis strain in poultry feed. J. Microbiol. Methods 2019, 163, 105646. [Google Scholar] [CrossRef] [PubMed]
- Ahn, Y.; Gibson, B.; Williams, A.; Alusta, P.; Buzatu, D.A.; Lee, Y.-J.; LiPuma, J.J.; Hussong, D.; Marasa, B.; Cerniglia, C.E. A comparison of culture-based, real-time PCR, droplet digital PCR and flow cytometric methods for the detection of Burkholderia cepacia complex in nuclease-free water and antiseptics. J. Ind. Microbiol. Biotechnol. 2020, 47, 475–484. [Google Scholar] [CrossRef]
- Voegel, T.M.; Larrabee, M.M.; Nelson, L.M. Development of droplet digital PCR assays to quantify genes involved in nitrification and denitrification, comparison with quantitative real-time PCR and validation of assays in vineyard soil. Can. J. Microbiol. 2021, 67, 174–187. [Google Scholar] [CrossRef]
- Taylor, S.C.; Laperriere, G.; Germain, H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: From variable nonsense to publication quality data. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Kline, M.C.; Romsos, E.L.; Duewer, D.L. Evaluating Digital PCR for the Quantification of Human Genomic DNA: Accessible Amplifiable Targets. Anal. Chem. 2016, 88, 2132–2139. [Google Scholar] [CrossRef] [Green Version]
- Ramírez, J.D.; Herrera, G.; Muskus, C.; Mendez, C.; Duque, M.C.; Butcher, R. Development of a Digital Droplet Polymerase Chain Reaction (ddPCR) assay to detect Leishmania DNA in samples from Cutaneous Leishmaniasis patients. Int. J. Infect. Dis. 2019, 79, 1–3. [Google Scholar] [CrossRef]
- Galluzzi, L.; Ceccarelli, M.; Diotallevi, A.; Menotta, M.; Magnani, M. Real-time PCR applications for diagnosis of leishmaniasis. Parasites Vectors 2018, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.S. Digital Assays Part I: Partitioning Statistics and Digital PCR. SLAS Technol. Transl. Life Sci. Innov. 2017, 22, 369–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Zhao, Z.; Avillan, J.J.; Call, D.R.; Davis, M.; Sischo, W.M.; Zhang, A. Dairy farm soil presents distinct microbiota and varied prevalence of antibiotic resistance across housing areas. Environ. Pollut. 2019, 254, 113058. [Google Scholar] [CrossRef] [PubMed]
- Arvia, R.; Sollai, M.; Pierucci, F.; Urso, C.; Massi, D.; Zakrzewska, K. Droplet digital PCR (ddPCR) vs quantitative real-time PCR (qPCR) approach for detection and quantification of Merkel cell polyomavirus (MCPyV) DNA in formalin fixed paraffin embedded (FFPE) cutaneous biopsies. J. Virol. Methods 2017, 246, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Koepfli, C.; Nguitragool, W.; Hofmann, N.E.; Robinson, L.J.; Ome-Kaius, M.; Sattabongkot, J.; Felger, I.; Mueller, I. Sensitive and accurate quantification of human malaria parasites using droplet digital PCR (ddPCR). Sci. Rep. 2016, 6, 39183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.-R.; Guo, X.-P.; Feng, J.-N.; Lu, D.-P.; Niu, Z.-S.; Tou, F.-Y.; Hou, L.-J.; Liu, M.; Yang, Y. Impact of ZnO nanoparticles on the antibiotic resistance genes (ARGs) in estuarine water: ARG variations and their association with the microbial community. Environ. Sci. Nano 2019, 6, 2405–2419. [Google Scholar] [CrossRef]
- Fujimoto, M.; Carey, D.E.; McNamara, P.J. Metagenomics reveal triclosan-induced changes in the antibiotic resistome of anaerobic digesters. Environ. Pollut. 2018, 241, 1182–1190. [Google Scholar] [CrossRef]
- Gaviria-Figueroa, A.; Preisner, E.C.; Hoque, S.; Feigley, C.E.; Norman, R.S. Emission and dispersal of antibiotic resistance genes through bioaerosols generated during the treatment of municipal sewage. Sci. Total Environ. 2019, 686, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.; Yu, S.; Rysz, M.; Luo, Y.; Yang, F.; Li, F.; Hou, J.; Mu, Q.; Alvarez, P. Prevalence and proliferation of antibiotic resistance genes in two municipal wastewater treatment plants. Water Res. 2015, 85, 458–466. [Google Scholar] [CrossRef]
- Gerdes, L.; Iwobi, A.; Busch, U.; Pecoraro, S. Optimization of digital droplet polymerase chain reaction for quantification of genetically modified organisms. Biomol. Detect. Quantif. 2016, 7, 9–20. [Google Scholar] [CrossRef] [Green Version]
- Noh, E.S.; Park, Y.J.; Kim, E.-M.; Park, J.Y.; Shim, K.B.; Choi, T.-J.; Kim, K.-H.; Kang, J.-H. Quantitative analysis of Alaska pollock in seafood products by droplet digital PCR. Food Chem. 2018, 275, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Talarico, S.; Korson, A.S.; Leverich, C.K.; Park, S.; Jalikis, F.G.; Upton, M.P.; Broussard, E.; Salama, N.R. High prevalence of Helicobacter pylori clarithromycin resistance mutations among Seattle patients measured by droplet digital PCR. Helicobacter 2018, 23, e12472. [Google Scholar] [CrossRef]
- Liao, Y.; Chen, Y.; Kou, X.; Xiao, Y.; Ye, J.; Wu, A. Diagnostic test accuracy of droplet digital PCR for the detection of EGFR mutation (T790M) in plasma: Systematic review and meta-analysis. Clin. Chim. Acta 2019, 503, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Überbacher, C.; Obergasteiger, J.; Volta, M.; Venezia, S.; Müller, S.; Pesce, I.; Pizzi, S.; Lamonaca, G.; Picard, A.; Cattelan, G.; et al. Application of CRISPR/Cas9 editing and digital droplet PCR in human iPSCs to generate novel knock-in reporter lines to visualize dopaminergic neurons. Stem Cell Res. 2019, 41, 101656. [Google Scholar] [CrossRef] [PubMed]
- Hulme, J. Recent advances in the detection of methicillin resistant Staphylococcus aureus (MRSA). BioChip J. 2017, 11, 89–100. [Google Scholar] [CrossRef]
- Koch, H.; Jeschke, A.; Becks, L. Use of dd PCR in experimental evolution studies. Methods Ecol. Evol. 2015, 7, 340–351. [Google Scholar] [CrossRef] [Green Version]
- Ram, M.R.; Teh, X.; Rajakumar, T.; Goh, K.L.; Leow, A.H.R.; Poh, B.H.; Mariappan, V.; Shankar, E.M.; Loke, M.F.; Vadivelu, J. Polymorphisms in the host CYP2C19 gene and antibiotic-resistance attributes of Helicobacter pyloriisolates influence the outcome of triple therapy. J. Antimicrob. Chemother. 2018, 74, 11–16. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).