Recombinant Protein Production with Escherichia coli in Glucose and Glycerol Limited Chemostats
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains
2.2. Growth Media and Culture Conditions
2.2.1. Growth Media
2.2.2. Culture Conditions
2.3. Sample Analysis
2.3.1. Biomass
2.3.2. Substrate Measurements
2.3.3. Volatile Fatty Acids
2.3.4. Flow Cytometry
2.3.5. Data Acquisition and Analysis
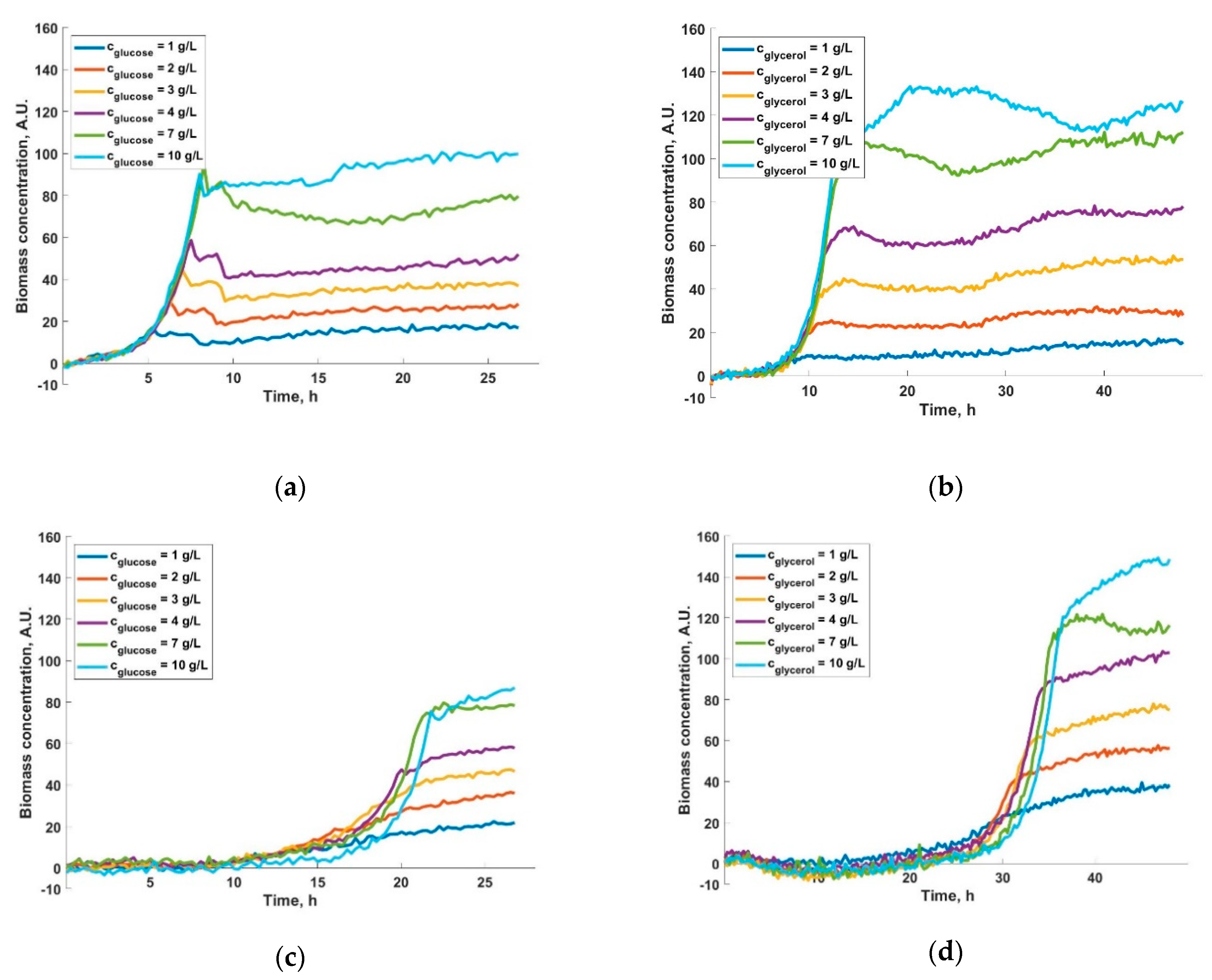
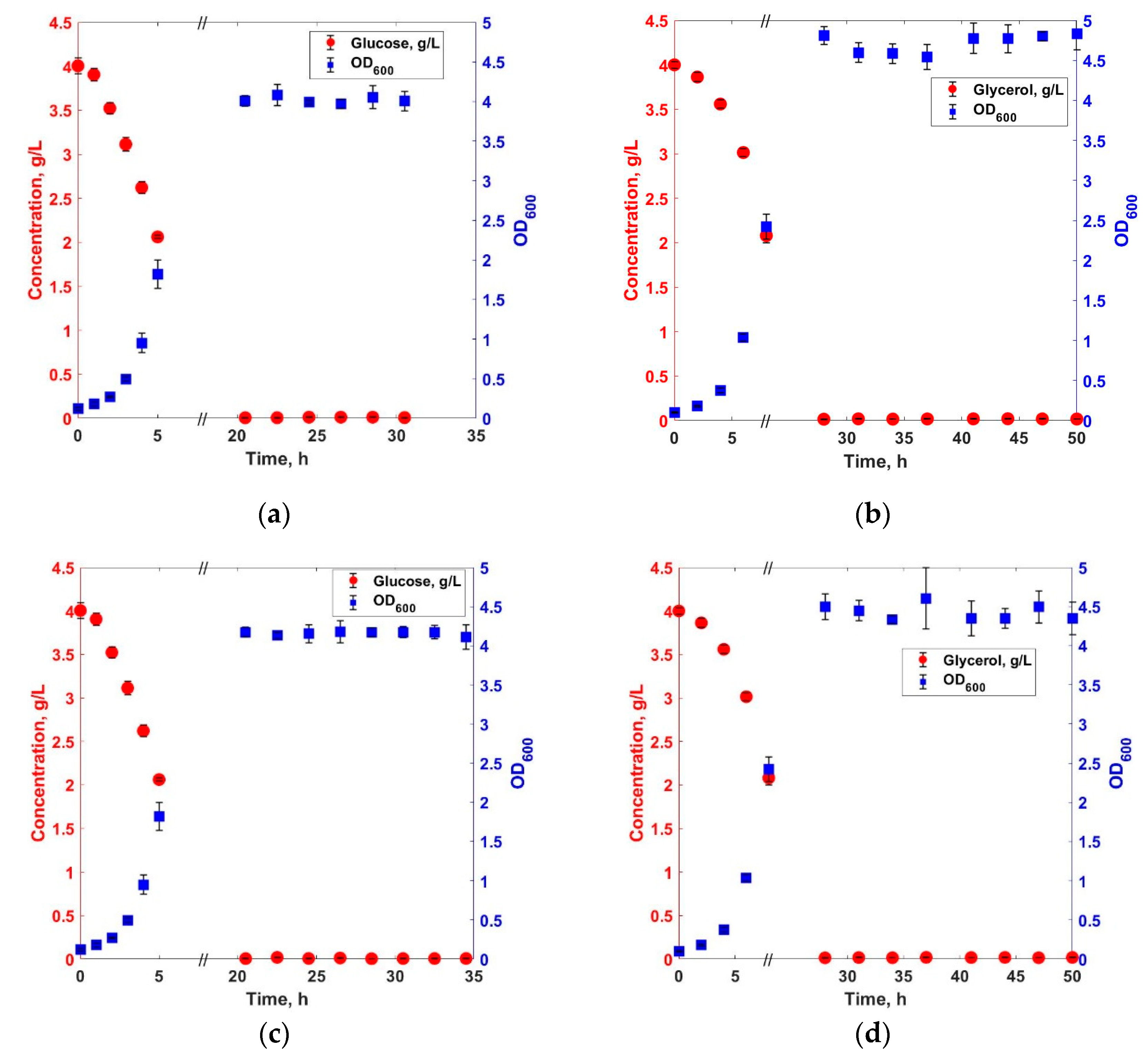
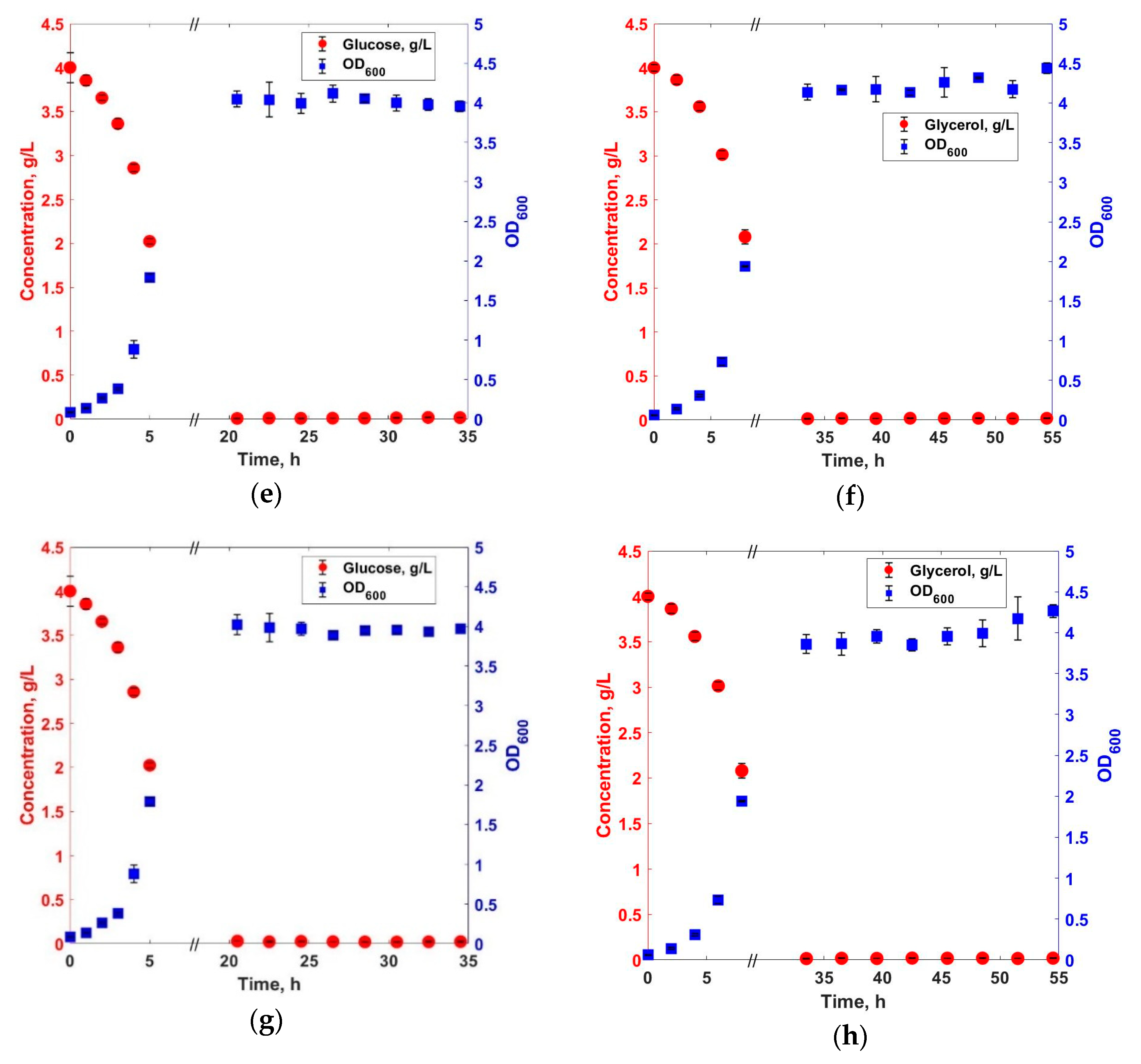
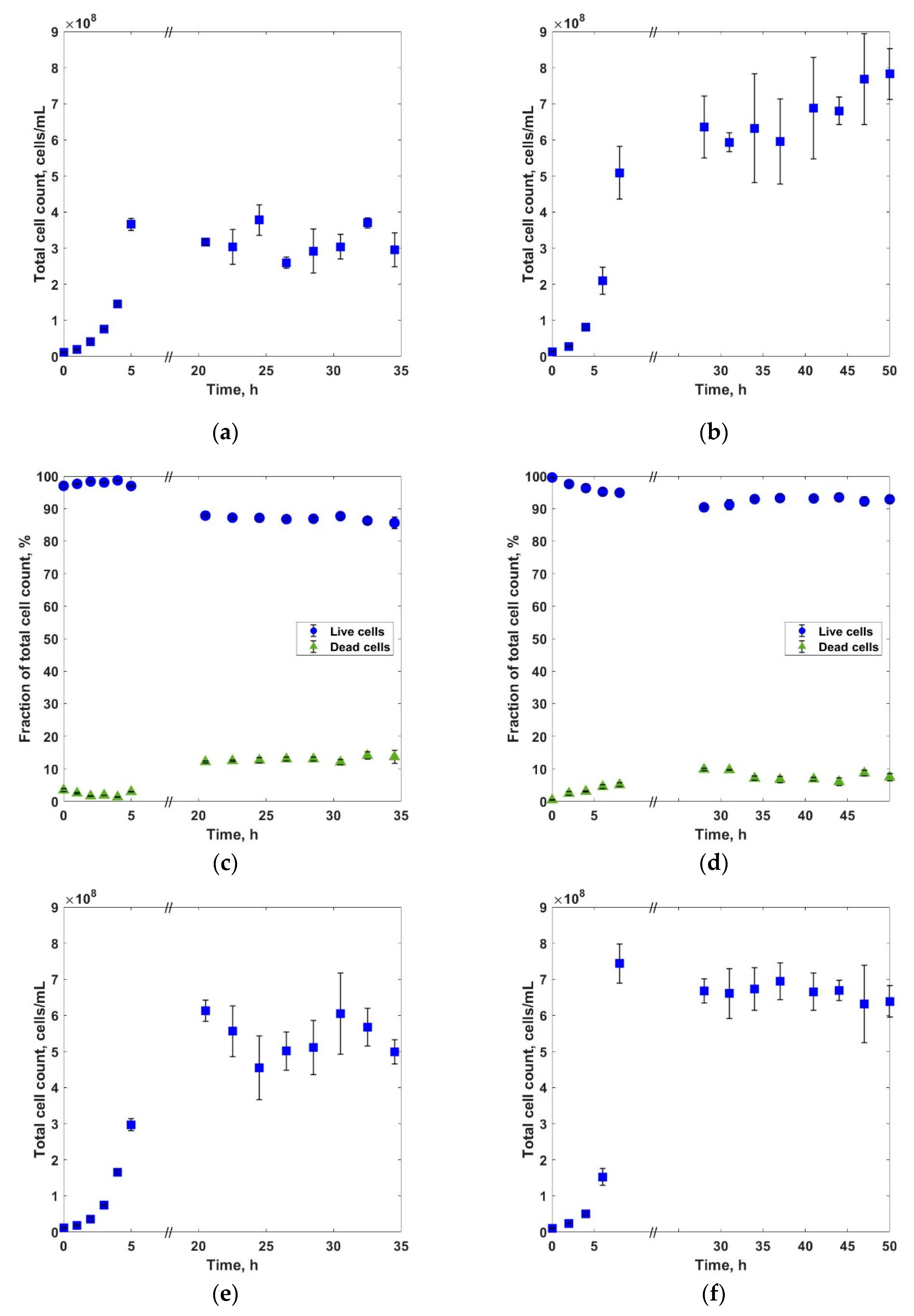
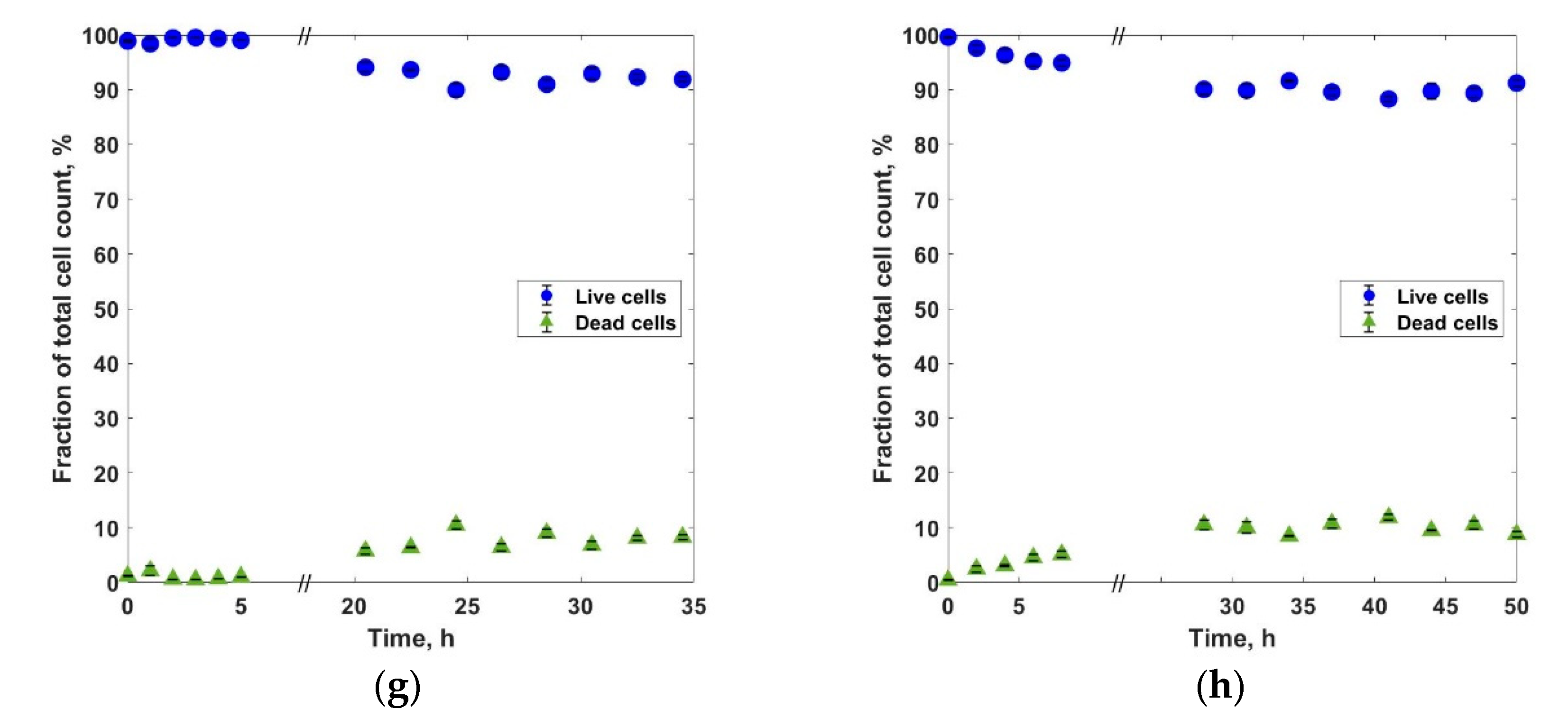
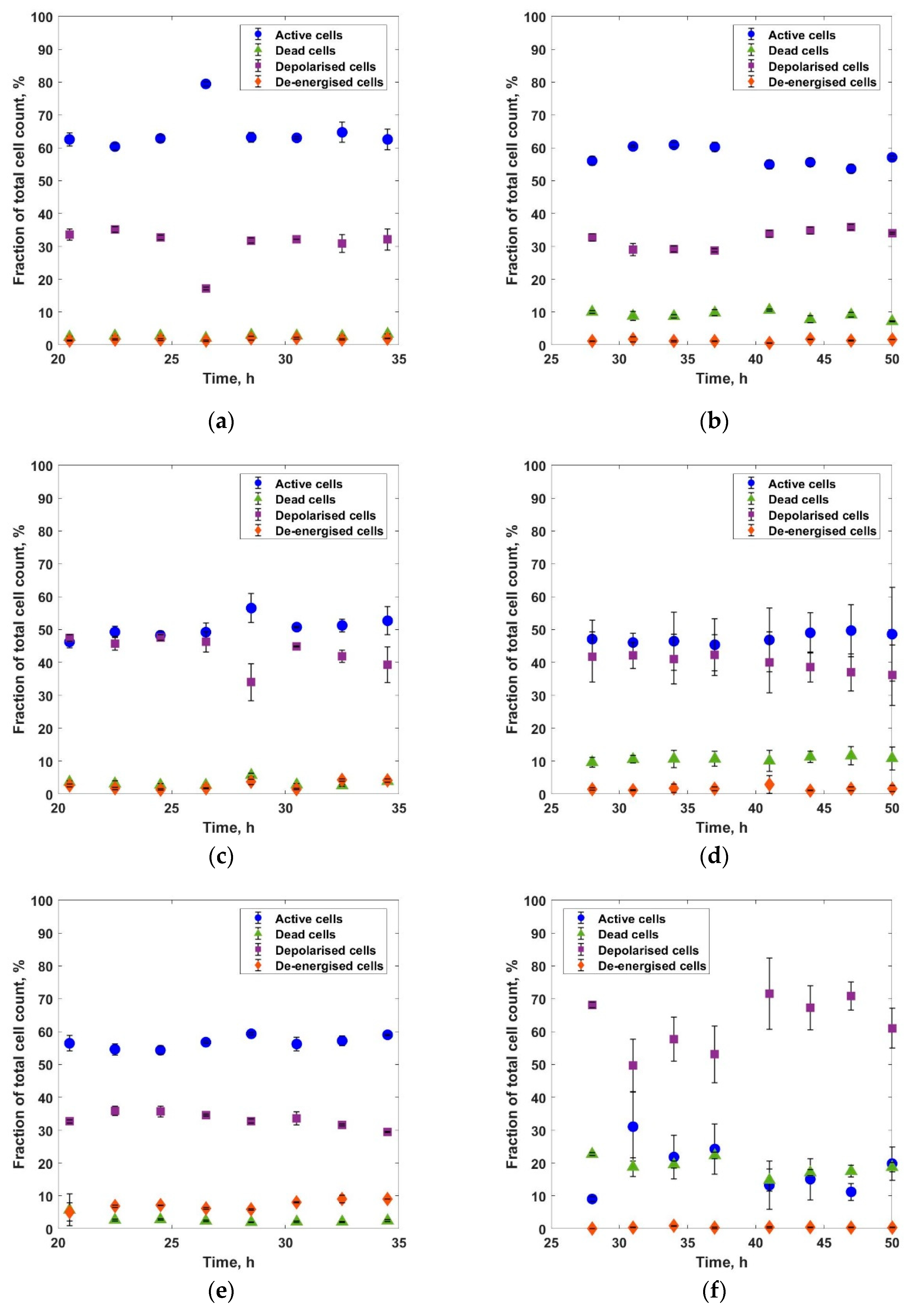
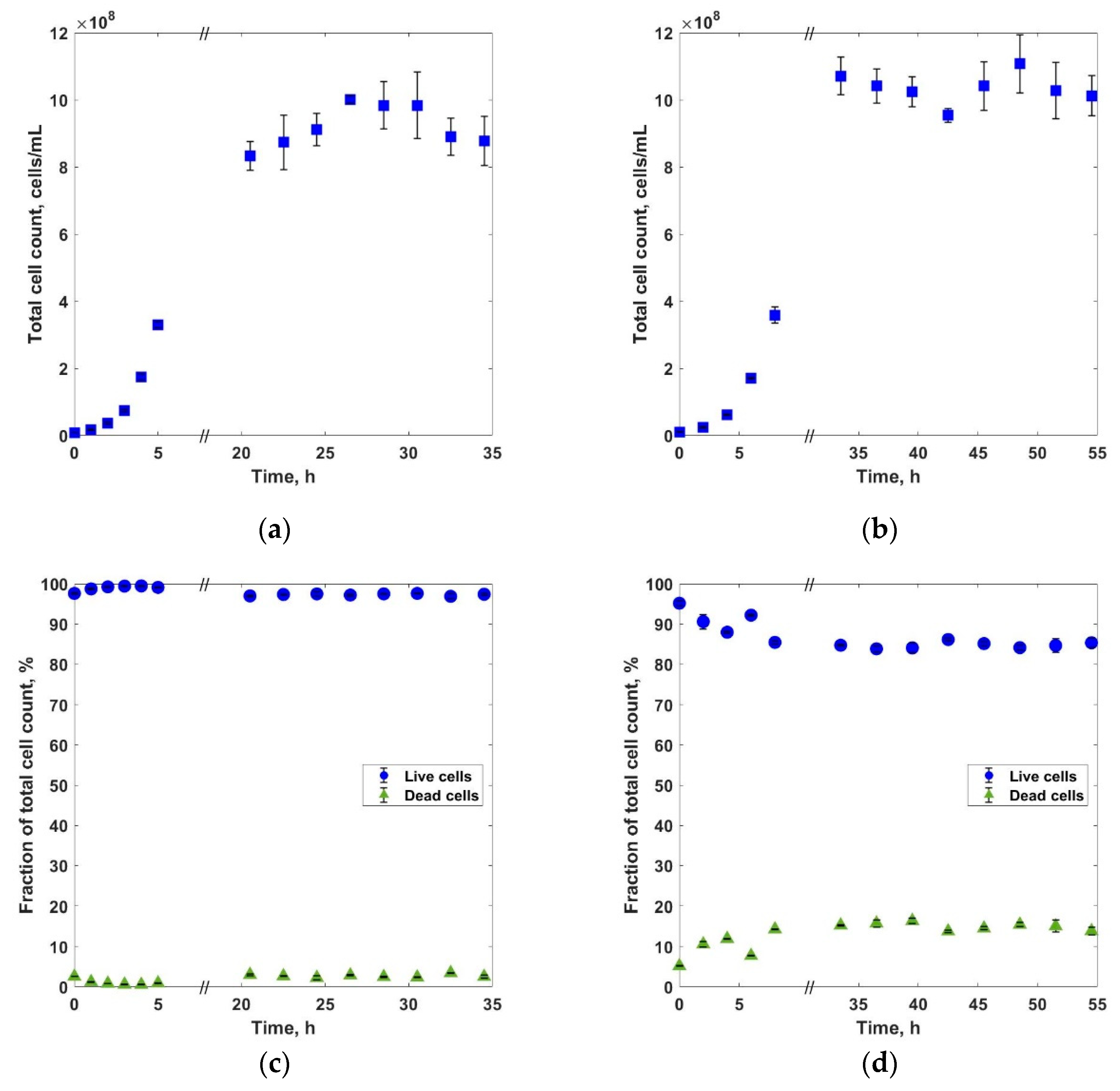
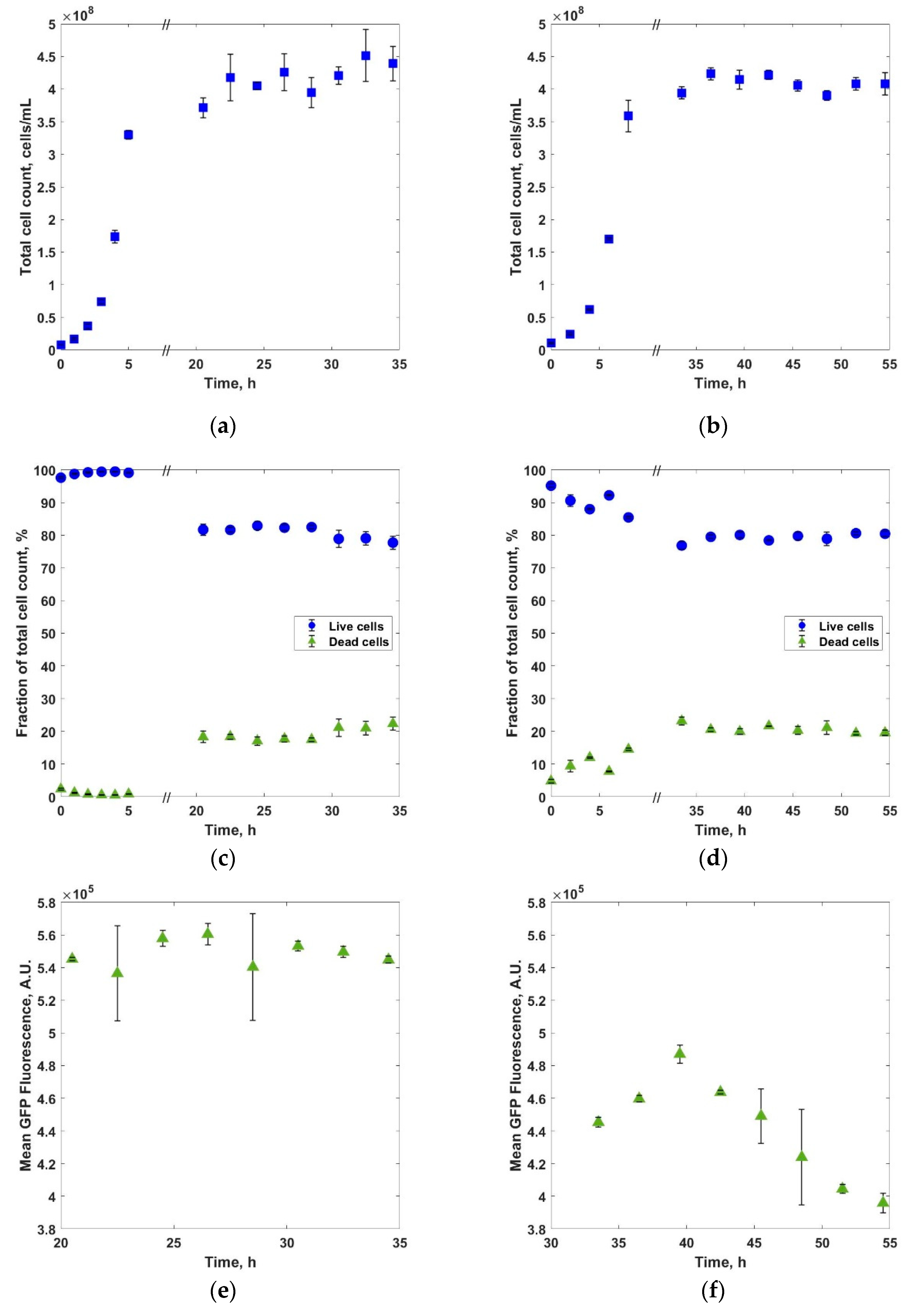
3. Results and Discussions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tripathi, N.K.; Shrivastava, A. Recent developments in bioprocessing of recombinant proteins: Expression hosts and process development. Front. Bioeng. Biotechnol. 2019, 7, 420. [Google Scholar] [CrossRef] [Green Version]
- Baeshen, M.N.; Al-Hejin, A.M.; Bora, R.S.; Ahmed, M.M.M.; Ramadan, H.A.I.; Saini, K.S.; Baeshen, N.A.; Redwan, E.M. Production of biopharmaceuticals in E. coli: Current scenario and future perspectives. J. Microbiol. Biotechnol. 2015, 25, 953–962. [Google Scholar] [CrossRef]
- Steinebach, F.; Ulmer, N.; Wolf, M.; Decker, L.; Schneider, V.; Wälchli, R.; Karst, D.; Souquet, J.; Morbidelli, M. Design and operation of a continuous integrated monoclonal antibody production process. Biotechnol. Prog. 2017, 33, 1303–1313. [Google Scholar] [CrossRef]
- Zhang, X.; Tervo, C.J.; Reed, J.L. Metabolic assessment of E. coli as a Biofactory for commercial products. Metab. Eng. 2016, 35, 64–74. [Google Scholar] [CrossRef] [Green Version]
- Diers, I.V.; Rasmussen, E.; Larsen, P.H.; Kjaersig, I.L. Yeast fermentation processes for insulin production. Bioprocess Technol. 1991, 13, 166–176. [Google Scholar]
- Egli, T. Microbial growth and physiology: A call for better craftsmanship. Front. Microbiol. 2015, 6, 287. [Google Scholar] [CrossRef]
- Walther, J.; Lu, J.; Hollenbach, M.; Yu, M.; Hwang, C.; McLarty, J.; Brower, K. Perfusion cell culture decreases process and product heterogeneity in a head-to-head comparison with fed-batch. Biotechnol. J. 2019, 14, 1700733. [Google Scholar] [CrossRef]
- Warikoo, V.; Godawat, R.; Brower, K.; Jain, S.; Cummings, D.; Simons, E.; Johnson, T.; Walther, J.; Yu, M.; Wright, B.; et al. Integrated continuous production of recombinant therapeutic proteins. Biotechnol. Bioeng. 2012, 109, 3018–3029. [Google Scholar] [CrossRef]
- Peebo, K.; Neubauer, P. Application of Continuous Culture Methods to Recombinant Protein Production in Microorganisms. Microorganisms 2018, 6, 56. [Google Scholar] [CrossRef] [Green Version]
- Croughan, M.S.; Konstantinov, K.B.; Cooney, C. The future of industrial bioprocessing: Batch or continuous? Biotechnol. Bioeng. 2015, 112, 648–651. [Google Scholar] [CrossRef]
- Fragoso-Jiménez, J.C.; Baert, J.; Nguyen, T.M.; Liu, W.; Sassi, H.; Goormaghtigh, F.; Van Melderen, L.; Gaytán, P.; Hernández-Chávez, G.; Martinez, A.; et al. Growth-dependent recombinant product formation kinetics can be reproduced through engineering of glucose transport and is prone to phenotypic heterogeneity. Microb. Cell Factories 2019, 18, 1–16. [Google Scholar] [CrossRef]
- Scott, M.; Gunderson, C.W.; Mateescu, E.M.; Zhang, Z.; Hwa, T. Interdependence of Cell Growth and Gene Expression: Origins and Consequences. Science 2010, 330, 1099–1102. [Google Scholar] [CrossRef]
- Schmideder, A.; Weuster-Botz, D. High-performance recombinant protein production with Escherichia coli in continuously operated cascades of stirred-tank reactors. J. Ind. Microbiol. Biotechnol. 2017, 44, 1021–1029. [Google Scholar] [CrossRef]
- Schmideder, A.; Cremer, J.H.; Weuster-Botz, D. Parallel steady state studies on a milliliter scale accelerate fed-batch bioprocess design for recombinant protein production with Escherichia coli. Biotechnol. Prog. 2016, 32, 1426–1435. [Google Scholar] [CrossRef]
- Kopp, J.; Slouka, C.; Spadiut, O.; Herwig, C. The rocky road from fed-batch to continuous processing with E. coli. Front. Bioeng. Biotechnol. 2019, 7, 328. [Google Scholar] [CrossRef] [PubMed]
- Slouka, C.; Kopp, J.; Strohmer, D.; Kager, J.; Spadiut, O.; Herwig, C. Monitoring and control strategies for inclusion body production in E. coli based on glycerol consumption. J. Biotechnol. 2019, 296, 75–82. [Google Scholar] [CrossRef]
- Martínez-Gómez, K.; Flores, N.; Castañeda, H.M.; Martínez-Batallar, G.; Hernández-Chávez, G.; Ramírez, O.T.; Gosset, G.; Encarnación, S.; Bolivar, F. New insights into Escherichia coli metabolism: Carbon scavenging, acetate metabolism and carbon recycling responses during growth on glycerol. Microb. Cell Factories 2012, 11, 46. [Google Scholar] [CrossRef] [Green Version]
- Monteiro, M.R.; Kugelmeier, C.L.; Pinheiro, R.S.; Batalha, M.O.; César, A. Glycerol from biodiesel production: Technological paths for sustainability. Renew. Sustain. Energy Rev. 2018, 88, 109–122. [Google Scholar] [CrossRef]
- Cabaleiro, R.G.; Mitchell, A.M.; Smith, W.; Wipat, A.; Ofiţeru, I.D. Heterogeneity in Pure Microbial Systems: Experimental Measurements and Modeling. Front. Microbiol. 2017, 8, 1813. [Google Scholar] [CrossRef] [Green Version]
- Heins, A.-L.; Johanson, T.; Han, S.; Lundin, L.; Carlquist, M.; Gernaey, K.V.; Sørensen, S.J.; Lantz, A.E. Quantitative Flow Cytometry to Understand Population Heterogeneity in Response to Changes in Substrate Availability in Escherichia coli and Saccharomyces cerevisiae Chemostats. Front. Bioeng. Biotechnol. 2019, 7, 187. [Google Scholar] [CrossRef] [Green Version]
- Ihssen, J.; Egli, T. Specific growth rate and not cell density controls the general stress response in Escherichia coli. Microbiology 2004, 150, 1637–1648. [Google Scholar] [CrossRef] [Green Version]
- Beal, J.; Farny, N.G.; Haddock-Angelli, T.; Selvarajah, V.; iGEM Interlab Study Contributors. Robust estimation of bacterial cell count from optical density. Commun. Biol. 2020, 3, 512. [Google Scholar] [CrossRef]
- McHugh, I.O.L.; Tucker, A.L. Flow cytometry for the rapid detection of bacteria in cell culture production medium. Cytom. Part A 2007, 71, 1019–1026. [Google Scholar] [CrossRef]
- Davey, H.M.; Kell, D.B. Flow cytometry and cell sorting of heterogeneous microbial populations: The importance of single-cell analyses. Microbiol. Rev. 1996, 60, 641–696. [Google Scholar] [CrossRef]
- Caron, G.N.-V.; Stephens, A.W. Badley Assessment of bacterial viability status by flow cytometry and single cell sorting. J. Appl. Microbiol. 1998, 84, 988–998. [Google Scholar] [CrossRef]
- Schaechter, M. A brief history of bacterial growth physiology. Front. Microbiol. 2015, 6, 289. [Google Scholar] [CrossRef] [Green Version]
- Kopp, J.; Slouka, C.; Ulonska, S.; Kager, J.; Fricke, J.; Spadiut, O.; Herwig, C. Impact of Glycerol as Carbon Source onto Specific Sugar and Inducer Uptake Rates and Inclusion Body Productivity in E. coli BL21(DE3). Bioengineering 2017, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- Eiteman, M.; Altman, E. Overcoming acetate in Escherichia coli recombinant protein fermentations. Trends Biotechnol. 2006, 24, 530–536. [Google Scholar] [CrossRef]
- Kosinski, M.J.; Rinas, U.; Bailey, J.E. Isopropyl-β-d-thiogalactopyranoside influences the metabolism of Escherichia coli. Appl. Microbiol. Biotechnol. 1992, 36, 782–784. [Google Scholar] [CrossRef]
- Dvorak, P.; Chrast, L.; Nikel, P.I.; Fedr, R.; Soucek, K.; Sedlackova, M.; Chaloupkova, R.; De Lorenzo, V.; Prokop, Z.; Damborsky, J. Exacerbation of substrate toxicity by IPTG in Escherichia coli BL21(DE3) carrying a synthetic metabolic pathway. Microb. Cell Factories 2015, 14, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Malakar, P.; Venkatesh, K.V. Effect of substrate and IPTG concentrations on the burden to growth of Escherichia coli on glycerol due to the expression of Lac proteins. Appl. Microbiol. Biotechnol. 2012, 93, 2543–2549. [Google Scholar] [CrossRef]
- Gomes, L.; Monteiro, G.; Mergulhão, F. The impact of IPTG induction on plasmid stability and heterologous protein expression by Escherichia coli biofilms. Int. J. Mol. Sci. 2020, 21, 576. [Google Scholar] [CrossRef] [Green Version]
- Benarroch, J.; Asally, M. The Microbiologist’s Guide to Membrane Potential Dynamics. Trends Microbiol. 2020, 28, 304–314. [Google Scholar] [CrossRef] [Green Version]
- Diaz, M.; Herrero, M.; Garcia, L.; Quirós, C. Application of flow cytometry to industrial microbial bioprocesses. Biochem. Eng. J. 2010, 48, 385–407. [Google Scholar] [CrossRef]
- Nebe-Von-Caron, G.; Stephens, P.; Hewitt, C.; Powell, J.; Badley, R. Analysis of bacterial function by multi-colour fluorescence flow cytometry and single cell sorting. J. Microbiol. Methods 2000, 42, 97–114. [Google Scholar] [CrossRef]
- Bridier, A.; Hammes, F.; Canette, A.; Bouchez, T.; Briandet, R. Fluorescence-based tools for single-cell approaches in food microbiology. Int. J. Food Microbiol. 2015, 213, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Davey, H.; Guyot, S. Estimation of Microbial Viability Using Flow Cytometry. Curr. Protoc. Cytom. 2020, 93, e72. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.T.; Choi, H.J.; Kim, K.H. Flow cytometric analysis of Salmonella enterica serotype Typhimurium inactivated with supercritical carbon dioxide. J. Microbiol. Methods 2009, 78, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, C.J.; Nebe-Von-Caron, G. An industrial application of multiparameter flow cytometry: Assessment of cell physiological state and its application to the study of microbial fermentations. Cytometry 2001, 44, 179–187. [Google Scholar] [CrossRef]
- Liu, M.; Durfee, T.; Cabrera, J.E.; Zhao, K.; Jin, D.J.; Blattner, F.R. Global Transcriptional Programs Reveal a Carbon Source Foraging Strategy by Escherichia coli. J. Biol. Chem. 2005, 280, 15921–15927. [Google Scholar] [CrossRef] [Green Version]
- Kittler, S.; Kopp, J.; Veelenturf, P.G.; Spadiut, O.; Delvigne, F.; Herwig, C.; Slouka, C. The Lazarus Escherichia coli Effect: Recovery of Productivity on Glycerol/Lactose Mixed Feed in Continuous Biomanufacturing. Front. Bioeng. Biotechnol. 2020, 8, 993. [Google Scholar] [CrossRef] [PubMed]
- Delvigne, F.; Goffin, P. Microbial heterogeneity affects bioprocess robustness: Dynamic single-cell analysis contributes to understanding of microbial populations. Biotechnol. J. 2013, 9, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Binder, D.; Drepper, T.; Jaeger, K.-E.; Delvigne, F.; Wiechert, W.; Kohlheyer, D.; Grünberger, A. Homogenizing bacterial cell factories: Analysis and engineering of phenotypic heterogeneity. Metab. Eng. 2017, 42, 145–156. [Google Scholar] [CrossRef] [PubMed]
| E.coli Strain | IPTG | Substrate | μmax | Sampling Times |
|---|---|---|---|---|
| W3110 | 0 mM | Glucose | 0.6 h−1 | 2 h |
| W3110 | 0 mM | Glycerol | 0.5 h−1 | 3 h |
| CLD1301 | 1 mM | Glucose | 0.67 h−1 | 2 h |
| CLD1301 | 1 mM | Glycerol | 0.39 h−1 | 3 h |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mitchell, A.M.; Gogulancea, V.; Smith, W.; Wipat, A.; Ofiţeru, I.D. Recombinant Protein Production with Escherichia coli in Glucose and Glycerol Limited Chemostats. Appl. Microbiol. 2021, 1, 239-254. https://doi.org/10.3390/applmicrobiol1020018
Mitchell AM, Gogulancea V, Smith W, Wipat A, Ofiţeru ID. Recombinant Protein Production with Escherichia coli in Glucose and Glycerol Limited Chemostats. Applied Microbiology. 2021; 1(2):239-254. https://doi.org/10.3390/applmicrobiol1020018
Chicago/Turabian StyleMitchell, Anca Manuela, Valentina Gogulancea, Wendy Smith, Anil Wipat, and Irina Dana Ofiţeru. 2021. "Recombinant Protein Production with Escherichia coli in Glucose and Glycerol Limited Chemostats" Applied Microbiology 1, no. 2: 239-254. https://doi.org/10.3390/applmicrobiol1020018
APA StyleMitchell, A. M., Gogulancea, V., Smith, W., Wipat, A., & Ofiţeru, I. D. (2021). Recombinant Protein Production with Escherichia coli in Glucose and Glycerol Limited Chemostats. Applied Microbiology, 1(2), 239-254. https://doi.org/10.3390/applmicrobiol1020018







