An Update on Root Lesion Nematode Species Infecting Cereal Crops in the Southwest of Western Australia
Abstract
1. Introduction
2. Materials and Methods
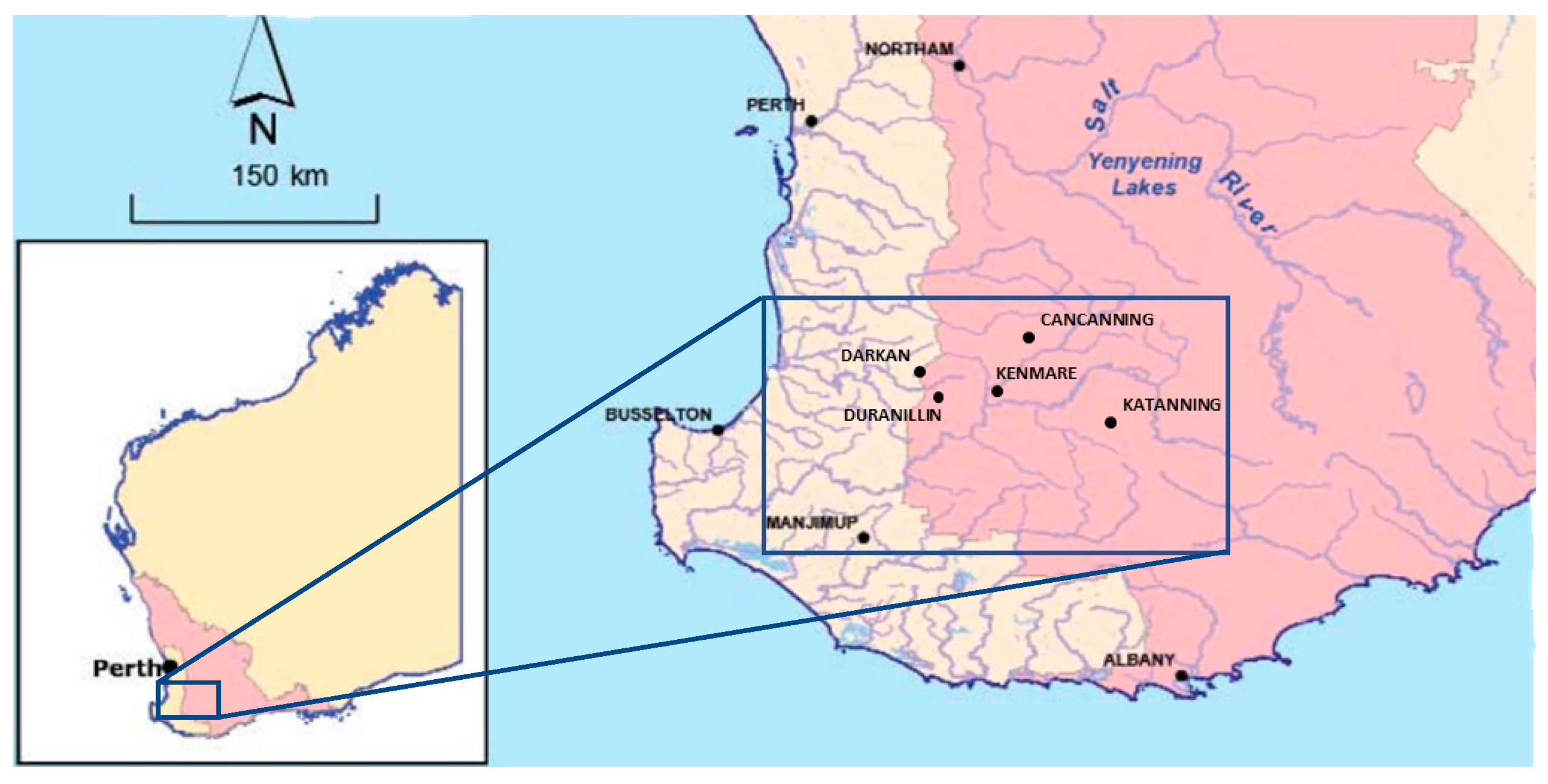
2.1. Field Sampling of Commercial Cereal Fields
2.2. Sampling of Seed Bulk-Up Nursery
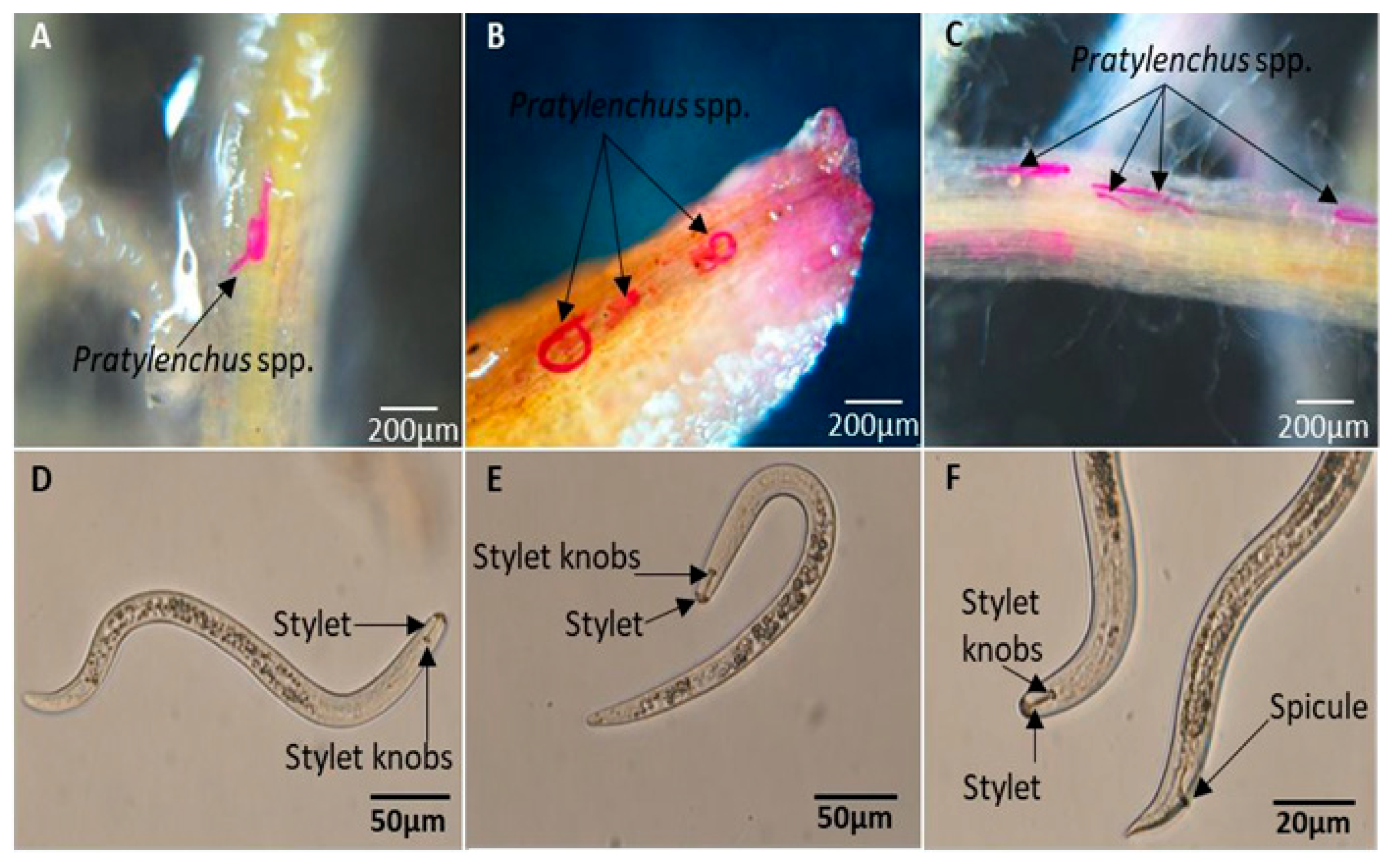
2.3. Confirmation of Pratylenchus spp. Infestation
2.4. Isolation and Quantification of Pratylenchus spp. in Roots and Soil Samples
2.5. Amplification of Partial rDNA of Pratylenchus spp.
2.6. Cloning and Sequencing of Clones of D2-D3 Amplicons
2.7. Analyses of Sequences and Phylogenetic Relationships
2.8. Statistical Analysis
3. Results
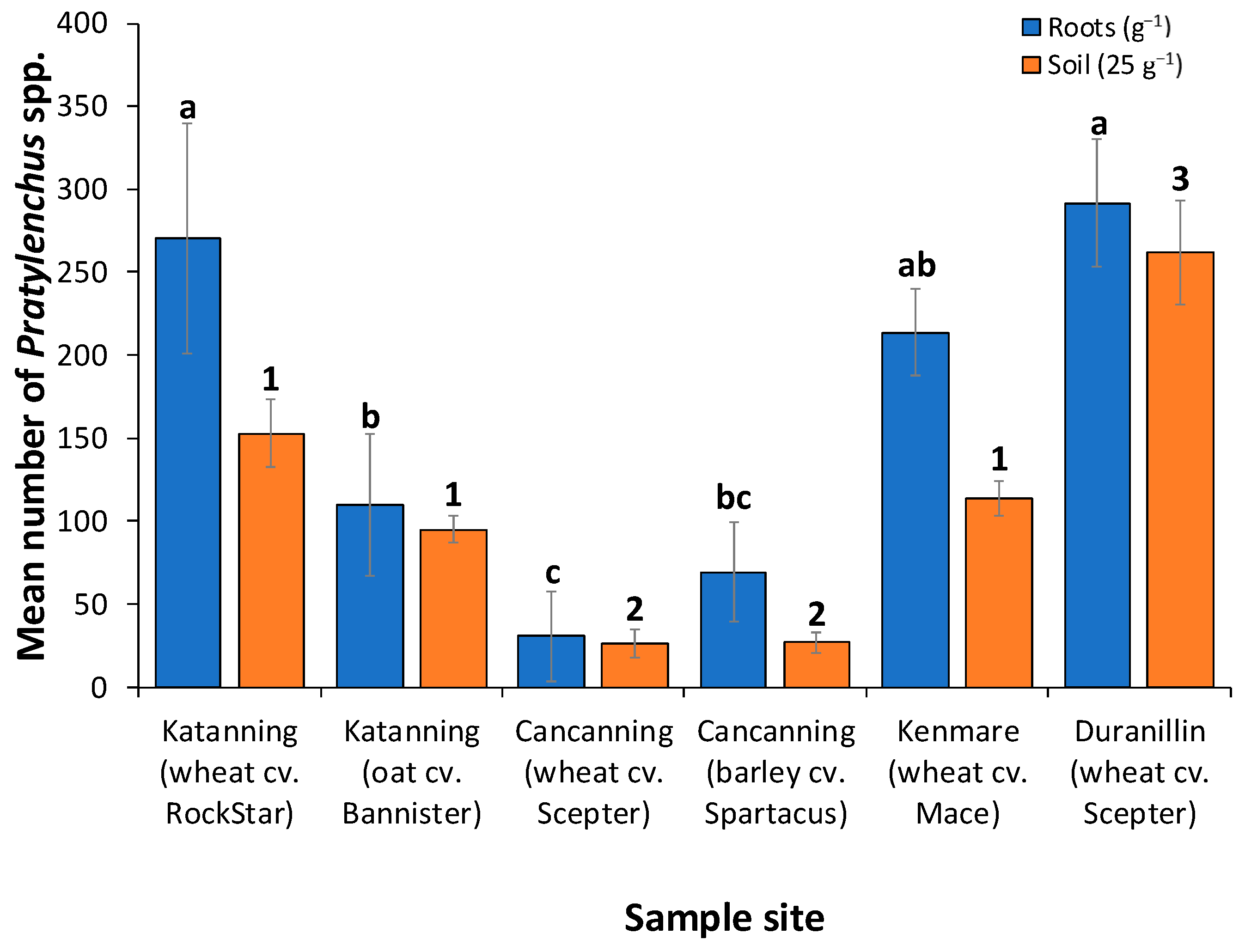
3.1. Root-Lesion Nematodes Were Present in All Fields Surveyed
3.2. The Levels of Infestation in the Fields Differed Significantly
3.3. Molecular Confirmation of the Pratylenchus spp. Identified
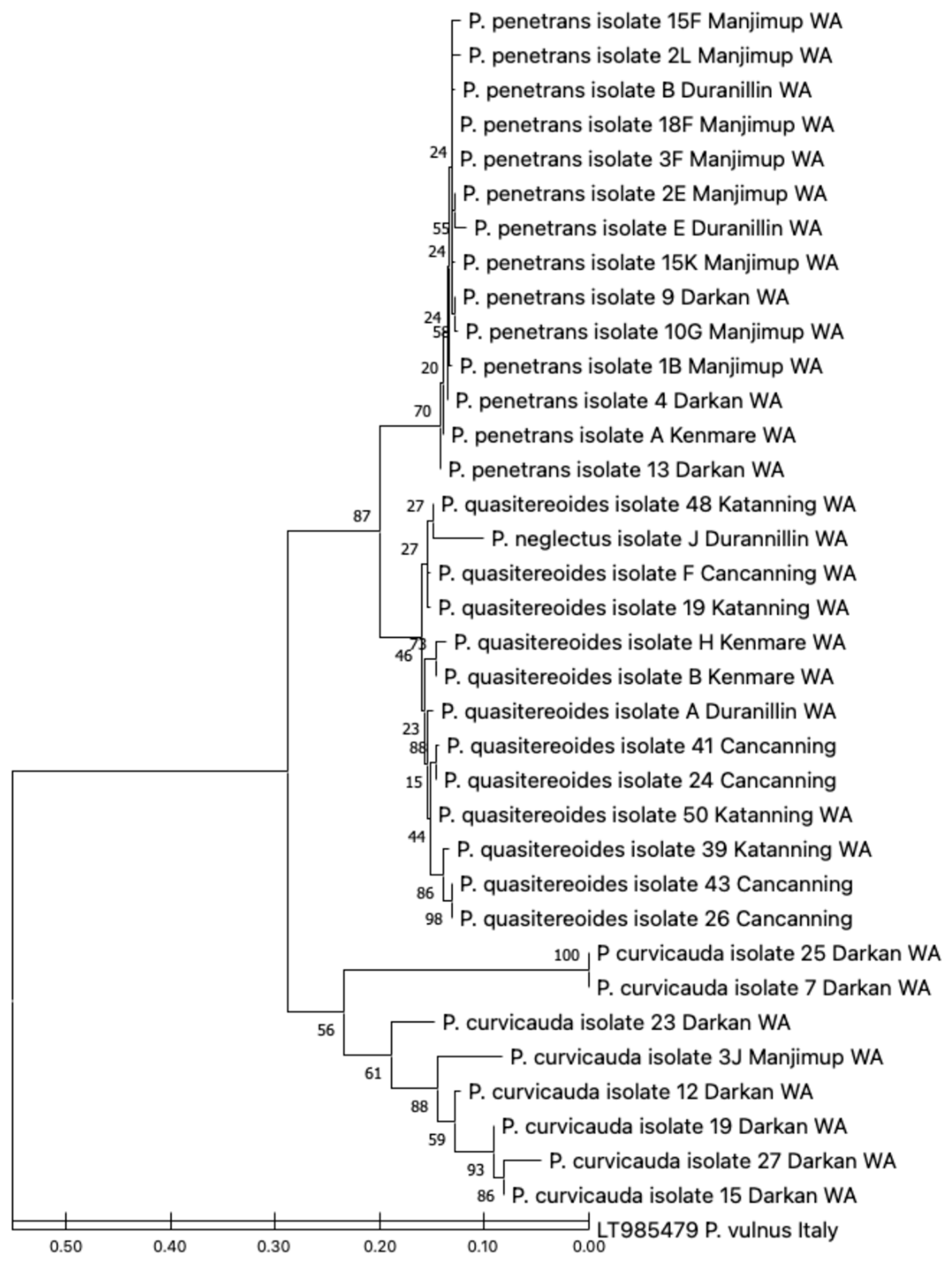
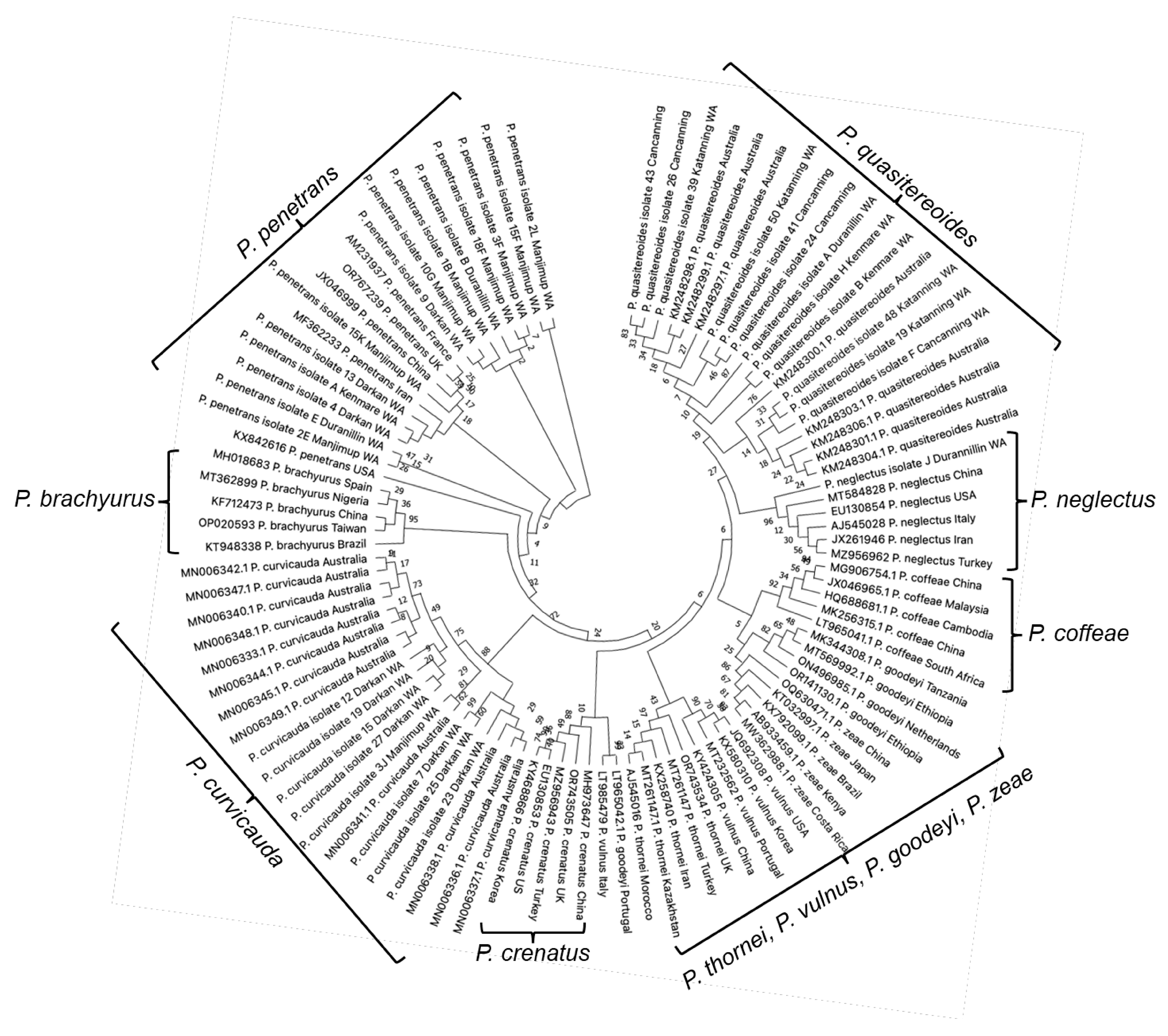
3.4. Phylogenetic Relationships of the Pratylenchus spp. Identified, with Reference to P. curvicauda and P. quasitereoides
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Castillo, P.; Vovlas, N. Pratylenchus (Nematoda: Pratylenchidae): Diagnosis, Biology, Pathogenicity and Management. Monographs and Perspectives; Hunt, D.J., Perry, R.N., Eds.; Koninklijke Brill NV: Leiden, The Netherlands, 2007; Volume 6, p. 599. [Google Scholar]
- Jones, J.T.; Haegeman, A.; Danchin, E.G.J.; Gaur, H.S.; Helder, J.; Jones, M.G.K.; Kikuchi, T.; Manzanilla-López, R.; Palomares-Rius, J.E.; Wesemael, W.M.L.; et al. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 2013, 14, 946–961. [Google Scholar] [PubMed]
- Hodda, M.; Nobbs, J. A review of current knowledge on particular taxonomic features of the Australasian nematode fauna, with special emphasis on plant feeders. Australas. Plant Pathol. 2008, 37, 308–317. [Google Scholar]
- Thompson, J.; Owen, K.; Stirling, G.; Bell, M. Root-lesion nematodes (Pratylenchus thornei and P. neglectus): A review of recent progress in managing a significant pest of grain crops in northern Australia. Australas. Plant Pathol. 2008, 37, 235–242. [Google Scholar]
- Vanstone, V.A.; Hollaway, G.J.; Stirling, G.R. Managing nematode pests in the southern and western regions of the Australian cereal industry: Continuing progress in a challenging environment. Australas. Plant Pathol. 2008, 37, 220–234. [Google Scholar]
- Power, S.; Shackley, B.; Paynter, B.; Seymour, M.; Dhammu, H.; Wackett, B. 2025 Western Australian Crop Sowing Guide. State of Western Australia (Department of Primary Industries and Regional Development), Perth. Bulletin Bulletin 4935. 2024. Available online: https://library.dpird.wa.gov.au/bulletins/292 (accessed on 7 March 2025).
- Begum, F.; Fosu-Nyarko, J.; Sharma, S.; Macleod, B.; Collins, S.; Jones, M.G.K. Serendipitous identification of Pratylenchus curvicauda from the grainbelt of Western Australia. J. Nematol. 2019, 51, e2019-46. [Google Scholar] [PubMed]
- Begum, F.; Jones, M.G.K.; Fosu-Nyarko, J. Assessment of the pest status of Pratylenchus curvicauda and ultrastructural changes in roots of infected wheat and barley. Plant Pathol. 2020, 69, 1574–1588. [Google Scholar]
- Fosu-Nyarko, J.; Jones, M.G. Advances in understanding the molecular mechanisms of root lesion nematode host interactions. Annu. Rev. Phytopathol. 2016, 54, 253–278. [Google Scholar]
- Hollaway, G.J.; Taylor, S.P.; Eastwood, R.F.; Hunt, C.H. Effect of field crops on density of Pratylenchus in Southeastern Australia; Part 2: P. thornei. J. Nematol. 2000, 32, 600. [Google Scholar] [PubMed]
- Taylor, S.P.; Hollaway, G.J.; Hunt, C.H. Effect of field crops on population densities of Pratylenchus neglectus and P. thornei in southeastern Australia; Part 1: P. neglectus. J. Nematol. 2000, 32, 591. [Google Scholar]
- Jones, M.G.K.; Iqbal, S.; Fosu-Nyarko, J. Belowground defence strategies against migratory nematodes. In Belowground Defence Strategies in Plants; Vos, C.M.F., Kazan, K., Eds.; Springer: Cham, Switzerland, 2016; pp. 253–278. [Google Scholar]
- Collins, S.; Wilkinson, C.; Kelly, S.; Hunter, H.; DeBrincat, L.; Reeves, K.; Chen, K. The Invisible Threat: Canola Yield Losses Caused by Root Lesion Nematode in WA. GRDC. 2017. Available online: https://grdc.com.au/resources-and-publications/grdc-update-papers/tab-content/grdc-update-papers/2017/02/the-invisible-threat-canola-yield-losses-caused-by-root-lesion-nematodes-in-wa (accessed on 15 January 2022).
- Australian Bureau of Statistics. Winter Broadacre Crops by Australia, State and Territory, Estimates Using New Data Sources and Methods—2023–2024, 2025. Available online: https://www.abs.gov.au/statistics/industry/agriculture/australian-agriculture-broadacre-crops/2023-24 (accessed on 11 March 2025).
- Begum, F. Biology and Molecular Charactersiation of the Root Lesion Nematode, Pratylenchus Curvicauda. Ph.D. Thesis, Murdoch University, Perth, Australia, 2017. [Google Scholar]
- Flower, K.; Hüberli, D.; Collins, S.; Thomas, G.; Ward, P.; Cordingley, N. Progression of plant-parasitic nematodes and foliar and root diseases under no-tillage with different crop rotations. Soil Tillage Res. 2019, 191, 18–28. [Google Scholar]
- Owen, K.J.; Clewett, T.G.; Thompson, J.P. Pre-cropping with canola decreased Pratylenchus thornei populations, arbuscular mycorrhizal fungi, and yield of wheat. Crop Pasture Sci. 2010, 61, 399–410. [Google Scholar] [CrossRef]
- Owen, K.J.; Clewett, T.G.; Bell, K.L.; Thompson, J.P. Cereal and Pulse Crops with Improved Resistance to Pratylenchus thornei are needed to maximize wheat production and expand crop sequence options. Agronomy 2022, 12, 573. [Google Scholar] [CrossRef]
- Harries, M.; Flower, K.C.; Renton, M.; Collins, S.J.; Hüberli, D. Links between soilborne pathogens, plant parasitic nematodes, farm management and biophysical constraints in a southern Australian rainfed cropping system. Crop Pasture Sci. 2022, 73, 1291–1307. [Google Scholar] [CrossRef]
- Owen, K.J.; Clewett, T.G.; Bell, K.L.; Thompson, J.P. Wheat biomass and yield increased when populations of the root-lesion nematode (Pratylenchus thornei) were reduced through sequential rotation of partially resistant winter and summer crops. Crop Pasture Sci. 2014, 65, 227–241. [Google Scholar] [CrossRef]
- Fanning, J.P.; Reeves, K.L.; Forknall, C.R.; McKay, A.C.; Hollaway, G.J. Pratylenchus thornei: The relationship between presowing nematode density and yield loss in wheat and barley. Phytopathology 2020, 110, 674–683. [Google Scholar] [CrossRef] [PubMed]
- Murray, G.M.; Brennan, J.P. Estimating disease losses to the Australian wheat industry. Australas. Plant Pathol. 2009, 38, 558–570. [Google Scholar] [CrossRef]
- Kelly, S.; Riley, I. Radopholus nativus (Nematoda: Pratylenchidae), a potential economic pest of wheat in Western Australia. Nematology 2001, 3, 25–30. [Google Scholar] [CrossRef]
- Riley, I.T.; Kelly, S.J. Endoparasitic nematodes in cropping soils of Western Australia. Aust. J. Exp. Agric. 2002, 42, 49–56. [Google Scholar] [CrossRef]
- Vanstone, V.A.; Kelly, S.J.; Hunter, H.F.; Gilchrist, M.C. Rotations for nematode management. In Crop Updates; Perth, Australia, 2005; p. 72. Available online: https://library.dpird.wa.gov.au/crop_up/22 (accessed on 17 February 2025).
- Hodda, M.; Collins, S.J.; Vanstone, V.A.; Hartley, D.; Wanjura, W.; Kehoe, M. Pratylenchus quasitereoides n. sp. from cereals in Western Australia. Zootaxa 2014, 3866, 277–288. [Google Scholar] [CrossRef]
- Khangura, R.K.; MacNish, G.C.; MacLeod, W.J.; Vanstone, V.A.; Hanbury, C.D.; Loughman, R.; Speijers, J.E. Current status of cereal root diseases in Western Australia under intensive cereal production and their comparison with the historical survey conducted during 1976–1982. J. Phytopathol. 2013, 161, 828–840. [Google Scholar] [CrossRef]
- Chambers, K.; McKay, A.; Hüberli, D.; Evans, M.; Vadakuttu, G.; Holloaway, G. Characterising Soil Borne Disease Risk in the Eastern Wheat Belt of Western Australia and National Significance of Major Diseases; GRDC Research Updates: Canberra, Australia, 2018. [Google Scholar]
- Siddiqi, M.R.; Dabur, K.R.; Bajaj, H.K. Descriptions of three new species of Pratylenchus filipjev, 1936 (Nematoda: Pratylenchidae). Nematol. Mediterr. 1991, 19, 1–7. [Google Scholar]
- Davis, J.; Braimbridge, M. The Ecology of Wheatbelt Lakes; Water Notes; Department of Environment and Conservation: Waigani, Papua New Guinea, 2005; Volume 33. [Google Scholar]
- Bybd, D.W.; Kirkpatrick, T.; Barker, K.R. An improved technique for clearing and staining plant tissues for detection of nematodes. J. Nematol. 1983, 15, 142. [Google Scholar] [PubMed]
- Tan, J.-A.C.; Jones, M.G.; Fosu-Nyarko, J. Gene silencing in root lesion nematodes (Pratylenchus spp.) significantly reduces reproduction in a plant host. Exp. Parasitol. 2013, 133, 166–178. [Google Scholar] [PubMed]
- Orlando, V.; Edwards, S.G.; Neilson, R.; Prior, T.; Roberts, D.; Back, M. Comparing the efficiency of six common methods for DNA extraction from root-lesion nematodes (Pratylenchus spp.). Nematology 2020, 23, 415–423. [Google Scholar]
- De Luca, F.; Fanelli, E.; Di vito, M.; Reyes, A.; De giorgi, C. Comparison of the sequences of the D3 expansion of the 26S ribosomal genes reveals different degrees of heterogeneity in different populations and species of Pratylenchus from the Mediterranean region. Eur. J. Plant Pathol. 2004, 110, 949–957. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [PubMed]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar]
- Wang, H.; Zhuo, K.; Ye, W.; Liao, J. Morphological and molecular charaterisation of Pratylenchus parazeae n. sp. (Nematoda: Pratylenchidae) parasitizing sugarcane in China. Eur. J. Plant Pathol. 2015, 143, 173–191. [Google Scholar]
- Handoo, Z.A.; Yan, G.; Kantor, M.R.; Huang, D.; Chowdhury, I.A.; Plaisance, A.; Bauchan, G.R.; Mowery, J.D. Morphological and molecular characterization of Pratylenchus dakotaensis n. sp. (Nematoda: Pratylenchidae), a new root-lesion nematode species on soybean in North Dakota, USA. Plants 2021, 10, 168. [Google Scholar] [CrossRef]
- Mehmood, N.; Khanum, T.A. Description of Pratylenchus siddiqi N. SP. from banana (Musa acuminata L.) Malir Cantt, Karachi, Sindh, Pakistan. Plant Prot. 2021, 5, 89–93. [Google Scholar]
- Nguyen, T.D.; Le, T.M.L.; Nguyen, H.T.; Nguyen, T.A.D.; Liebanas, G.; Trinh, Q.P. Morphological and molecular characteristics of Pratylenchus haiduongensis sp. n., a new species of root–lesion nematodes associated with carrot in Vietnam. J. Nematol. 2017, 49, 276. [Google Scholar]
- Qing, X.; Bert, W.; Gamliel, A.; Bucki, P.; Duvrinin, S.; Alon, T.; Braun Miyara, S. Phylogeography and molecular species delimitation of Pratylenchus capsici n. sp., a new root-lesion nematode in Israel on pepper (Capsicum annuum). Phytopathology 2019, 109, 847–858. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.R.; Nyiragatare, A.; Janssen, T.; Couvreur, M.; Decraemer, W.; Bert, W. Morphological and molecular characterisation of Pratylenchus rwandae n. sp.(Tylenchida: Pratylenchidae) associated with maize in Rwanda. Nematology 2018, 20, 781–794. [Google Scholar] [CrossRef]
- Troccoli, A.; Fanelli, E.; Castillo, P.; Liébanas, G.; Cotroneo, A.; De Luca, F. Pratylenchus vovlasi sp. Nov. (Nematoda: Pratylenchidae) on raspberries in North Italy with a morphometrical and molecular characterization. Plants 2021, 10, 1068. [Google Scholar] [CrossRef] [PubMed]
- Fosu-Nyarko, J.; Tan, J.A.C.; Gill, R.; Agrez, V.G.; Rao, U.; Jones, M.G.K. De novo analysis of the transcriptome of Pratylenchus zeae to identify transcripts for proteins required for structural integrity, sensation, locomotion and parasitism. Mol. Plant Pathol. 2015, 17, 532–552. [Google Scholar] [CrossRef] [PubMed]
- Nicol, P.; Gill, R.; Fosu-Nyarko, J.; Jones, M.G.K. De novo analysis and functional classification of the transcriptome of the root lesion nematode, Pratylenchus thornei, after 454 GS FLX sequencing. Int. J. Parasitol. 2012, 42, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.G.K.; Fosu-Nyarko, J. Molecular biology of root lesion nematodes (Pratylenchus spp.) and their interaction with host plants. Ann. Appl. Biol. 2014, 164, 163–181. [Google Scholar] [CrossRef]

| Sample Location | Clones Sequenced | Identified Pratylenchus spp. | Sequence Characteristics | ||||
|---|---|---|---|---|---|---|---|
| Unique Groups of Sequences Among the Population | Length (Nucleotides) | Positions with Insertions or Deletions | Positions with Substitutions | NCBI Genbank Accession Numbers | |||
| Darkan | 26 | P. curvicauda | 7 | 312–316 | 8 | 83 | PP203078-84 |
| P. penetrans | 3 | 344 | 0 | 4 | PP203085-87 | ||
| Manjimup | 141 | P. penetrans | 8 | 343–344 | 3 | 11 | PP203088-95 |
| P. curvicauda | 1 | 240 | NA | NA | PP203096 | ||
| Cancanning | 15 | P. quasitereoides | 5 | 345 | 0 | 17 | PP203097-101 |
| Katanning | 13 | P. quasitereoides | 4 | 344–346 | 2 | 12 | PP203102-105 |
| Kenmare | 21 | P. penetrans | 1 | 344 | NA | NA | PP203106 |
| P. quasitereoides | 2 | 344–345 | 1 | 2 | PP203107-108 | ||
| Duranillin | 40 | P. penetrans | 2 | 344 | 0 | 6 | PP203109-110 |
| P. quasitereoides | 1 | 192 | NA | NA | PP203111 | ||
| P. neglectus | 1 | 347 | NA | NA | PP203112 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Copeland, R.G.R.; Iqbal, S.; Angessa, T.T.; Collins, S.J.; Jones, M.G.K.; Fosu-Nyarko, J. An Update on Root Lesion Nematode Species Infecting Cereal Crops in the Southwest of Western Australia. Crops 2025, 5, 19. https://doi.org/10.3390/crops5020019
Copeland RGR, Iqbal S, Angessa TT, Collins SJ, Jones MGK, Fosu-Nyarko J. An Update on Root Lesion Nematode Species Infecting Cereal Crops in the Southwest of Western Australia. Crops. 2025; 5(2):19. https://doi.org/10.3390/crops5020019
Chicago/Turabian StyleCopeland, Rhys G. R., Sadia Iqbal, Tefera T. Angessa, Sarah J. Collins, Michael G. K. Jones, and John Fosu-Nyarko. 2025. "An Update on Root Lesion Nematode Species Infecting Cereal Crops in the Southwest of Western Australia" Crops 5, no. 2: 19. https://doi.org/10.3390/crops5020019
APA StyleCopeland, R. G. R., Iqbal, S., Angessa, T. T., Collins, S. J., Jones, M. G. K., & Fosu-Nyarko, J. (2025). An Update on Root Lesion Nematode Species Infecting Cereal Crops in the Southwest of Western Australia. Crops, 5(2), 19. https://doi.org/10.3390/crops5020019







